Abstract
Purpose
ZMYND8 is closely correlated with cancerous proliferation and invasiveness. However, its prognostic value has not been estimated in a nasopharyngeal carcinoma (NPC). The purpose of this study was to elucidate the status of ZMYND8 expression and its prognostic significance in NPCs.
Methods
The status of ZMYND8 expression was investigated by immunohistochemistry for NPC samples in the study. The cutoff value of ZMYND8 expression was confirmed in NPCs using ROC-curve analysis. Correlations between ZMYND8 expression and clinicopathological variables and patient prognosis were analyzed by various statistical methods.
Results
Our study showed that low expression of ZMYND8 strongly correlated with late T stage in NPCs (P<0.05). Kaplan–Meier survival analysis revealed a significant association between low ZMYND8 expression and worse overall survival (P<0.05). Most importantly, Cox regression analysis confirmed ZMYND8 expression in NPC could be an independent prognostic factor.
Conclusion
Low expression of ZMYND8 could be of importance, due to its displaying more aggressive behavior in NPC. Therefore, ZMYND8 expression might serve as an independent prediction factor in patients with NPCs.
Keywords:
Introduction
Nasopharyngeal carcinoma (NPC) is the most common cancer in the nasopharynx, endemic in Southeast Asia, especially southern China, and seriously affects human health.Citation1 Most NPCs are undifferentiated nonkeratinized carcinoma by World Health Organization (WHO) pathological classification, with characteristics of fast growth, metastasis of regional lymph nodes, and/or metastasis to distant organs. Once the metastasis occurs, the prognosis for these patients is not extremely optimistic. The locpregional control rate for NPC has improved significantly in the past few years with improvements in therapeutic strategy, but the incidence of distant metastasis remains ineffectively controlled.Citation2 Although these advanced approaches attempting to resolve the dilemma have resulted in the development of NPC, there remain a considerable number of underdetermined factors were paid attention working for progression of NPC. To the best of our knowledge, it is closely related to genetic susceptibility, living customs, and Epstein–Barr virus infection,Citation3 which induces tumorigenesis by multiple steps involved with genetic and epigenetic events.Citation1,Citation4 However, the mechanism in detail has yet to be addressed. In clinical practice, the TNM staging system is commonly used to predict the clinical outcome of NPC patients. However, patients at same clinical stage might manifest different prognoses. Therefore, there must be several unknown factors that play a crucial role in regulating the development of NPC. Consequently, there is an urgent need to find the signaling pathways associated with molecular and biological changes in NPC, and further to generate new prognostic biomarkers to determine the prognosis of patients more precisely, which would be beneficial for tailored treatment for NPC patients. As such, it is essential to explore novel biomarkers for predicting prognosis and investigate new effective therapeutic targets for NPC.
Research has indicated that ZMYND8 is initially employed as an activated protein kinase C– binding protein (also called PRKCBP1 or RACK7),Citation5 which has a completely different domain architecture consisting of an MYND domain close to its C terminus and a PHD finger, with bromo- and PWWP domains located at its N terminus. The presence of a triple PHD–bromo–PWWP domain structure for ZMYND8 indicates that this protein has the potential to combine with chromatin.Citation6,Citation7 Further studies have demonstrated that ZMYND8 is the crucial regulatory factor for histone lysine 4 trimethyl (H3K4me3)-specific demethylase KDM5C binding to enhancers as their suppressor. Deficiency of ZMYND8 or KDM5C results in downregulation of S100A oncogenes and various cancer-related phenotypes,Citation8 and ZMYND8 has also been identified as a transcriptional corepressor for downregulation of metastasis-linked genes via JARID1D mediation.Citation9 Other research has identified that the MYND domain of ZMYND8 facilitates recruitment of GATAD2A/NuRD to sites of DNA damage and further promotes its repair.Citation10 Delgado et al found that ZMYND8 combines with superenhancers of B lymphocytes to achieve efficient somatic hypermutation of IgH-variable regions associated with immunity regulation.Citation11 Another recent study reported that ZMYND8 harbors an alteration of somatic copy number in high-grade serous ovarian cancer in the Cancer Genome Atlas Research Network,Citation12 and ZMYND8 with a high mutation frequency of 19% was shown in mismatch repair–deficient colorectal cancers, which supports ZMYND8 playing the role of tumor oncogene.Citation13 More recently, another study showed that ZMYND8 inhibits tumor growth, proliferation, and tumorigenicity in prostate cancer, cervical cancer, and breast cancer cell lines.Citation8,Citation14 However, there have been no relevant studies on the prognostic value of ZMYND8 in NPCs. Therefore, the purpose of this study was to elucidate the status of ZMYND8 expression by immunohistochemistry and its prognostic significance in NPCs.
Methods
Ethics statement
The study was supported by the Institute Research Medical Ethics Committee of Sun Yat-Sen University and conducted in accordance with the Declaration of Helsinki. As all samples were anonymized, patient consent was waived.
Patients and tissue specimens
We collected paraffin samples from September 2002 to November 2004 in the Department of Pathology, Sun Yat-Sen University Cancer Center. Diagnostic criteria for all tissue samples was based on the 2000 WHO criteria for tumor classification and tumor staging on the basis of the International Union Against Cancer and the American Joint Committee on Cancer.
Immunohistochemistry
ZMYND8 protein was stained by immunohistochemistry in accordance with standard EnVision procedures. We sliced paraffin blocks into 3 μm sections and placed dry sections in an incubator at 60°C. Slides were deparaffinized with xylene, rehydrated in a graded ethanol series, immersed in 3% hydrogen peroxide for 10 minutes to inhibit endogenous peroxidase activity, and subsequently placed in boiled citric acid buffer for antigen retrieval. We incubated slides with antibody ZMYND8 (EPR16924, operative solution concentration 1:1,000; Abcam) at 37°C for 50 minutes. Thereafter, they were incubated with secondary antibody (K5007; Dako) at 37°C for 30 minutes. Then, staining was applied with 3,3-diaminobenzidine and the degree of staining observed by microscopy. Finally, slides were counterstained with hematoxylin, dehydrated in a graded ethanol series, cleared in xylene, and mounted with neutral gum. Positive and negative controls were set in the staining procedure.
Immunohistochemistry evaluation
ZMYND8 expression was assessed. The presence of dark-brown nuclei was taken to be positive for ZMYND8 expression, and scoring criteria were applied: each sample tissue harbored an intensity score (I score) of 0–3 (I0 means negative expression, I1 weak expression, I2 moderate expression, and I3 strong expression). Subsequently, ZMYND8 was elucidated according to the percentage of positively stained cells in 5% increments of 0–100%, obtaining a percentage score (P score). Finally, the total H score (0–300) was calculated by multiplying each I score and P score. (eg, [I1 × P1] + [I2 × P2] + [I3 × P3]).
Selection of cutoff value
The plot of true-positive fraction (sensitivity) versus false-negative fraction (1 – specificity) by various cutoffs generates an ROC curve in the unit square. ROC-curve analysis can determine the optimal cutoff value by point (eg, 0 or 1).Citation15,Citation16 The sensitivity and specificity for every clinicopathological variable was plotted, and constructing corresponding ROC curves for the ZMYND8 score, revealing both maximum sensitivity and specificity. Scores less than or equal to cutoff served as low expression of ZMYND8 and more than cutoff as high expression. Clinicopathological characteristics in ROC-curve analysi were T stage, N stage, M stage, clinical stage, relapse and survival status.
Statistical analysis
Statistical analyses were performed using SPSS 16.0. Correlations between ZMYND8 expression and clinicopathological variables of NPC patients were analyzed by χ2. Univariate analysis was used to determine associations between clinicopathological variables and overall survival and relapse-free survival of patients by the Kaplan–Meier method. Multivariate analysis was performed with the Cox regression model to identify independent prognostic factors. Two-tailed P<0.05 served as statistical significance.
Results
Patient characteristics
Clinicopathological features of NPC patients are described in . There were 137 (72.1%) men and 53 (27.9%) women, with median age of 46 years. Follow-up was 3–185 months (median 81.3 months). A total of 157 patients (82.6%) at late stages (III and IV) were diagnosed, and the other 33 patients (17.4%) were at early stages (I and II). Seven of 190 patients had metastasis to distant organs.
Table 1 Correlation between the ZMYND8 expression and clincopathological variables in nasopharyngeal carcinoma patients
Choice of cutoff score for ZMYND8 expression
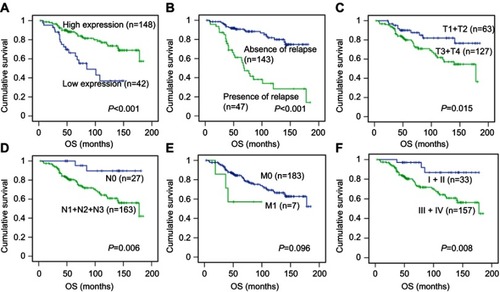
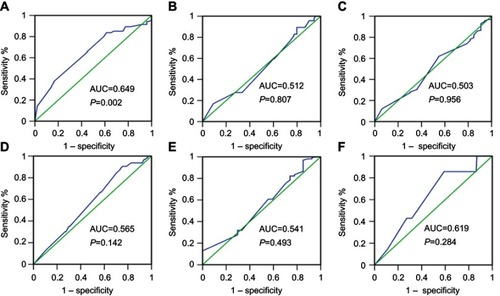
The status of expression of ZMYND8 protein in NPC tissues was displayed in . To choose a suitable cutoff score of ZMYND8 for further analysis.Citation15,Citation17 Tumor tissue samples with scores more than the obtained cutoff value were identified as high ZMYND8 expression and those less or equal to cutoff as low ZMYND8 expression. As shown in , ROC-curve points for of survival status closest to 0 and 1). On the basis of this outcome, we choose a score of 155 as the cutoff value. Low expression of ZMYND8 was found in 42 of 190 (22.1%) patients with NPCs, and 148 of 190 (77.9%) patients had high expression.
Figure 1 Expression of ZMYND8 protein in nasopharyngeal carcinoma tissue. Negative expression: (A) 10×); (E) 20×.; Weak expression: (B) 10×; (F) 20×.; Moderate expression: (C) 10×;, (G) 20×). ; Strong expression: (D) 10×; (H) 20×), .
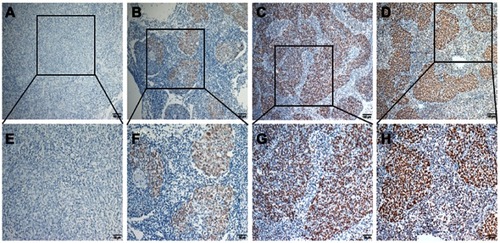
Figure 2 ROC-curve analysis of cutoff for high and low expression of ZMYND8 protein in nasopharyngeal carcinoma by various clinical parameters.
Notes: Sensitivity and specificity for each clinical parameter is plotted, with corresponding ROC curves for ZMYND8 scores. Cutoff values corresponding to points on curves closest to 0 and 1), meaning that the point had both maximum sensitivity and specificity. Green lines are reference lines and blue lines are movements in different cutoff values of corresponding curves by sensitivity (y-axis and 1 – specificity (y-axis). Survival status (A); clinical stage (B); relapse (C); T stage (D); M stage (E); N stage (F).
Relationship between ZMYND8 expression and clinicopathological variables of NPC patients
Low and high expression of ZMYND8 in NPC patients linked to several clinicopathological variables are described in . The data demonstrated that low expression of ZMYND8 was significantly correlated with late T stage (P<0.05), and no significant association was found between ZMYND8 expression and other clinicopathological variables, such as N stage, M stage, clinical stage, age, sex, relapse and therapy regimen. (P>0.05).
Relationship between ZMYND8 expression and NPC-patient survival
Univariate analysis showed a significant impact of well-known clinicopathological prognostic factors on NPC patients’ survival rates, such as T stage, N stage, clinical stage, and relapse (P<0.05, ). Univariate analysis also demonstrated that low expression of ZMYND8 was closely associated with worse overall survival (P<0.001, ), and presence of relapse, late T stage (T3 + T4), presence of lymph-node metastasis (N1 + N2 + N3) and advanced clinical stage (III + IV) were significantly correlated with poor overall survival (P<0.05, –). No statistical significance was found between expression of ZMYND8 and relapse-free survival of patients with NPC (HR 1.091, 95% CI 0.526–2.264; P=0.815;, ). Cox regression showed that low expression of ZMYND8 was an independent prognostic factor for overall survival (HR 0.300, 95% CI 0.164–0.547; P<0.001; ).
Table 2 Univariate analysis of different prognostic factors in 190 patients with nasopharyngeal carcinoma
Table 3 Univariate and multivariate analysis of ZMYND8 associated with relapse-free survival in nasopharyngeal carcinoma patients
Table 4 Univariate and multivariate analysis of ZMYND8 associated with overall survival in nasopharyngeal carcinoma patients
Discussion
NPC is characterized by high sensitivity to radiotherapy and chemotherapy. Tumor metastases lead to elevated mortality rates, despite new therapeutic strategies combining with conventional radiotherapy and chemotherapy.Citation18 Different clinical outcomes are revealed for patients with the same clinical stage after receiving the same treatment. Therefore, it is necessary to exploit new objective evidence that could effectively distinguish patients with different prognoses that belong to the same clinical stage. Reports have show abnormal gene expression in NPC,Citation19,Citation20 but exploring novel molecular biomarkers that might predict tumor progression or relapse remains an urgent task.
ZMYND8 was initially identified as a RACK protein that was able to interact with activated PKCβI,Citation5 which has recently been found to be tumor-suppressive in various cancer types.Citation21 ZMYND8 participates in DNA-damage response by binding to the NuRD chromatin- remodeling complex, which promotes transcriptional silencing induced by DNA damage and furthermore facilitates repair of DNA double-strand breaks.Citation22 ZMYND8 links to H3K36me2 to further regulate genes that induced by retinoic acid for neuronal differentiation.Citation23 Interestingly, several other studies have revealed ZMYND8 acts as an important regulator in controlling cell growth, invasion, and migration, possibly through regulating enhancer functions and interacting with JARID1D.Citation8,Citation9 Research has uncovered ZMYND8 to be a tumor suppressor in human breast cancer, cervical cancer, and prostate cancer by retinoic acid mediation or functioning as a transcriptional corepressor.Citation9,Citation14 However, in recent years, reports appeared pointing to acetylation of ZMYND8 mediating progression and metastasis of breast cancer by interacting with HIF-dependent transcriptional program,Citation24 and a ZMYND8–RELA fusion gene was found to increase dramatically via the NFκB signal pathway in acute erythroid leukaemia.Citation25 Copy numbers of ZMYND8 are also increased two- to threefold fold in several cancer cell lines according to the UK Institute of Cancer Research and Catalogue of Somatic Mutations in Cancer (COSMIC). Mismatch repair–deficient colorectal cancers harbor a high inactivating mutation frequency (19%) of ZMYND8.Citation13 These studies implied that ZMYND8 can play a critical role in tumor progression and development. However, expression of the ZMYND8 protein and its prognostic significance in NPC remain unknown. In our study, we investigated ZMYND8 protein expression in 190 NPC tissue samples. Our results showed that low ZMYND8 expression was intensively correlated with late T stage, indicating that ZMYND8 inhibited differentiation and proliferation of NPC, in agreement with Shen et al.Citation8 Our data suggest that a crucial role of ZMYND8 is as a tumor suppressor during the development and progression of NPC. In addition, these results support ZMYND8 acting as a suppressor of malignant tumor transformation, possibly though its inactivated role in human cancers. The inactivated mechanism of ZMYND8 might be correlated with its inactivating mutations (nonsense and frame shift; 9.5%; COSMIC), which have been reported in various cancer subtypes. Moreover, low ZMYND8 expression was significantly associated with unfavorable survival statistics for NPC patients by univariate and multivariate analysis revealed that it is as an independent prognostic factor for predicting NPC patients’ clinical outcomes. These findings indicated that inactivated ZMYND8 might result in aggressive proliferation of tumors and could be used as an important biomarker for the evaluation of prognosis in NPC.
A previous study reported that the IGH gene was regulated by the chromatin reader ZMYND8 in mature B lymphocytes to achieve antibody diversity.Citation11 In combination with our findings, we speculate that ZMYND8 is essential for the modulation of an effective immunoresponse to maintain a somewhat degree of level in NPC, and that it would activate the silencing sequence and further lead to loss of its expression in NPC when the immunity microenvironment is abnormal. Other studies have demonstrated that ZMYND8 interacts with members of H3K4me3-specific KDM5 family, such as KDM5C and KDM5D, to suppress cancer-linked genes across regulating transcription.Citation8,Citation9 ZMYND8 is also target gene of all-trans retinoic acid (ATRA), and can participate in ATRA-mediated inhibition of cancer-cell proliferation and invasion.Citation14,Citation23 These molecular studies provided a multitude of underlying mechanisms that resulted in downregulation of ZMYND8. However, potential mechanisms by which ZMYND8 affects prognosis are still unclear and need to be further investigated. Therefore, we will deeply explore the potential mechanisms of ZMYND8-linked gene-mediated progression and metastasis of NPC in future experiments.
In a word, our study revealed that ZMYND8 expression might be an effective tool for assessment of those NPC patients at increased risk of tumor invasiveness and proliferation. Low expression of ZMYND8 acted as a new independent prognostic factor in NPC, and if we can upregulate ZMYND8 expression by activating its upstream regulatory factor in the associated signal pathway by a molecular biology approach, these patients with NPC would be suitable for individual treatment, possibly resulting in better prognoses.
Conclusion
Low ZMYND8 expression of could be of importance, due to displaying more aggressive behavior in NPC. Therefore, ZMYND8 expression might serve as an independent prediction factor in patients with NPCs.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgment
This work was supported by the Youth Foundation of the National Natural Science Foundation of China (81702755).
References
- Fandi A, Altun M, Azli N, Armand JP, Cvitkovic E. Nasopharyngeal cancer: epidemiology, staging, and treatment. Semin Oncol. 1994;21:382–397.8209270
- Lee AW, Ng WT, Chan YH, Sze H, Chan C, Lam TH. The battle against nasopharyngeal cancer. Radiother Oncol. 2012;104:272–278. doi:10.1016/j.radonc.2012.08.00122938727
- Al-Sarraf M, Reddy MS. Nasopharyngeal carcinoma. Curr Treat Options Oncol. 2002;3:21–32.12057084
- Endo K, Kondo S, Shackleford J, et al. Phosphorylated ezrin is associated with EBV latent membrane protein 1 in nasopharyngeal carcinoma and induces cell migration. Oncogene. 2009;28:1725–1735. doi:10.1038/onc.2009.2019234486
- Fossey SC, Kuroda S, Price JA, Pendleton JK, Freedman BI, Bowden DW. Identification and characterization of PRKCBP1, a candidate RACK-like protein. Mamm Genome. 2000;11:919–925. doi:10.1007/s00335001017411003709
- Ansieau S, Sergeant A. [BS69 and RACK7, a potential novel class of tumor suppressor genes]. Pathologie-biologie. 2003;51:397–399.12948759
- Gong F, Miller KM. Double duty: ZMYND8 in the DNA damage response and cancer. Cell Cycle. 2018;17:414–420. doi:10.1080/15384101.2017.137615029393731
- Shen H, Xu W, Guo R, et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016;165:331–342. doi:10.1016/j.cell.2016.02.06427058665
- Li N, Li Y, Lv J, et al. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol Cell. 2016;63:470–484. doi:10.1016/j.molcel.2016.06.03527477906
- Spruijt CG, Luijsterburg MS, Menafra R, et al. ZMYND8 Co-localizes with NuRD on target genes and regulates poly(ADP-Ribose)-dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Rep. 2016;17:783–798. doi:10.1016/j.celrep.2016.09.03727732854
- Delgado-Benito V, Rosen DB, Wang Q, et al. The chromatin reader ZMYND8 regulates Igh enhancers to promote immunoglobulin class switch recombination. Mol Cell. 2018;72:636–649 e638. doi:10.1016/j.molcel.2018.08.04230293785
- Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi:10.1038/nature1016621720365
- Park J, Betel D, Gryfe R, et al. Mutation profiling of mismatch repair-deficient colorectal cncers using an in silico genome scan to identify coding microsatellites. Cancer Res. 2002;62:1284–1288.11888892
- Basu M, Khan MW, Chakrabarti P, Das C. Chromatin reader ZMYND8 is a key target of all trans retinoic acid-mediated inhibition of cancer cell proliferation. Biochim Biophys Acta Gene Regul Mech. 2017;1860:450–459. doi:10.1016/j.bbagrm.2017.02.00428232094
- Cai MY, Zhang B, He WP, et al. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Sci. 2010;101:1543–1549. doi:10.1111/j.1349-7006.2010.01560.x20367640
- Greiner M, Pfeiffer D, Smith RD. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev Vet Med. 2000;45:23–41.10802332
- Luo RZ, Cai PQ, Li M, et al. Decreased expression of PTPN12 correlates with tumor recurrence and poor survival of patients with hepatocellular carcinoma. PLoS One. 2014;9:e85592. doi:10.1371/journal.pone.008559224475046
- Al-Sarraf M, LeBlanc M, Giri PG, et al. Chemoradiotherapy versus radiotherapy in patients with advanced nasopharyngeal cancer: phase III randomized Intergroup study 0099. J Clin Oncol. 1998;16:1310–1317. doi:10.1200/JCO.1998.16.4.13109552031
- Guo X, Zeng Y, Deng H, et al. Genetic polymorphisms of CYP2E1, GSTP1, NQO1 and MPO and the risk of nasopharyngeal carcinoma in a Han Chinese population of Southern China. BMC Res Notes. 2010;3:212. doi:10.1186/1756-0500-3-21220663217
- Luo DH, Chen QY, Liu H, et al. The independent, unfavorable prognostic factors endothelin A receptor and chemokine receptor 4 have a close relationship in promoting the motility of nasopharyngeal carcinoma cells via the activation of AKT and MAPK pathways. J Transl Med. 2013;11:203. doi:10.1186/1479-5876-11-20323987636
- Antal CE, Hudson AM, Kang E, et al. Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell. 2015;160:489–502. doi:10.1016/j.cell.2015.01.00125619690
- Gong F, Chiu LY, Cox B, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197–211. doi:10.1101/gad.252189.11425593309
- Adhikary S, Sanyal S, Basu M, et al. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of All-trans-retinoic Acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J Biol Chem. 2016;291:2664–2681. doi:10.1074/jbc.M115.67998526655721
- Chen Y, Wang Y, Luo W. ZMYND8 is a primary HIF coactivator that mediates breast cancer progression. Mol Cell Oncol. 2018;5:e1479619. doi:10.1080/23723556.2018.147961930250924
- Panagopoulos I, Micci F, Thorsen J, et al. Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS One. 2013;8:e63663. doi:10.1371/journal.pone.006366323667654