Abstract
Purpose
Although the roles of lncRNA FOXD2-AS1 have been investigated in many types of cancers including colorectal cancer (CRC), its functionality remains to be further investigated. Analysis of the TCGA data set revealed that FOXD2-AS1 was up-regulated in CRC tissues. This study aimed to analyze the function of FOXD2-AS1 in CRC.
Methods
FOXD2-AS1 expression was detected by qPCR. A 5-year follow-up study was performed to analyze the prognostic value of FOXD2-AS1 for CRC. Overexpression experiments were performed to analyze the interactions among FOXD2-AS1, miR-25-3p and Sema4C. Transwell assays were performed to analyze cell invasion and migration.
Results
In this study, we further confirmed the up-regulation of FOXD2-AS1 in CRC patients and showed that high FOXD2-AS1 level predicted poor survival. Bioinformatics analysis showed that miR-25-3p may bind FOXD2-AS1, while over-expression experiments showed no effects on each other’s expression. Instead, FOXD2-AS1 over-expression led to the up-regulate Sema4C, which is a target of miR-25-3p. Transwell assay showed that FOXD2-AS1 and Sema4C over-expression led to the increased invasion and migration rates of CRC cells. MiR-25-3p plays the opposite role and attenuated the effects of FOXD2-AS1 and Sema4C over-expression.
Conclusion
FOXD2-AS1 may regulate the miR-25-3p/Sema4C axis to promote the invasion and migration of CRC cells.
Introduction
Colorectal cancer (CRC), also referred to as colon cancer or bowel cancer, is a type of malignancy develops from rectum or colon.Citation1 CRC is the 3rd most commonly diagnosed cancer and a leading cause of cancer deaths.Citation1 In 2018, CRC caused a total number of 551,269 deaths, which were 5.8% of all cancer deaths.Citation2 During the same year, CRC affected 1,096,601 new cases, accounting for 6.1% of all new cancer cases.Citation2 Therefore, the reduction of the incidence and mortality rates of CRC is of great clinical significance. CRC screening is an effective approach to reduce mortality. However, a CRC screening program is only applied to a small portion of the whole population.Citation3,Citation4 Due to direct medical cost and indirect cost from reduced productivity, CRC is a heavy burden on public health worldwide.Citation5 Therefore, novel therapeutic and preventative approaches are still needed.
Obesity, physical inactivity, alcohol consumption, and tobacco smoking are major risk factors for CRC.Citation6,Citation7 However, avoidance of these risk factors requires the intensive intervention for people’s lifestyle, which is not practical for the majority of the population.Citation6,Citation7 Studies on the molecular pathogenesis of CRC have identified a considerable number of genetic alterations involved in CRC.Citation8 The identification of critical genetic players in CRC provided novel targets for the development of targeted therapies.Citation9,Citation10 MiR-25-3p is a well-characterized tumor-suppressive miRNA in cervical cancer,Citation11 while its roles in CRC are unknown. Through bioinformatics analysis, we predicted that miR-205 may target FOXD2-AS1, which has been characterized as an oncogenic long (>200 nt) non-coding RNA (lncRNA) in several types of cancers including CRC.Citation12,Citation13 However, the roles of the interactions between FOXD2-AS1 and miR-205 in CRC remain unclear. This study aimed to investigate the interactions between FOXD2-AS1 and miR-205 in CRC.
Materials And Methods
CRC Patients
We enrolled 32 male and 28 female CRC patients (38 to 66 years, 52.1± 5.5 years) at The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University between July 2012 and July 2015. Before the admission of patients, this study passed the review of The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University Ethics Committee. Inclusion criteria: 1) CRC patients diagnosed by histopathological exam; 2) newly diagnosed cases; 3) completed treatment and 5-year follow-up. Exclusion criteria: 1) CRC recurrence after therapies; 2) any therapies initiated within 3 months before admission; 3) patients combined with multiple diseases. All the 60 CRC patients were informed of details of experiments and they all signed informed consent.
Biopsy
Before the initiation of therapies, a biopsy was performed under Magnetic Resonance Imaging (MRI) to collect both tumor (CRC) and adjacent non-tumor tissues. All tissue samples were confirmed by performing histopathological exams. Before the following experiments, all samples were stored in a sink containing liquid nitrogen.
Therapies And Follow-Up
The patients were first staged based on the standard established by The American Joint Committee on Cancer (AJCC). The 60 patients included 10, 19, 17 and 14 cases at clinical stage I, II, III and IV, respectively. Based on clinical stages and the health conditions of each patient, different treatment approaches, such as chemotherapy, radiation therapy, targeted therapy, and surgery were performed. Before the initiation of therapies, a 5-year follow-up was carried out to monitor the survival of the 60 patients. All the 60 patients completed the follow-up or died of CRC during follow-up.
CRC Cell Line And Transient Transfection
CR4 human CRC cell line from Sigma-Aldrich (USA) was used. Cell culture conditions were 95% humidity, 5% CO2 and 37 °C. The cell culture medium was composed of 10% FBS and 90% Eagle’s Minimum Essential Medium. Transfections were performed using cells harvested at 75–85% confluence.
Expression vectors of FOXD2-AS1 and Sema4C were constructed using pcDNA3.1 (GenePharma, Shanghai, China) vector as the backbone. MiR-25-3p mimic and NC (negative control) miRNA, as well as FOXD2-AS1 siRNA and NC siRNA, were also synthesized by GenePharma. Lipofectamine 2000 (Thermo Fisher Scientific) was used to transfected 50 mM miRNA or siRNA (NC miRNA or siRNA as NC group) or 10 mM vector (empty vector as NC group) into 4×106 cells. To avoid cytotoxicity, cells were washed with fresh medium after incubation with a transfection mixture for 6h. The following assays were performed using cells collected at 24h post-transfection. Control (C) cells were un-transfected cells in all cases.
Luciferase Assay
Basic pGL3 Luciferase Reporter Vector (Promega Corporation) was used to construct the FOXD2-AS1 vector. Luciferase assay was performed by transfecting FOXD2-AS1 vector + NC miRNA (NC group) or FOXD2-AS1 vector + miR-25-3p mimic (miR-25-3p group) into cells. Luciferase activity was measured at 24h post-transfection using the Luciferase Assay System (Promega Corporation).
RNA Extractions And DNA Removal
TRIZOL reagent (Invitrogen, USA) was used for all RNA extractions from both biopsies and CR4 cells. During RNA precipitation, 85% of ethanol was used to harvest miRNAs. All RNA samples were incubated with gDNA Eraser (TaKaRa, Japan) to digest gDNA.
RT-PCR
To measure the expression levels of FOXD2-AS1 and Sema4C mRNA, SSRT III system (Thermo Fisher Scientific) and QuantiTect SYBR Green PCR Kit (Qiagen) were used for reverse transfections (RTs) and qPCR mixture preparation, respectively. GAPDH was used as an endogenous control.
To measure the expression levels of miR-25-3p, poly (A) was added to mature miRNAs, RTs were performed and qPCR reactions were carried out. All steps were completed using All-in-OneTM miRNA qRT-PCR Detection Kit (GeneCopoeia, Guangzhou, China). The 2−ΔΔCT method was used to analyze relative fold changes.
Primer sequences were: 5ʹ-TGGACCTAGCTGCAGCTCCA-3ʹ (forward) and 5ʹ-AGTTGAAGGTGCACACACTG-3ʹ (reverse) for FOXD2-AS1; 5ʹ-ACCTTGTGCCGCGTAAGACA-3ʹ (forward) and 5ʹ-CGTCAGCGTCAGTGTCAGGA-3ʹ (reverse) for Sema4C; 5ʹ-GTCTCCTCTGACTTCAACAGC-3ʹ (forward) and 5ʹ-ACCACCCTGTTGCTGTAGCCA-3ʹ (reverse) for GAPDH. Forward primer of miR-25-3p was 5ʹ-CATTGCACTTGTCTCGGTC-3ʹ. Reverse primer of primer of miR-25-3p and U6 primers were included in the kit.
Western Blot
RIPA buffer (Sigma-Aldrich, USA) was used to prepare cell lysate of CR4 cells with protease inhibitors. SDS-PAGE (10% running gel following 5% stacking) was used to separate proteins. Pre-stained SeeBlue rainbow marker (Invitrogen, USA) was used to find the position of target gene bands. Gel transferred was performed using PVDF membranes and blocking was performed in 5% non-fat milk for 2h at room temperature. GAPDH (ab38168, Abcam) and Sema4C (ab135856, Abcam) rabbit polyclonal primary antibodies were used to incubate the membranes for 18h at 4ºC. HRP (IgG) (Goat Anti-Rabbit, ab6721, Abcam) secondary antibody was then used to further incubate the membranes for 2 h at room temperature. Signals were produced using secondary antibody and data were normalized using ImageJ v.148 software.
Cell Migration And Invasion Assay
For cell migration assay, the upper chambers of a 24-well Transwell plate of 8 μm (BD Biosciences) were filled with a serum-free medium containing 105 cells. 20% FBS-containing medium (0.6 mL) was added into the lower chamber to serve as a chemoattractant. Cells were cultivated under the aforementioned conditions for 24h. After that, cotton swabs were used to remove the cells on the upper surface, while cells migrated to the lower surface were fixed and stained with 0.1% Crystal Violet (Sigma-Aldrich). Five visual fields were randomly selected under a light microscope to calculate the number of migrating cells. Cell invasion assay was performed in the same way except that Matrigel Invasion Chambers (BD Bioscience) were used.
Data Analysis
Three independent biological replicates were included in each experiment and mean values were calculated. Exploration of differences between CRC and non-tumor tissues was performed using a paired t-test. One-way ANOVA followed by the post hoc Tukey’s test was to analyze differences among multiple groups. Survival curves were performed following steps: 1) dividing the patients into high and low FOXD2-AS1 level groups (n=30) with the median expression level in CRC as cutoff value; 2) survival curve plotting by Kaplan-Meier plotter; 3) survival curve comparison by log rank test. P<0.05 was statistically significant.
Results
Up-Regulation Of FOXD2-AS1 In CRC Predicts Poor Survival
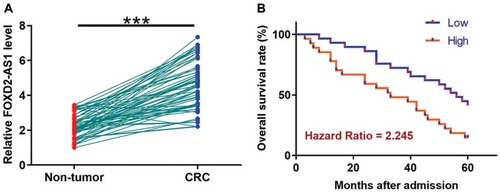
TCGA dataset was analyzed using an online program name GEPIA (http://gepia.cancer-pku.cn/) to explore the differential expression of FOXD2-AS1 in CRC. It was observed that FOXD2-AS1 was significantly up-regulated in CRC comparing to non-tumor tissues (4.4 vs. 1.69). To further confirm the up-regulation of FOXD2-AS1 in CRC, qPCR was performed to measure the expression levels of FOXD2-AS1 in both CRC and non-tumor tissues derived from the 60 CRC patients included in this study. Comparing to non-tumor samples, expression levels of FOXD2-AS1 were significantly higher in CRC tissue samples (, p<0.0001). Survival curves were plotted and compared following the aforementioned methods. It was observed that the survival rate of patients in the high FOXD2-AS1 level group was significantly lower than that of patients in the low FOXD2-AS1 level group ().
Figure 1 Up-regulation of FOXD2-AS1 in CRC predicts poor survival. qPCR was performed to measure the expression levels of FOXD2-AS1 in both CRC and non-tumor tissues derived from the 60 CRC patients included in this study (A). Survival curves were performed following steps: 1) dividing the patients into high and low FOXD2-AS1 level groups (n=30) with the median expression level in CRC as cutoff value; 2) survival curve plotting by Kaplan-Meier plotter; 3) survival curve comparison by log rank test (B). PCR reactions were repeated 3 times and mean values were presented. ***p<0.0001.
MiR-25-3p May Bind FOXD2-AS1 But Failed To Affect Its Expression
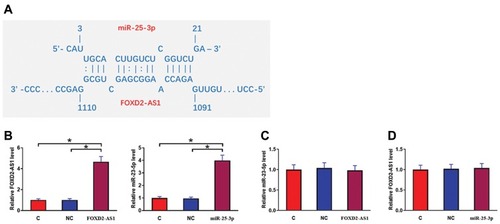
IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) was used to predict the interaction between miR-25-3p and FOXD2-AS1. It was observed that miR-25-3p can bind FOXD2-AS1 and form strong base pairing (). Luciferase assay was performed by transfecting FOXD2-AS1 vector + NC miRNA (NC group) or FOXD2-AS1 vector + miR-25-3p mimic (miR-25-3p group) into cells. Comparing to the NC group, relative luciferase activity was significantly lower in the miR-25-3p group (Supplemental Figure 1, p<0.05), supporting the direct interaction between them. To further analyze the relationship between them, CR4 cells were transfected with FOXD2-AS1 vector or miR-25-3p mimic. Over-expression of FOXD2-AS1 and miR-25-3p was confirmed at 24h post-transfection by qPCR (, p<0.05). Comparing to the C and NC group, cells with FOXD2-AS1 over-expression showed no significantly altered expression level of miR-25-3p (). Similarly, cells with miR-25-3p over-expression also showed no significantly changed FOXD2-AS1 expression ().
Figure 2 MiR-25-3p may bind FOXD2-AS1 but failed to affect its expression. IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) was used to predict the interaction between miR-25-3p and FOXD2-AS1. It was observed that miR-25-3p can bind FOXD2-AS1 and form strong base pairing (A). To further analyze the relationship between them, CR4 cells were transfected with FOXD2-AS1 vector or miR-25-3p mimic. Over-expression of FOXD2-AS1 and miR-25-3p was confirmed at 24h post-transfection by qPCR (B). Moreover, the effects of FOXD2-AS1 over-expression on miR-25-3p (C) and the effects of miR-25-3p over-expression on FOXD2-AS1 (D) were analyzed by qPCR. Experiments were repeated 3 times and data were expressed as mean values. *p<0.05.
FOXD2-AS1 Over-Expression Up-Regulated miR-25-3p Target Gene Sema4C
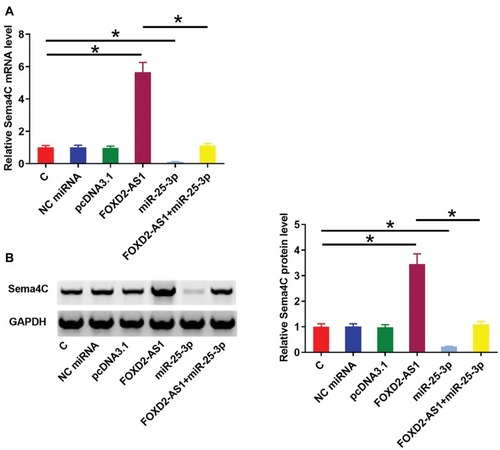
Sema4C is a target gene of miR-25-3p. Therefore, qPCR and Western blot experiments were performed to analyze the effects of miR-25-3p and FOXD2-AS1 over-expression on the expression of Sema4C at both mRNA () and protein () levels. Comparing to C and NC groups, cells with FOXD2-AS1 over-expression showed up-regulated Sema4C (p<0.05). In contrast, cells with miR-25-3p over-expression showed down-regulated Sema4C and reduced effects of FOXD2-AS1 over-expression on Sema4C expression (p<0.05). It is worth noting that FOXD2-AS1 siRNA silencing was also achieved (Supplemental Figure 2A), and FOXD2-AS1 siRNA silencing led to the down-regulation of Sema4C at both mRNA and protein levels (Supplemental Figure 2B, p<0.05.)
Figure 3 FOXD2-AS1 over-expression up-regulated miR-25-3p target gene Sema4C. Sema4C is a target gene of miR-25-3p. Therefore, qPCR and Western blot experiments were performed to analyze the effects of miR-25-3p and FOXD2-AS1 over-expression on the expression of Sema4C at both mRNA (A) and protein (B) levels. Experiments were repeated 3 times and data were expressed as mean values. *p<0.05.
FOXD2-AS1 Regulated miR-25-3p/Sema4c Axis To Promote CR4 Cell Invasion And Migration
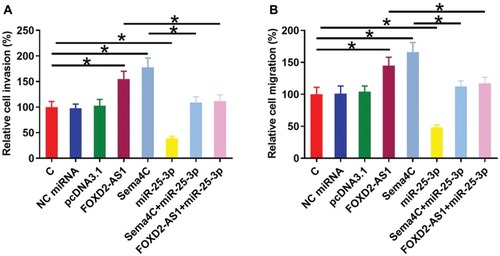
Transwell invasion and migration assays were performed to analyze the effects of FOXD2-AS1, miR-25-3p and Sema4C over-expression on the invasion () and migration () of CR4 cells. Comparing to C and NC groups, FOXD2-AS1 and Sema4C over-expression led to the increased invasion and migration rates of CRC cells (p<0.05). MiR-25-3p plays the opposite role and attenuated the effects of FOXD2-AS1 and Sema4C over-expression (p<0.05).
Figure 4 FOXD2-AS1 regulated miR-25-3p/Sema4C axis to promote CR4 cell invasion and migration. Transwell invasion and migration assays were performed to analyze the effects of FOXD2-AS1, miR-25-3p and Sema4C over-expression on the invasion (A) and migration (B) of CR4 cells. Experiments were repeated 3 times and data were expressed as mean values. *p<0.05.
Discussion
This study analyzed the interactions between FOXD2-AS1, miR-25-3p, and Sema4C in CRC. We observed that FOXD2-AS1 was up-regulated in CRC and its high expression level in CRC predicted the poor survival of CRC patients. Moreover, FOXD2-AS1 may regulate miR-25-3p/Sema4C to promote cancer cell invasion and migration.
The up-regulation of FOXD2-AS1 has been observed in many types of cancers, which is consistent with the reported oncogenic roles of FOXD2-AS1 in those types of cancers.Citation12–Citation14 In a recent study, Yang et al reported the up-regulation of FOXD2-AS1 in CRC.Citation13 They also showed that FOXD2-AS1 was able to target the Notch signaling pathway to suppress EMT in CRC.Citation13 Consistency, two datasets in this study, TCGA dataset and expression data of FOXD2-AS1 in both CRC and non-tumor tissues derived from CRC patients included in this study, both showed the up-regulation of FOXD2-AS1 in CRC. Moreover, our study showed the enhancing effects of FOXD2-AS1 over-expression on CRC cell invasion and migration. Our data further confirmed the oncogenic roles of FOXD2-AS1 in CRC.
By analyzing the TCGA dataset we observed the up-regulation of FOXD2-AS1 in most types of cancers. However, we also observed the obviously low levels of FOXD2-AS1 in acute myeloid leukemia (AML) comparing to non-tumor tissues (1.65 vs. 10.17), indicating the potential tumor-suppressive roles of FOXD2-AS1 in AML. Therefore, it will be interesting for future studies to analyze the functionality of FOXD2-AS1 in AML.
MiR-25-3p has been characterized as a tumor-suppressive miRNA or oncogenic miRNA in many types of cancers. For instance, miR-25-3p was down-regulated in tongue squamous cell carcinoma and its over-expression suppressed cancer cell proliferation.Citation15 In contrast, miR-25-3p was up-regulated in triple-negative breast cancer and was able to target BTG2 to promote the proliferation of cancer cells.Citation16 In a recent study, miR-25-3p was reported to reverse EMT by targeting Sema4C in cervical cancer cells.Citation11 In this study, we first reported the tumor-suppressive of miR-25-3p in CRC by targeting Sema4C to suppress tumor cell invasion and migration. Interestingly, our data showed that FOXD2-AS1 may sponge miR-25-3p to attenuate its effects on Sema4C expression as well as cancer cell invasion and migration. However, more studies may be needed to further explore the other possible mechanism.
Conclusion
In conclusion, FOXD2-AS1 was up-regulated in CRC and may play an oncogenic role in CRC by sponging miR-25-3p to up-regulate Sema4C, therefore suppressing cancer cell invasion and migrations.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgment
We thank the financial support from the Graduate research and innovation program of Jiangsu Province, No: KYCX18_1504.
References
- Schreuders EH, Ruco A, Rabeneck L, et al. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10 ):1637–1649. doi:10.1136/gutjnl-2014-30908626041752
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6 ):394–424. doi:10.3322/caac.v68.630207593
- Wolf AMD, Fontham ETH, Church TR, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68(4 ):250–281. doi:10.3322/caac.2145729846947
- Force USPST, Bibbins-Domingo K, Grossman DC, et al. Screening for colorectal cancer: US preventive services task force recommendation statement. JAMA. 2016;315(23 ):2564–2575. doi:10.1001/jama.2016.598927304597
- Favoriti P, Carbone G, Greco M, Pirozzi F, Pirozzi RE, Corcione F. Worldwide burden of colorectal cancer: a review. Updates Surg. 2016;68(1 ):7–11. doi:10.1007/s13304-016-0359-y27067591
- Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4 ):191–197. doi:10.1055/s-0029-124245821037809
- Jeon J, Du M, Schoen RE, et al. Determining risk of colorectal cancer and starting age of screening based on lifestyle, environmental, and genetic factors. Gastroenterology. 2018;154(8 ):2152–2164 e2119. doi:10.1053/j.gastro.2018.02.02129458155
- Grasso CS, Giannakis M, Wells DK, et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8(6 ):730–749. doi:10.1158/2159-8290.CD-17-132729510987
- Bahrami A, Khazaei M, Hasanzadeh M, et al. Therapeutic potential of targeting PI3K/AKT pathway in treatment of colorectal cancer: rational and progress. J Cell Biochem. 2018;119(3 ):2460–2469. doi:10.1002/jcb.2595028230287
- Ross JS, Fakih M, Ali SM, et al. Targeting HER2 in colorectal cancer: the landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer. 2018;124(7 ):1358–1373.29338072
- Song J, Li Y. miR-25-3p reverses epithelial-mesenchymal transition via targeting Sema4C in cisplatin-resistance cervical cancer cells. Cancer Sci. 2017;108(1 ):23–31. doi:10.1111/cas.2017.108.issue-127743413
- Liu X, Fu Q, Li S, et al. LncRNA FOXD2-AS1 functions as a competing endogenous RNA to regulate TERT expression by sponging miR-7-5p in thyroid cancer. Front Endocrinol (Lausanne). 2019;10:207. doi:10.3389/fendo.2019.0020731024447
- Yang X, Duan B, Zhou X. Long non-coding RNA FOXD2-AS1 functions as a tumor promoter in colorectal cancer by regulating EMT and Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(16 ):3586–3591.28925486
- Dong H, Cao W, Xue J. Long noncoding FOXD2-AS1 is activated by CREB1 and promotes cell proliferation and metastasis in glioma by sponging miR-185 through targeting AKT1. Biochem Biophys Res Commun. 2019;508(4 ):1074–1081. doi:10.1016/j.bbrc.2018.12.05030553445
- Xu JY, Yang LL, Ma C, Huang YL, Zhu GX, Chen QL. MiR-25-3p attenuates the proliferation of tongue squamous cell carcinoma cell line Tca8113. Asian Pac J Trop Med. 2013;6(9 ):743–747. doi:10.1016/S1995-7645(13)60130-323827155
- Chen H, Pan H, Qian Y, Zhou W, Liu X. MiR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer. 2018;17(1 ):4. doi:10.1186/s12943-017-0754-029310680
