Abstract
Background
Breast cancer (BC) is the most commonly diagnosed cancer in women. Tumor recurrence and metastasis are the key causes of death in BC patients. Long non-coding RNA (lncRNA) is closely associated with BC progression. lncRNA nuclear-enriched abundant transcript (NEAT)1 has been reported to regulate the proliferation and mobility of several types of cancer cells. However, how lncRNA NEAT1 affects the proliferation and invasion of BC cells is not known.
Methods
Quantitative real time-polymerase chain reaction (qRT-PCR) was used to measure expression of lncRNA NEAT1 and microRNA (miR)-146b-5p in BC tissues and cell lines. Cell Counting Kit (CCK)-8, cell colony-formation, wound-healing, and Transwell™ assays were undertaken to determine the effects of lncRNA NEAT1 and miR-146b-5p on progression of BC cells. The interaction between lncRNA NEAT1 and miR-146b-5p was examined by luciferase reporter, RNA-binding protein immunoprecipitation (RIP), and RNA-pulldown assays.
Results
Expression of lncRNA NEAT1 was upregulated in BC tissues and cell lines. High expression of lncRNA NEAT1 predicted poor overall survival in BC patients. Silencing of expression of lncRNA NEAT1 inhibited epithelial–mesenchymal transition (EMT) and suppressed the proliferation, migration and invasion of BC cells. Ectopic expression of lncRNA NEAT1 induced EMT and promoted BC progression. Mechanistic investigations revealed that miR-146b-5p was a direct target of lncRNA NEAT1, and its expression was correlated negatively with expression of lncRNA NEAT1 in BC tissues.
Conclusion
lncRNA NEAT1 could (i) serve as a novel prognostic marker for BC and (ii) be a potential therapeutic target for BC.
Introduction
Breast cancer (BC) is a leading cause of cancer-related death worldwide.Citation1 Resection, radiotherapy, chemotherapy and targeted therapy are the common treatment strategies for BC.Citation1,Citation2 However, most patients obtain only modest survival benefits and die from tumor recurrence and distant metastasis.Citation3–Citation5 Therefore, understanding the mechanisms of the growth and metastasis of BC cells and identifying more efficacious therapeutic targets is extremely important.
Long non-coding RNAs (lncRNAs) are RNAs of length >200 nt incapable of coding proteins.Citation6,Citation7 Studies have revealed that multiple lncRNAs are dysregulated in many types of cancers, and that lncRNAs are closely associated with the diagnosis and prognosis in cancer patients,Citation8–Citation12 including BC.Citation7,Citation13,Citation14 lncRNAs have been implicated in the regulation of various cancer hallmarks, including proliferation, metastasis, apoptosis, and chemoresistance.Citation6,Citation12,Citation15
lncRNA LOXL1-AS1 has been reported to promote the invasion and metastasis of BC cells by antagonizing the expression and activity of microRNA (miR)-708-5p.Citation16 LINC02273 can epigenetically increase the transcription of AGR2 and lead to metastasis of BC cells.Citation17 In addition, nuclear-enriched abundant transcript (NEAT)1 is a nuclear-restricted lncRNA and is a critical architectural part of gene paraspeckles.Citation18 lncRNA NEAT1 can promote the growth and metastasis of tumor cells in several types of cancers, such as colorectal cancer,Citation19 pancreatic cancer,Citation20 and hepatocellular carcinoma.Citation21 lncRNA NEAT1 has been reported to regulate the growth and mobility of BC cells by targeting CBX7 and RTCB.Citation22 Also, lncRNA NEAT1 confers chemoresistance and cell “stemness” in triple-negative breast cancer (TNBC) cells.Citation23 Epithelial–mesenchymal transition (EMT) is a key inducer of tumor metastasis,Citation24 but the effect of lncRNA NEAT1 on EMT in BC cells is not known.
We explored the role of lncRNA NEAT1 in BC progression. We found that overexpression of lncRNA NEAT1 was associated with reduced overall survival in BC patients. Moreover, lncRNA NEAT1 induced EMT and promoted the proliferation, migration and invasion of BC cells by inhibiting miR-146b-5p expression.
Materials and Methods
Ethical Approval of the Study Protocol
The study protocol was approved by the Ethics Review Board of Guangzhou Panyu Central Hospital (FR-EE-2017415) in Guangzhou, China. Patients provided written informed consent. Our study was conducted in accordance with the Declaration of Helsinki 1975 and its subsequent amendments.
Clinical Specimens
Human BC tissues (n = 56) and adjacent non-tumor tissues (n = 56) were obtained from patients undergoing surgery at Guangzhou Panyu Central Hospital. Before surgery, no patient received chemotherapy or radiotherapy. Tissues were stored in liquid nitrogen and were confirmed by pathologic examination. Information regarding age, premenopausal status, postmenopausal status, treatment or BC subtype of enrolled BC patients is summarized in .
Table 1 Patients’ Information
Cells and Cell Culture
Human normal breast cells (MCF10A) and human BC cell lines (BT474, MCF-7, MDA-MB-231, MDA-MB-453, SK-BR-3) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, P.R. China). Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin, and maintained in a humidified atmosphere of 5% CO2 at 37°C.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA from BC tissues and cell lines was extracted with E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek, Atlanta, GA, USA). Reverse transcription was undertaken with the Transcriptor First Strand cDNA Synthesis kit (Roche, Basel, Switzerland). For miRNA, total RNA was extracted with the PrimeScript miRNA cDNA Synthesis kit (TaKaRa Biotechnology, Shiga, Japan). Then, complimentary (c)DNA was amplified and quantified using a CFX96 real-time thermocycler (Bio-Rad Laboratories, Hercules, CA, USA) with SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad Laboratories). Expression of lncRNA and miRNA was normalized to that of ACTB and U6, respectively. Relative gene expression was analyzed by the 2−ΔΔCT method.
Cell Transfection
Cells (2×105) were seeded in six-well plates and cultured overnight. Then, cells underwent transfection assays. Small interfering (si)RNA targeting lncRNA NEAT1, miR-146b-5p mimics, scrambled oligonucleotides and pcDNA3.1-NEAT1 were obtained from Shanghai GenePharma (Shanghai, China). Transfection was undertaken with Lipofectamine™ 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Western Blotting
Total protein in BC cells after transfections was extracted using RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA) containing a protease inhibitor cocktail (Roche). The protein concentration was determined using a BCA™ Protein Assay Kit (Invitrogen). Then, proteins (50 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 8% gels and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories). Next, PVDF membranes were blocked and incubated with primary antibodies against E-cadherin (1:1000 dilution), vimentin (1:1000), and β-actin (1:1000) (Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:2000 dilution; Cell Signaling Technology) for 1 h at room temperature. Then, protein bands were detected by an enhanced chemiluminescence system according to manufacturer (Bio-Rad Laboratories) protocols.
Cell-Proliferation Assay
The effects of lncRNA NEAT1 and miR-146b-5p on proliferation of BCs cells were examined by the Cell Counting Kit (CCK)-8 assay (Beyotime Biotechnology, Beijing, China) and cell colony-formation assay. In brief, cells (2×103) seeded in 96-well plates were transfected with miRNA mimic, siRNA or plasmids. For the CCK-8 assay, absorbance at 450 nm was measured with a microplate reader when cells were transfected for 0, 24, 48, and 72 h, respectively. For the cell colony-formation assay, cells were seeded into six-well plates at 2000 cells per well. Then, cells cultured for 12 days were fixed with 4% paraformaldehyde and stained with 0.05% Crystal Violet. Images of cell colonies were photographed and the number of cell colonies quantified.
Wound-Healing Assay
A wound-healing assay was conducted to determine the effect of lncRNA NEAT1 and miR-146b-5p on migration of BC cells. In brief, cells (2×105) seeded in six-well plates were transfected as indicated and cultured to ~90% confluence. Then, cells were scratched with 200-μL pipette tips and washed with phosphate-buffered saline to discard cell debris. After 24-h incubation, the wound width of monolayers of BC cells was observed and wound area quantified.
Transwell™ Assay
The effects of lncRNA NEAT1 and miR-146b-5p on the invasion of BC cells were determined by Transwell assays (Corning, Corning, NY, USA). Cells (4×104) were resuspended in serum-free medium and added to the upper filters (which had been pre-coated with the diluent Matrigel™ (Corning)). The bottom chambers were filled with 600 μL of culture medium containing 10% fetal bovine serum. After 24-h incubation, invasive cells were fixed with 4% paraformaldehyde and stained with 0.2% Crystal Violet. Then, cotton swabs were used to rub away the cells on the inner sides of the upper filters, and the number of invasive cells was counted.
Luciferase Reporter Assay
The 3ʹ-UTR of lncRNA NEAT1 with miR-146b-5p binding sites and its mutant were cloned into the pGL3 luciferase reporter vector (Invitrogen). Cells (5×103) were seeded in 96-well plates and cultured overnight. Then, MCF-7 cells were co-transfected with miR-146b-5p mimics and wild-type p-GL3-NEAT1 (NEAT1-WT) or mutant p-GL3-NEAT1 (NEAT1-Mut). After transfection for 48 h, a Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA) was used to measure the luciferase activity, which was normalized to the activity of Renilla luciferase driven by a constitutively expressed promoter in the phRL vector.
RNA-Binding Protein Immunoprecipitation (RIP) Assay
Cells (2×105) were seeded in six-well plates and cultured overnight. Then, the RIP assay was carried out using the Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) and Argonaute-2 antibody (Cell Signaling Technology) in accordance with the literature.Citation25,Citation26 Expression of co-precipitated RNAs was determined by qRT-PCR.
RNA-Pulldown Assay
Biotin-labeled NC miRNA (NC-Bio) and biotin-labeled miR-146b-5p (miR-146b-5p-Bio) were obtained from Shanghai GenePharma. Cells (2×105) were seeded in six-well plates and cultured overnight. Then, cells were transfected with bio-NC miRNA or bio-miR-146b-5p for 48 h. Next, an RNA-pulldown experiment using the Magnetic RNA Protein Pull-down kit (Thermo Fisher Scientific) was done according to manufacturer instructions. Expression of lncRNA NEAT1 in pulled-down RNAs was measured by qRT-PCR.
Statistical Analyses
Statistical analyses were processed using Prism 7.0 (GraphPad, San Diego, CA, USA). Data are the mean ± SD. Significance was determined by the two-tailed Student’s t-test (two groups) or one-way ANOVA followed by the Tukey’s test (more than two groups). P < 0.05 was considered significant.
Results
Expression of lncRNA NEAT1 is Upregulated in BC Tissues and Cell Lines
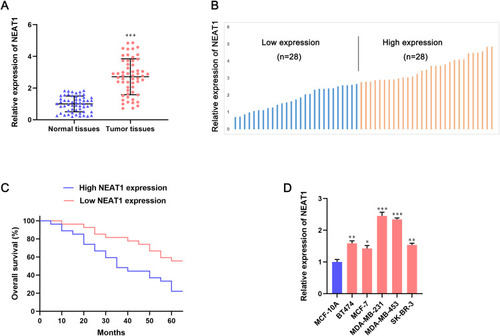
To investigate the role of lncRNA NEAT1 in BC progression, expression of lncRNA NEAT1 in human BC tissues and adjacent non-tumor tissues was measured by qRT-PCR. Expression of lncRNA NEAT1 was upregulated significantly in human BC tissues compared with that in adjacent non-tumor tissues (P < 0.001) (). Then, to analyze the correlation between expression of lncRNA NEAT1 and overall survival in patients with BC, 68 BC tissues were divided into two groups (high expression of NEAT1 and low expression of NEAT1) (). High expression of lncRNA NEAT1 was associated with reduced overall survival in patients with BC (P < 0.001) (). Moreover, compared with a human normal BC line (MCF10A), expression of lncRNA NEAT1 increased by 1.5-to-2.5-fold in five human BC cell lines (BT474, MCF-7, MDA-MB-231, MDA-MB-453, SK-BR-3) (). Taken together, these data suggested that lncRNA NEAT1 might function as a tumor-promoting lncRNA in progression of BC cells.
Figure 1 Expression of lncRNA NEAT1 is upregulated in breast-cancer tissues and predicts poor overall survival in breast-cancer patients. (A) Relative expression of lncRNA NEAT1 in breast-cancer tissues and adjacent tissues was determined by qRT-PCR. (B) Breast-cancer tissues (n = 56) were divided into two groups (high and low expression of lncRNA NEAT1) based on median expression. (C) Kaplan–Meier analyses of correlation between lncRNA NEAT1 expression and overall survival in patients with breast cancer. (D) Expression of lncRNA NEAT1 in human normal breast cells (MCF10A) and human breast cancer cells (BT474, MCF-7, MDA-MB-231, MDA-MB-453, SK-BR-3) was measured by qRT-PCR. Data are the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with normal tissues or MCF-10A cells.
Silencing Expression of lncRNA NEAT1 Inhibits EMT and Suppresses the Proliferation, Migration, and Invasion of BC Cells
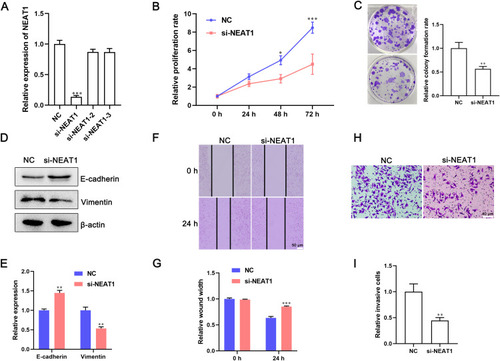
To investigate the effect of lncRNA NEAT1 on the proliferation and mobility of cells, MDA-MB-231 cells were transfected with lncRNA NEAT1-specific siRNA. qRT-PCR showed that expression of lncRNA NEAT1 was downregulated significantly in cells transfected with lncRNA NEAT1 siRNA (P < 0.001) (). Then, the proliferation of cells was determined by the CCK-8 assay and cell colony-formation assay. Silencing of expression of lncRNA NEAT1 inhibited proliferation dramatically () and led to a reduction in the number of cell colonies by ~50% () compared with that in cells transfected with NC siRNA. EMT has an important role in the invasion and metastasis of tumor cells. Western blotting and qRT-PCR revealed that expression of E-cadherin (marker of epithelial cells) was upregulated, whereas expression of vimentin (marker of mesenchymal cells) was downregulated significantly in cells transfected with lncRNA NEAT1 siRNA (). Then, cell mobility after transfection with lncRNA NEAT1 siRNA was determined by a wound-healing assay and Transwell assay. Knockdown of expression of lncRNA NEAT1 inhibited the closure of wound width and suppressed migration of MDA-MB-231 cells (). Moreover, the number of invaded cells was reduced by 50% after transfection with lncRNA NEAT1 siRNA (). Silencing of expression of lncRNA NEAT1 inhibited the proliferation, migration and invasion of MDA-MB-453 cells (Figure S1). Taken together, these results demonstrated that knockdown of expression of lncRNA NEAT1 inhibited EMT and suppressed the proliferation, migration and invasion of BC cells.
Figure 2 Knockdown of expression of lncRNA NEAT1 inhibits EMT and suppresses the proliferation, migration, and invasion of breast-cancer cells. MDA-MB-231 cells were transfected with lncRNA NEAT1 siRNA or negative control (NC) siRNA. (A) Expression of lncRNA NEAT1 in cells was measured by qRT-PCR. (B) Effect of lncRNA NEAT1 on proliferation of MDA-MB-231 cells was determined by the CCK-8 assay. (C) Representative images and quantification of cell colonies. (D and E) Expression of E-cadherin and vimentin was measured by (D) Western blotting and (E) qRT-PCR. (F and G) A wound-healing assay was used to detect migration of MDA-MB-231 cells. (F) Representative images of wound width and (G) quantification of cell migration. (H and I) Invasion capacity of cells was detected by the Transwell™ assay. (H) Representative images and (I) quantification of invasive cells after the indicated transfections. Data are the mean ± SD, n = 3. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with NC siRNA groups.
Overexpression of lncRNA NEAT1 Induces EMT and Promotes the Proliferation, Migration, and Invasion of BC Cells
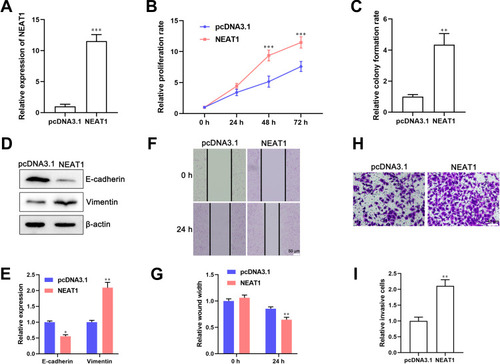
Next, we evaluated whether ectopic expression of lncRNA NEAT1 induced EMT and promoted the proliferation, migration and invasion of MCF-7 cells. Expression of lncRNA NEAT1 increased by more than 10-fold in cells transfected with a lncRNA NEAT1-overexpressing plasmid (). Overexpression of lncRNA NEAT1 promoted proliferation of BC cells significantly () and induced a 4-fold increase in the number of cell colonies (). Western blotting and qRT-PCR revealed that E-cadherin expression was downregulated, whereas vimentin expression was upregulated, in cells transfected with a lncRNA NEAT1-overexpressing plasmid compared with that in cells transfected with a vector (, ). NEAT1 overexpression promoted migration of MCF-7 cells (). Moreover, overexpression of lncRNA NEAT1 increased the number of invasive cells (). Taken together, these data suggested that ectopic expression of lncRNA NEAT1 induced EMT and promoted the proliferation, migration and invasion of BC cells.
Figure 3 Ectopic expression of lncRNA NEAT1 induces EMT and promotes the proliferation, migration, and invasion of breast-cancer cells. MCF-7 cells were transfected with a lncRNA NEAT1-overexpressing plasmid or pcDNA3.1 vector. (A) Expression of lncRNA NEAT1 in cells was determined by qRT-PCR. (B and C) Effect of lncRNA NEAT1 on proliferation of MCF-7 cells was examined by (B) the CCK-8 assay and (C) cell colony-formation assay. (D and E) Expression of E-cadherin and vimentin in breast-cancer cells was detected by (D) Western blotting and (E) qRT-PCR. (F and G) Wound-healing assay to ascertain the effect of lncRNA NEAT1 on migration of breast-cancer cells. (H) Representative images and (I) quantification of the number of invasive cells. Data are the mean ± SD, n = 3. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with the cells transfected with the pcDNA3.1 vector.
miR-146b-5p is a Target of lncRNA NEAT1
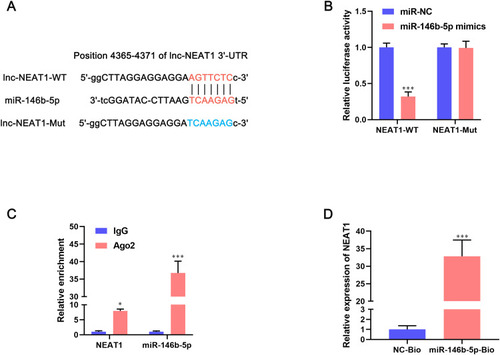
lncRNA can bind to miRNA and, thus, inhibit the expression and activity of miRNA. Hence, we further explored the regulatory mechanisms of lncRNA NEAT1 on BC progression. Starbase 2.0 (www.starbaserobins.org/) was used to predict the target of lncRNA NEAT1. We observed a potential binding site between lncRNA NEAT1 and miR-146b-5p (). Then, a dual-luciferase reporter gene assay was used to ascertain if miR-146b-4p was bound directly to the 3ʹ-UTR of lncRNA NEAT1. Co-transfection of NEAT1-WT and miR-146b-5p mimics reduced the luciferase activity significantly, whereas co-transfection with NEAT1-Mut and miR-146b-5p mimics had a negligible effect on the luciferase activity of BC cells (). These differences suggested that the 3ʹ-UTR of lncRNA NEAT1 was bound complementarily to miR-146b-5p. The RIP assay showed that lncRNA NEAT1 and miR-146b-5p were enriched in Ago2 pellets compared with that in IgG groups (). The RNA-pulldown assay showed that endogenous lncRNA NEAT1 could be pulled down in cells transfected with miR-146b-5p-Bio when compared with cells transfected with NC-Bio (P < 0.001) (). Collectively, these results suggested that miR-146b-5p was a direct target of lncRNA NEAT1.
Figure 4 miR-146b-5p is a direct target of lncRNA NEAT1 in breast-cancer cells. (A) The recognition sequences between lncRNA NEAT1 and miR-146b-5p were identified using Starbase 2.0. (B) MCF-7 cells were co-transfected with miR-146b-5p mimics and NEAT1-WT or NEAT1-Mut. Then, the luciferase activity was measured by the dual-luciferase reporter assay. Data are the mean ± SD, n = 3. ***P < 0.001 compared with miR-NC groups. (C) The interaction between lncRNA NEAT1 and miR-146b-5p was determined by an Ago2-RIP assay. Data are the mean ± SD, n = 3. ***P < 0.001 compared with IgG groups. (D) An RNA-pulldown assay was undertaken with MCF-7 cells transfected with biotin-labeled NC or miR-146b-5p mimics. Endogenous expression of lncRNA NEAT1 was measured by qRT-PCR. Data are the mean ± SD, n = 3. *P < 0.05 and ***P < 0.001 compared with NC-Bio groups.
miR-146b-5p Expression is Regulated Negatively by lncRNA NEAT1 in BC
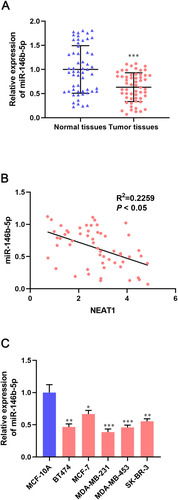
lncRNA NEAT1 was bound directly to miR-146b-5p, so we examined if expression of lncRNA NEAT1 was correlated negatively with miR-146b-5p expression in BC tissues and cell lines. qRT-PCR revealed that miR-146b-5p expression was downregulated significantly in BC tissues compared with that in adjacent non-tumor tissues (P < 0.001) (). Expression of lncRNA NEAT1 had a dramatically negative correlation with miR-146b-5p expression in BC tissues (R2 = 0.2259, P < 0.05) (). Compared with a human normal breast cell line (MCF10A), expression of lncRNA NEAT1 fell by 35% to 60% in five human BC cell lines (BT474, MCF-7, MDA-MB-231, MDA-MB-453, SK-BR-3 cells) (). Taken together, our data suggest that lncRNA NEAT1 had a negative correlation with miR-146b-5p in BC tissues and cell lines, and confirmed that miR-146b-5p could be a target of lncRNA NEAT1.
Figure 5 miR-146b-5p expression is downregulated in breast-cancer tissues and cell lines. (A) Relative expression of miR-146b-5p in breast-cancer tissues and adjacent tissues was determined by qRT-PCR. (B) Pearson correlation analysis of expression of lncRNA NEAT1 and miR-146b-5p in breast-cancer tissues. (C) qRT-PCR was done to measure expression of miR-146b-5p in normal breast cells (MCF10A) and breast cancer cell lines (BT474, MCF-7, MDA-MB-231, MDA-MB-453, SK-BR-3). Data are the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with normal tissues or MCF-10A cells.
miR-146b-5p Inhibits lncRNA NEAT1-Mediated Progression of BC Cells
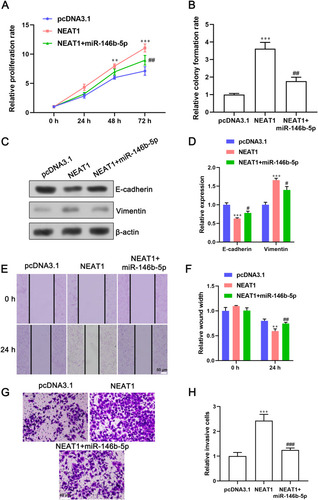
To further confirm the effect of miR-146b-5p and lncRNA NEAT1 on EMT and the proliferation, migration and invasion of BC cells, MCF-7 cells were co-transfected with miR-146b-5p mimics and a lncRNA NEAT1-overexpressing plasmid. miR-146b-5p mimics abrogated the ability of lncRNA NEAT1 to promote proliferation of MCF-7 cells () and formation of cell colonies (). Overexpression of lncRNA NEAT1 inhibited expression of E-cadherin and promoted expression of vimentin in BC cells, and this action could be reversed in cells co-transfected with a lncRNA NEAT1-overexpressing plasmid and miR-146b-5p mimics (). miR-146b-5p mimics impaired the ability of lncRNA NEAT1 to promote the migration and invasion of MCF-7 cells (–). Collectively, these results demonstrated that lncRNA NEAT1 induced EMT and promoted the proliferation, migration and invasion of BC cells by targeting miR-146b-5p.
Figure 6 miR-146b-5p overexpression inhibits lncRNA NEAT1-induced EMT, as well as the proliferation, migration, and invasion of breast-cancer cells. MCF-7 cells were co-transfected with a lncRNA NEAT1-overexpressing plasmid and miR-146b-5p mimics. (A and B) Effect of miR-146b-5p and lncRNA NEAT1 on proliferation of MCF-7 cells was determined by the (A) CCK-8 assay and (B) cell colony-formation assay. (C and D) Expression of E-cadherin and vimentin in cells after the indicated transfections was determined by (C) Western blotting and (D) qRT-PCR. (E and F) Representative image and quantification of the wound width in MCF-7-cell monolayers. (G and H) Representative images and quantification of invasive cells after indicated treatments. Data are the mean ± SD, n = 3. **P < 0.01 and ****P < 0.001 compared with the cells transfected with the pcDNA3.1 vector. #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with the cells transfected with a lncRNA NEAT1-overexpressing plasmid.
Discussion
BC is the major cause of cancer-related death in women worldwide. BC patients obtain only limited therapeutic outcomes, and most of them have poor improvements in survival benefits due to tumor recurrence and distant metastasis.Citation1–Citation5 Multiple factors regulate the growth and metastasis of BC cells, such as EMTCitation27 as well as genetic and epigenetic alternations in tumor cells.Citation17,Citation28,Citation29 An increasing number of studies has focused on lncRNA dysregulation in tumor cells. We found that the high expression of lncRNA NEAT1 in BC tissues and BC cell lines was correlated negatively with overall survival in BC patients. Further investigations showed that lncRNA NEAT1 induced EMT and promoted the proliferation, migration and invasion of BC cells by “sponging” miR-146b-5p. Our study indicated that lncRNA NEAT1 has a role in the growth and metastasis of BC cells.
lncRNA is a type of non-coding RNA, and has an important role in transcriptional control, chromatin remodeling, intracellular signaling and posttranscriptional regulation.Citation30,Citation31 Scholars have reported that lncRNA is closely associated with tumorigenesis,Citation32 tumor growth,Citation33 angiogenesis,Citation34 metastasis,Citation16 drug resistance,Citation35 and cell stemness.Citation36 Similar to the role of miRNA in tumor progression, lncRNA can be divided into “tumor-promoting lncRNA” and “tumor-suppressive lncRNA”. We demonstrated that lncRNA NEAT1 functioned as a tumor-promoting lncRNA in BC progression. Expression of lncRNA NEAT1 had a negative correlation with miR-146b-5p expression in BC tissues. Moreover, ectopic expression of miR-146b-5p inhibited lncRNA NEAT1-mediated EMT, as well as the proliferation, migration and mobility of BC cells, which suggested that miR-146b-5p might serve as a tumor-suppressive miRNA in BC. miRNAs can inhibit protein translation and/or degrade targeted mRNAs.Citation37 miR-146b-5p overexpression inhibited EMT of BC cells, as indicated by upregulation of E-cadherin expression and downregulation of vimentin expression. These changes suggested that miR-146b-5p might modulate the transcription and translation of EMT-related genes in BC cells. Clearly, more research is needed to explore further the mechanism of action of miR-146b-5p in BC progression. In addition to sponging miRNAs, lncRNA can also regulate transcription and translation. Whether lncRNA NEAT1 regulates expression of EMT- or metastasis-related genes directly in BC cells merits further investigation.
Expression of lncRNA NEAT1 in MDA-MB-231 and MDA-MB-453 cells (two TNBC cell lines) was higher than that in BT474 and MCF cells (two BC cell lines that are positive for estrogen receptor (ER) expression). TNBC is characterized by negative expression of the ER, progesterone receptor (PR) and human epidermal growth factor receptor (HER)-2. TNBC is the most aggressive type of BC, and has highly proliferative and metastatic features.Citation38–Citation40 TNBC accounts for about 10–20% of all types of BC.Citation38,Citation39 Different from ER-positive or HER-2-positive BC, efficacious agents to treat TNBC are lacking.Citation41,Citation42 However, whether expression of lncRNA NEAT1 is relevant to expression of HER-2, PR, or ER in BC tissues deserves further research.
Conclusions
lncRNA NEAT1 has a role in EMT as well as the proliferation, migration and invasion of BC cells probably because it can target miR-146b-5p. Our findings suggest that targeting lncRNA NEAT1 might be a promising approach to inhibit the growth and metastasis of BC cells.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflict of interest in this work.
References
- Dandawate PR, Subramaniam D, Jensen RA, et al. Targeting cancer stem cells and signaling pathways by phytochemicals: novel approach for breast cancer therapy. Semin Cancer Biol. 2016;40–41:192–208. doi:10.1016/j.semcancer.2016.09.001
- Chou J, Quigley DA, Robinson TM, et al. Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer Discov. 2020;10(3):351–370. doi:10.1158/2159-8290.CD-19-052832071145
- Scimeca M, Trivigno D, Bonfiglio R, et al. Breast cancer metastasis to bone: from epithelial to mesenchymal transition to breast osteoblast-like cells. Semin Cancer Biol. 2020. doi:10.1016/j.semcancer.2020.01.004
- Samadi P, Saki S, Dermani FK, et al. Emerging ways to treat breast cancer: will promises be met? Cell Oncol. 2018;41(6):605–621. doi:10.1007/s13402-018-0409-1
- Lim B, Hortobagyi GN. Current challenges of metastatic breast cancer. Cancer Metastasis Rev. 2016;35(4):495–514. doi:10.1007/s10555-016-9636-y27933405
- Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287–301. doi:10.1016/j.tcb.2017.11.00829274663
- Rodríguez Bautista R, Ortega Gómez A, Hidalgo Miranda A, et al. Long non-coding RNAs: implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non- and triple-negative breast cancer. Clin Epigenetics. 2018;10(1):88. doi:10.1186/s13148-018-0514-z29983835
- Wu Y, Yang L, Zhao J, et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14(1):191. doi:10.1186/s12943-015-0455-526552600
- Zhang X, Yan Z, Wang L, et al. STAT1-induced upregulation of lncRNA RHPN1-AS1 predicts a poor prognosis of hepatocellular carcinoma and contributes to tumor progression via the miR-485/CDCA5 axis. J Cell Biochem. 2020.
- Wang Y, Du L, Yang X, et al. A nomogram combining long non-coding RNA expression profiles and clinical factors predicts survival in patients with bladder cancer. Aging. 2020;12(3):2857–2879. doi:10.18632/aging.10278232047140
- Sun J, Zhang Z, Bao S, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;8(1):e000110. doi:10.1136/jitc-2019-00011032041817
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.01027070700
- Fan H, Yuan J, Li X, et al. LncRNA LINC00173 enhances triple-negative breast cancer progression by suppressing miR-490-3p expression. Biomed Pharmacother. 2020;125:109987. doi:10.1016/j.biopha.2020.10998732058222
- Du Y, Wei N, Hong J, et al. Long non-coding RNASNHG17 promotes the progression of breast cancer by sponging miR-124-3p. Cancer Cell Int. 2020;20(1):40. doi:10.1186/s12935-020-1129-y32042267
- Wang L, Cho KB, Li Y, et al. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20:5758.
- Dong HT, Liu Q, Zhao T, et al. Long non-coding RNA LOXL1-AS1 drives breast cancer invasion and metastasis by antagonizing miR-708-5p expression and activity. Mol Ther Nucleic Acids. 2019;19:696–705. doi:10.1016/j.omtn.2019.12.01631945728
- Xiu B, Chi Y, Liu L, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18(1):187. doi:10.1186/s12943-019-1115-y31856843
- Sasaki YT, Ideue T, Sano M, et al. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106(8):2525–2530. doi:10.1073/pnas.080789910619188602
- Liu H, Li A, Sun Z, et al. Long non-coding RNA NEAT1 promotes colorectal cancer progression by regulating miR-205-5p/VEGFA axis. Hum Cell. 2020;33:386–396.32065361
- Feng Y, Gao L, Cui G, et al. LncRNA NEAT1 facilitates pancreatic cancer growth and metastasis through stabilizing ELF3 mRNA. Am J Cancer Res. 2020;10(1):237–248.32064164
- Liu Q, Shi H, Yang J, et al. Long non-coding RNA NEAT1 promoted hepatocellular carcinoma cell proliferation and reduced apoptosis through the regulation of Let-7b-IGF-1R Axis. Onco Targets Ther. 2019;12:10401–10413. doi:10.2147/OTT.S21776331819522
- Yan L, Zhang Z, Yin X, et al. lncRNA NEAT1 facilitates cell proliferation, invasion and migration by regulating CBX7 and RTCB in breast cancer. Onco Targets Ther. 2020;13:2449–2458. doi:10.2147/OTT.S24076932273717
- Shin VY, Chen J, Cheuk IW, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10(4):270. doi:10.1038/s41419-019-1513-530894512
- Singh M, Yelle N, Venugopal C, et al. EMT: mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi:10.1016/j.pharmthera.2017.08.00928834698
- Krell J, Stebbing J, Carissimi C, et al. TP53 regulates miRNA association with AGO2 to remodel the miRNA-mRNA interaction network. Genome Res. 2016;26(3):331–341. doi:10.1101/gr.191759.11526701625
- Ottaviani S, Stebbing J, Frampton AE, et al. TGF-β induces miR-100 and miR-125b but blocks let-7a through LIN28B controlling PDAC progression. Nat Commun. 2018;9(1):1845. doi:10.1038/s41467-018-03962-x29748571
- Lourenço AR, Roukens MG, Seinstra D, et al. C/EBPɑ is crucial determinant of epithelial maintenance by preventing epithelial-to-mesenchymal transition. Nat Commun. 2020;11(1):785. doi:10.1038/s41467-020-14556-x32034145
- Lima ZS, Ghadamzadeh M, Arashloo FT, et al. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol. 2019;12:38.30975222
- Xu J, Wang Z, Li S, et al. Combinatorial epigenetic regulation of non-coding RNAs has profound effects on oncogenic pathways in breast cancer subtypes. Brief Bioinform. 2018;19(1):52–64. doi:10.1093/bib/bbw09927742663
- Nenad B, Maag Jesper LV, Dinger Marcel E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43.27233618
- Zhang Y, Tao Y, Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinform. 2018;19(5):930–945. doi:10.1093/bib/bbx04228449042
- Xia W, Liu Y, Cheng T, et al. Down-regulated lncRNA SBF2-AS1 inhibits tumorigenesis and progression of breast cancer by sponging microRNA-143 and repressing RRS1. J Exp Clin Cancer Res. 2020;39(1):18. doi:10.1186/s13046-020-1520-531952549
- Zhao L, Zhou Y, Zhao Y, et al. Long non-coding RNA TUSC8 inhibits breast cancer growth and metastasis via miR-190b-5p/MYLIP axis. Aging. 2020;12(3):2974–2991. doi:10.18632/aging.10279132039833
- Niu Y, Bao L, Chen Y, et al. HIF-2-induced long non-coding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res. 2020;80(5):964–975. doi:10.1158/0008-5472.CAN-19-153231900259
- Han M, Gu Y, Lu P, et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19(1):26. doi:10.1186/s12943-020-1145-532020881
- Xu Z, Liu C, Zhao Q, et al. Long non-coding RNA CCAT2 promotes oncogenesis in triple-negative breast cancer by regulating stemness of cancer cells. Pharmacol Res. 2020;152:104628. doi:10.1016/j.phrs.2020.10462831904506
- Zhang C, Zhang Y, Ding W, et al. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell. 2015;6(12):881–889. doi:10.1007/s13238-015-0223-826507842
- Carey L, Winer E, Viale G, et al. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7(12):683–692. doi:10.1038/nrclinonc.2010.15420877296
- Yeo SK, Guan JL. Breast cancer: multiple subtypes within a tumor? Trends Cancer. 2017;3(11):753–760. doi:10.1016/j.trecan.2017.09.00129120751
- Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15):4429–4434. doi:10.1158/1078-0432.CCR-06-304517671126
- Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232(2):142–150. doi:10.1002/path.428024114677
- Szekely B, Silber AL, Pusztai L. New therapeutic strategies for triple-negative breast cancer. Oncology (Williston Park). 2017;31(2):130–137.28205193
