Abstract
Background
Many studies have confirmed that circular RNAs (circRNAs) play a key role in the biological progression of cancers. However, the function of a novel circRNA, circ_0046599, in hepatocellular carcinoma (HCC) progression has not been explored.
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to measure the expression of circ_0046599, microRNA (miR)-1258 and Ribophorin II (RPN2). Subcellular fractionation location assay was used to localize circ_0046599 in HCC cells. The circular characteristic of circ_0046599 was verified using Ribonuclease R (RNase R) digestion assay. Besides, cell counting kit 8 (CCK8) assay, colony formation assay, wound healing assay and transwell assay were used to detect cell proliferation, migration and invasion, respectively. The lactate production and glucose level were determined by Lactate and Glucose Assay Kits. Furthermore, the protein levels of glycolysis, metastasis and proliferation-related marker proteins, as well as RPN2 were tested by Western blot (WB) analysis. Moreover, dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay were employed to confirm the interactions among circ_0046599, miR-1258 and RPN2. In addition, mice xenograft models were applied to observe the effect of circ_0046599 silencing on HCC tumor growth in vivo.
Results
Circ_0046599 was highly expressed in HCC tissues and cells, and its knockdown could suppress HCC cell proliferation, migration, invasion and glycolysis process. MiR-1258 could be targeted by circ_0046599, and its inhibitor could invert the suppressing effect of circ_0046599 knockdown on HCC progression. Further, RPN2 was a target of miR-1258. Overexpressed RPN2 could reverse the inhibiting effect of miR-1258 overexpression on HCC progression. Also, knockdown of circ_0046599 could restrain HCC tumor growth in vivo.
Conclusion
Our results provided new evidence that circ_0046599 could promote the progression of HCC by increasing RPN2 expression via sponging miR-1258.
Keywords:
Introduction
Hepatocellular carcinoma (HCC) is a common histological subtype of liver cancer, accounting for 90% of primary liver cancer.Citation1,Citation2 Genetic factors, hepatitis and obesity are considered to be major risk factors for HCC.Citation3,Citation4 The treatment of HCC includes surgical treatment, medical treatment, radiotherapy and so on.Citation5,Citation6 However, due to the high recurrence rate and metastasis rate of HCC, the prognosis of HCC is still poor.Citation7,Citation8 Therefore, it is necessary to study the pathogenesis of HCC to explore feasible therapeutic options for HCC.
Circular RNAs (circRNAs) are a class of non-coding RNAs with a special circular structure that function like microRNA (miRNA) sponges.Citation9,Citation10 In recent years, the research on the function of circRNA has reached a white-hot stage. Researchers have found that circRNA plays an important role in the progression of many types of cancer, including HCC.Citation11,Citation12 More importantly, the hypothesis that circRNA can act as a competitive endogenous RNA (ceRNA) to sponge miRNA provides a reliable way to elucidate the molecular mechanism of circRNA.Citation13,Citation14 For example, circABCB10 could promote HMG20A expression to enhance the proliferation and invasion of HCC by sponging miR-670-3p.Citation15 Also, circ_0001955 could accelerate the tumorigenesis of HCC via regulating the miR-516a-5p/TRAF6-MAPK11 network.Citation16 Therefore, circRNA may be an important biomarker for cancer therapy and may provide an important basis for elucidating the pathogenesis of cancers.
In our research, we screened the differentially expressed circRNA in HCC tumor tissues and hepar normal tissues based on the GSE97332 database, and discovered that circ_0046599 was remarkably higher expressed in HCC tumor tissues. However, there have no studies on the function of circ_0046599 in HCC at present. Therefore, the purpose of this study is to explore the role of circ_0046599 in the progression of HCC, and further analyze the potential molecular mechanism to provide a new biomarker for HCC treatment.
Materials and Methods
Clinical Tissues
Our study was approved by the Ethics Committee of Jiangxi Provincial People’s Hospital Affiliated to Nanchang University and was conducted in accordance with the Declaration of Helsinki. A total of 38 patients with HCC who were treated in Jiangxi Provincial People’s Hospital Affiliated to Nanchang University from March 2010 to December 2013 were recruited. After the operation, 38 paired HCC tissues and adjacent normal tissues were collected and frozen in liquid nitrogen. Patients were followed up from the date of surgery until the end of the study (December 2018) or the date of death. For this study, all patients signed written informed consent.
Cell Culture and Transfection
HCC cell lines (HCCLM3 and Huh7) and human liver epithelial cell line (THLE-2) were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China). HCCLM3 and Huh7 cells were grown in Dulbecco’s modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA), and THLE-2 cells were cultured in BEGM Bullet Kit (Lonza, Walkersville, MD, USA). In addition, all the media should be supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) and 1% penicillin/streptomycin (Invitrogen). All cells were incubated at 37°C with 5% CO2.
Circ_0046599 small interference RNA and overexpression plasmid (si-circ_0046599#1/#2 and circ_0046599) or negative controls (si-NC and circ-NC), miR-1258 mimic and inhibitor (miR-1258 and anti-miR-1258) or negative controls (NC and anti-NC), Ribophorin II (RPN2) overexpression plasmid (RPN2) and negative control (vector) were synthesized by GenePharma (Shanghai, China). Lentiviral short hairpin RNA against circ_0046599 (sh-circ_0046599) and negative control (sh-NC) were obtained from RiboBio (Guangzhou, China). Lipofectamine 3000 (Invitrogen) was used to transfect the above plasmids and oligonucleotides into cells.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TransZol (Transgen Biotech, Beijing, China) and reverse-transcribed into cDNA using First-Strand cDNA Synthesis SuperMix (Transgen Biotech). Finally, SYBR Green (Solarbio, Beijing, China) was employed to perform qRT-PCR on PCR system. The qRT-PCR results were analyzed with the 2−ΔΔCt method, and relative expression was normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6. All primer sequences were listed as below: circ_0046599, F 5ʹ-CTCCACCGAACGATACACACGTT-3ʹ, R 5ʹ-CTCACCCGCTGGGGCATC-3ʹ; miR-1258, F 5ʹ-CTGCGAGTCCCTGGAGTTAG-3ʹ, R 5ʹ-CGGTCCCCTAACTACCCATT-3ʹ; RPN2, F 5ʹ-AGGAAGTGGTGTTTGTTGCC-3ʹ, R 5ʹ-ACAGTCGAGGGAGCTTCTTC-3; GAPDH, F 5ʹ-TCAAGAAGGTGGTGAAGCAG-3ʹ, R 5ʹ-GAGGGGAGATTCAGTGTGGT-3ʹ; U6, F 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ, R 5ʹ-ACGCTTCACGAATTTGCGTGTC-3ʹ.
Subcellular Fractionation Location Assay
The cytoplasm and nucleus RNAs of HCCLM3 and Huh7 cells were isolated by Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Thorold, ON, Canada). Afterward, the expression levels of circ_0046599, GAPDH and U6 in the cytoplasm and nucleus of cells were examined by qRT-PCR. GAPDH and U6 were used as the cytoplasm control and nucleus control, respectively.
Ribonuclease R (RNase R) Digestion Assay
After extracted RNA from HCCLM3 and Huh7 cells, 3 U/μg RNase R (Epicentre, Madison, WI, USA) was added into 5 μg RNA for incubation 15 min at 37°C. Then, qRT-PCR was performed to assess the expression of circ_0046599 and GAPDH with or without RNase R digestion. GAPDH was served as the representative linear RNA.
Cell Counting Kit 8 (CCK8) Assay
HCCLM3 and Huh7 cells (1 × 103) were seeded in 96-well plates. At the indicated time point, CCK8 solution (Beyotime, Shanghai, China) was added into cells and incubated for 4 h. The absorbance at 450 nm was recorded by a microplate reader to evaluate cell viability.
Colony Formation Assay
HCCLM3 and Huh7 cells (110 per well) were seeded into 6-well plates and incubated for 2 weeks. Cells were then fixed with paraformaldehyde and stained with crystal violet. The microscope was used to count the number of colonies (> 50 cells).
Wound Healing Assay
HCCLM3 and Huh7 cells (5 × 105) were seeded in 6-well plates. After 24 h, 20 μL pipette tip was used to evenly draw 5 straight lines in the petri dish. Then, the cells were washed with phosphate buffer solution (PBS) and added with serum-free medium. After that, the cells were photographed under a microscope, and the wound area was calculated. After incubated for 24 h, the wound area was calculated again to evaluate the migration ability of cells.
Transwell Assay
Cell invasion was detected by the transwell chambers (Corning Inc., Corning, NY, USA). Serum-free medium was added into the upper chamber, while serum medium was filled into the lower chamber. HCCLM3 and Huh7 cells were seeded on the upper chamber, which was pre-coated with a Matrigel (Corning Inc.). All cells were cultured at 37°C for 24 h. After that, the lower chamber cells were with paraformaldehyde and stained with crystal violet. Then, the number of invaded cells was counted with a microscope.
Lactate Production and Glucose Level Measurement
The cell medium was collected 48 h after transfection. After centrifugation, the medium supernatant was collected in the centrifuge tube, and then the lactate production and glucose level were measured by Lactate Assay Kit and Glucose Assay Kit (BioVision, Milpitas, CA, USA) according to the manufacturer’s agreement.
Western Blot (WB) Analysis
Protein was extracted using RIPA buffer (Beyotime), and then protein samples were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Next, the membrane was blocked with 5% nonfat milk and incubated with primary antibody and secondary antibody. The protein signals were analyzed using enhanced chemiluminescent (ECL) substrate (Solarbio). All antibody was obtained from Abcepta (San Diego, CA, USA) and listed as below: hexokinase 2 (HK2, 1:1000, AP8140a), matrix metalloproteinase 2 (MMP2, 1:1000, AP53416), MMP9 (1:250, AM1975b), RPN2 (1:1000, AP2410a), proliferating cell nuclear antigen (PCNA, 1:2000, AM8545b), GAPDH (1:500, AP50811), anti-rabbit (1:2000, ASR1038) or anti-mouse (1:2000, ASR2846).
Dual-Luciferase Reporter Assay
The sequences of wild-type (wt) or mutant-type (mut) circ_0046595 and RPN2 3ʹUTR (circ_0046599-wt/mut and RPN2-wt/mut) were inserted into the pGL3 promoter vector (GenePharma). HCCLM3 and Huh7 cells were seeded in 24-well plates, and the above vectors and miR-1258 mimic or NC were co-transfected into cells. After 48 h, the ratio of firefly luciferase activity to Renilla luciferase activity was determined by Dual-Lucy Assay Kit (Solarbio) to count the relative luciferase activity.
RNA Immunoprecipitation (RIP) Assay
HCCLM3 and Huh7 cells were lysed in RIP lysis buffer (Sigma-Aldrich. Louis, MO, USA). Antibodies against immunoglobulin G (Anti-IgG) or argonaute 2 (Anti-Ago2) conjugated with magnetic beads (Sigma-Aldrich) were incubated with cell lysates overnight at 4°C. Then, qRT-PCR was used to measure the enrichment of circ_0046599, miR-1258 and RPN2 in Anti-Ago2 or Anti-IgG.
Mice Xenograft Models
All animal assays were approved by the Animal Research Committee of Jiangxi Provincial People’s Hospital Affiliated to Nanchang University and performed according to the Guide for the Care and Use of Laboratory Animals. Male BALB/c nude mice were bought from Sebiona (Guangzhou, China) and randomly divided into 2 groups (N = 3 per group). Huh7 cells (1 × 107) stably transfected with sh-circ_0046599 or sh-NC were subcutaneously injected into the right flank of mice. Tumor volume was calculated by detecting the tumor every week. Five weeks later, the mice were euthanized by isoflurane inhalation followed by cervical dislocation at the endpoint stage. The tumor was removed for further testing.
Statistical Analysis
GraphPad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA, USA) was used for data analysis. The results were presented by the mean ± standard deviation. Student’s t-test or one-way analyses of variance followed by Tukey’s test were performed for assessing the significance between 2 groups or among multiple groups, respectively. Correlation analysis was performed using Pearson correlation coefficient analysis. P < 0.05 was considered as significant.
Results
Circ_0046599 Was Highly Expressed in HCC Tissues and Cells
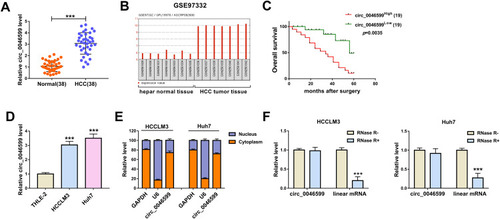
The expression of circ_0046599 was first detected by qRT-PCR, and the results revealed that circ_0046599 was upregulated in HCC tissues compared to adjacent normal tissues (). Then, in GSE97332 database, we found that circ_0046599 had elevated expression in HCC tumor tissues compared with that in hepar normal tissues (). Kaplan-Meier analysis showed that HCC patients with high circ_0046599 expression had lower overall survival than those with low circ_0046599 expression (). Additionally, we also measured circ_0046599 expression in HCC cell lines and discovered that circ_0046599 expression was higher in HCC cell lines (HCCLM3 and Huh7) than that in THLE-2 cells (). Subcellular fractionation location assay results suggested that circ_0046599 was mainly distributed in the cytoplasm of HCCLM3 and Huh7 cells (). To confirm the circular characteristic of circ_0046599, we treated with RNase R in extracted RNA and then detected the expression of circ_0046599 and GAPDH in HCCLM3 and Huh7 cells. The results showed that RNase R could degrade linear RNA (GAPDH) rather than circRNA (circ_0046599) (). Therefore, our results confirmed that the circ_0046599 was a circRNA and might have a vital function in HCC.
Figure 1 The expression of circ_0046599 was increased in HCC. (A) QRT-PCR results showed that circ_0046599 was upregulated in HCC tissues (HCC) compared with adjacent normal tissues (Normal). (B) GSE97332 database analyzed that circ_0046599 was highly expressed in HCC tumor tissues compared to hepar normal tissues. (C) Kaplan-Meier analysis suggested that the overall survival of HCC patients was associated with circ_0046599 expression. (D) QRT-PCR results indicated that circ_0046599 expression was elevated in HCC cell lines (HCCLM3 and Huh7) compared to THLE-2 cells. (E) Subcellular fractionation location assay showed that circ_0046599 was mainly distributed in the cytoplasm of HCCLM3 and Huh7 cells. (F) RNase R digestion assay revealed that circ_0046599 was resistant to the digestion of RNase R in HCCLM3 and Huh cells. ***P < 0.001.
Silenced Circ_0046599 Restrained the Proliferation, Migration, Invasion and Glycolysis Process of HCC Cells
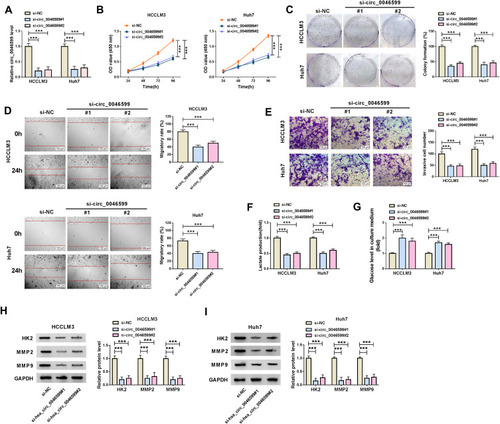
For exploring the function of circ_0046599 in HCC, we transfected with the siRNA of circ_0046599 in HCC cells. The downregulated circ_0046599 after transfected with si-circ_0046599#1/#2 indicated that the transfection efficiency of them was good (). Subsequently, we evaluated the biological functions of HCC cells. CCK8 and colony formation assays revealed that circ_0046599 knockdown markedly suppressed the viabilities and colony formation rates of HCCLM3 and Huh7 cells, which suggested that the proliferation of HCC cells was inhibited by circ_0046599 silencing (). Besides, the results of wound healing assay showed that the interference of circ_0046599 remarkably hindered the migratory rates of HCCLM3 and Huh7 cells (). Also, the number of invaded HCCLM3 and Huh7 cells was reduced by circ_0046599 knockdown, as demonstrated by transwell assay (). In addition, we also found that after circ_0046599 silencing, the lactate production of HCCLM3 and Huh7 cells was decreased significantly, while glucose level was increased significantly, indicating that the glycolysis process of HCC cells was repressed by circ_0046599 silencing (). At the same time, we also discovered that the protein expression levels of glycolysis rate-limiting enzyme (HK2) and metastasis-related proteins (MMP2 and MMP9) were obviously restrained by silenced circ_0046599, which once again confirmed the above results (). Hence, our results revealed that knockdown of circ_0046599 could inhibit the progression of HCC.
Figure 2 Effects of circ_0046599 knockdown on HCC progression. (A) The transfection efficiency of si-circ_0046599#1/#2 was confirmed by qRT-PCR. The proliferation of HCCLM3 and Huh cells was measured by CCK8 assay (B) and colony formation assay (C). (D) Wound healing assay was used to detect the migration of HCCLM3 and Huh cells. (E) The invasion of HCCLM3 and Huh cells was determined by transwell assay. The lactate production (F) and glucose level (G) of HCCLM3 and Huh cells were tested by corresponding Assay Kits. (H and I) WB analysis was performed to assess the protein levels of HK2, MMP2 and MMP9 in HCCLM3 and Huh cells. ***P < 0.001.
Circ_0046599 Directly Interacted with miR-1258
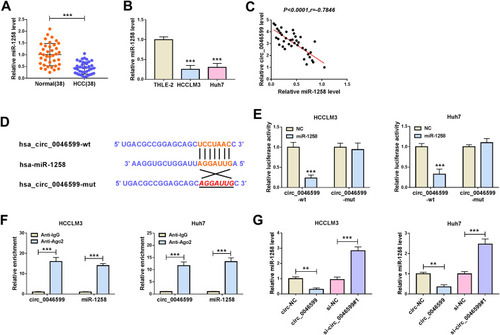
To explore the potential molecular mechanism of circ_0046599, we used the Circular RNA Interactome tool to predict miRNAs that could interact with circ_0046599. Through screening, we focused on miR-1258. QRT-PCR indicated that miR-1258 was significantly lower expressed in HCC tissues and cells (), and correlation analysis showed a significant negative correlation between miR-1258 and circ_0046599 expression in HCC tissues (). Subsequently, circ_0046599-wt and circ_0046599-mut reporter vectors were constructed based on the complementary sequences between miR-1258 and circ_0046599 (). Dual-luciferase reporter assay results showed that miR-1258 overexpression remarkably reduced the luciferase activity of circ_0046599-wt reporter rather than circ_0046599-mut reporter (). Furthermore, the results of RIP assay uncovered that the expression levels of circ_0046599 and miR-1258 were obviously enriched in Anti-Ago2 compared with that in Anti-IgG (). Additionally, we also found that miR-1258 expression was repressed by circ_0046599 overexpression, while promoted by circ_0046599 knockdown in HCCLM3 and Huh7 cells (). All data indicated that circ_0046599 could serve as a sponge of miR-1258 in HCC.
Figure 3 MiR-1258 could be targeted by circ_0046599 in HCC. QRT-PCR results revealed that miR-1258 was downregulated in HCC tissues (A) and cells (B) compared with matched controls. (C) Pearson correlation coefficient analysis was employed to analyze the correlation between circ_0046599 and miR-1258. (D) The binding and mutant sequences between circ_0046599 and miR-1258 were presented. The interaction between circ_0046599 and miR-1258 was verified by dual-luciferase reporter assay (E) and RIP assay (F). (G) QRT-PCR was used to detect miR-1258 expression in HCCLM3 and Huh cells to evaluate the effect of circ_0046599 on miR-1258 expression. **P < 0.01, ***P < 0.001.
Inhibition of miR-1258 Could Reverse the Suppression Effect of Circ_0046599 Knockdown on HCC Progression
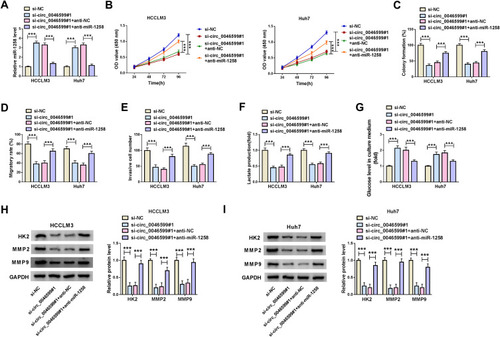
To investigate whether circ_0046599 regulated HCC progression by sponging miR-1258, we performed the rescue experiments using co-transfected with si-circ_0046599#1 and anti-miR-1258 into HCCLM3 and Huh7 cells. The detection results of miR-1258 expression suggested that miR-1258 inhibitor could invert the promotion effect of circ_0046599 silencing on miR-1258 expression, indicating that the transfections of si-circ_0046599#1 and anti-miR-1258 were successful (). Then, the results of CCK8 and colony formation assays confirmed that miR-1258 inhibitor could reverse the inhibition effect of circ_0046599 knockdown on the viabilities and colony formation rates of HCCLM3 and Huh7 cells (). Further, the suppression influence of silenced circ_0046599 on the migration and invasion of HCCLM3 and Huh7 cells also could be inverted by miR-1258 inhibitor (). Moreover, through detecting the lactate production and glucose level of HCCLM3 and Huh7 cells, we found that the inhibitory effect of circ_0046599 interference on the glycolysis process of HCC cells also could be reversed by miR-1258 inhibitor (). In addition, miR-1258 inhibitor could invert the decreasing effect of circ_0046599 silencing on the protein levels of HK2, MMP2 and MMP9 in HCCLM3 and Huh7 cells (). These results confirmed that circ_0046599 regulated HCC progression by targeting miR-1258.
Figure 4 Circ_0046599 regulated HCC progression by sponging miR-1258. (A) The expression of miR-1258 was detected by qRT-PCR to confirm the transfection efficiency of si-circ_0046599#1 and anti-miR-1258. CCK8 assay (B) and colony formation assay (C) were performed to assess the proliferation of HCCLM3 and Huh cells. (D) The migration of HCCLM3 and Huh cells was determined by wound healing assay. (E) Transwell assay was employed to measure the invasion of HCCLM3 and Huh cells. The glycolysis process of HCCLM3 and Huh cells was detected by testing the lactate production (F) and glucose level (G) using corresponding Assay Kits. (H and I) The protein levels of HK2, MMP2 and MMP9 in HCCLM3 and Huh cells were evaluated by WB analysis. ***P < 0.001.
MiR-1258 Directly Targeted RPN2 in HCC
To perfect the hypothesis of the circRNA-miRNA-mRNA regulatory network in HCC, the Targetscan tool was used to predict the mRNAs that could bind to miR-1258 in a complementary manner. It was found that RPN2 was a target of miR-1258. By detecting the mRNA expression of RPN2, we found that it was highly expressed in HCC tissues compared with adjacent normal tissues (). The protein level results of RPN2 in HCC tissues were consistent with the mRNA results (). Meanwhile, we also uncovered that RPN2 protein expression was upregulated in HCC cells compared to THLE-2 cells (). Additionally, correlation analysis results suggested that RPN2 expression was significantly positively correlated with circ_0046599 and negatively correlated with miR-1258 in HCC (). Then, we built the RPN2-wt and RPN2-mut reporter vectors to perform the dual-luciferase reporter assay, and the results showed that the luciferase activity of RPN2-wt reporter could be repressed by miR-1258 mimic, while the luciferase activity of RPN2-mut had not any changed in HCCLM3 and Huh7 cells (). Besides, RIP assay results revealed that the enrichment of miR-1258 and RPN2 was markedly increased in Anti-Ago2 (). To further confirm the regulation of miR-1258 and circ_0046599 on RPN2 expression, we co-transfected with miR-1258 mimic and circ_0046599 overexpression plasmids into HCCLM3 and Huh7 cells. Through measuring RPN2 protein expression, we discovered that miR-1258 overexpression could inhibit RPN2 expression, while the addition of circ_0046599 could reverse this effect (). Therefore, all data confirmed that circ_0046599 indirectly regulated RPN2 expression by sponging miR-1258 in HCC.
Figure 5 MiR-1258 directly targeted RPN2 in HCC. (A) QRT-PCR results indicated that the mRNA expression of RPN2 was increased in HCC tissues (HCC) compared with adjacent normal tissues (Normal). WB analysis results suggested that the protein expression levels of RPN2 were higher in HCC tissues (B) and cells (C) than that in matched controls. Pearson correlation coefficient analysis was employed to analyze the correlation between RPN2 and circ_0046599 (D) or miR-1258 (E). (F) The binding sites and mutant binding sites between RPN2 and miR-1258 were shown. Dual-luciferase reporter assay (G) and RIP assay (H) were performed to confirm the interaction between RPN2 and miR-1258. (I) The RPN2 protein expression in HCCLM3 and Huh cells was measured by WB analysis to assess the effect of miR-1258 and circ_0046599 expression on RPN2 expression. ***P < 0.001.
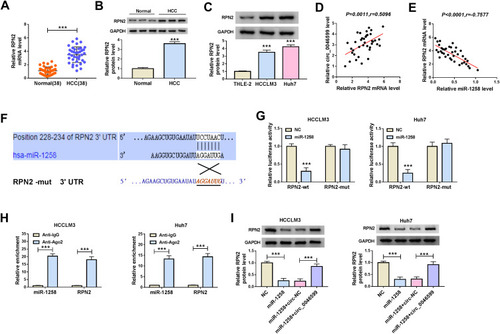
Overexpression of RPN2 Could Invert the Inhibitory Effect of miR-1258 Mimic on HCC Progression
To confirm that miR-1258 regulated HCC progression by targeting RPN2, we co-transfected with miR-1258 mimic and RPN2 overexpression plasmid into HCC cells. The detection results of RPN2 protein level indicated that overexpressed RPN2 could reverse the decreasing effect of miR-1258 overexpression on RPN2 expression in HCCLM3 and Huh7 cells, confirming the transfection efficiencies of miR-1258 mimic and RPN2 overexpression plasmid were excellent (). Subsequently, by measuring the viabilities and colony formation rates of HCCLM3 and Huh7 cells, we uncovered that miR-1258 overexpression could restrain the proliferation of HCC cells, whereas this effect could be inverted by RPN2 overexpression (). Furthermore, the results of wound healing assay and transwell assay also indicated that overexpressed RPN2 could reverse the suppression effect of miR-1258 mimic on the migration and invasion of HCCLM3 and Huh7 cells (). Besides, we also evaluated the glycolysis process of HCC cells through detecting the lactate production and glucose level of cells, and the results suggested that miR-1258 overexpression could hinder the lactate production and enhance the glucose level of HCCLM3 and Huh7 cells, while the addition of RPN2 could invert this influence (). Additionally, the inhibition effect of miR-1258 overexpression on the protein levels of HK2, MMP2 and MMP9 also could be recovered by overexpressed RPN2 in HCCLM3 and Huh7 cells (). Hence, we concluded that miR-1258 regulated the progression of HCC by targeting RPN2.
Figure 6 MiR-1258 regulated HCC progression by targeting RPN2. (A) The protein expression of RPN2 was determined by WB analysis to confirm the transfection efficiency of miR-1258 mimic and RPN2 overexpression plasmid. CCK8 assay (B) and colony formation assay (C) were used to detect the proliferation of HCCLM3 and Huh cells. (D) Wound healing assay was employed to test the migration of HCCLM3 and Huh cells. (E) Transwell assay was performed to assess the invasion of HCCLM3 and Huh cells. The lactate production (F) and glucose level (G) of HCCLM3 and Huh cells were measured using corresponding Assay Kits. (H and I) The protein levels of HK2, MMP2 and MMP9 were detected by WB analysis in HCCLM3 and Huh cells. ***P < 0.001.
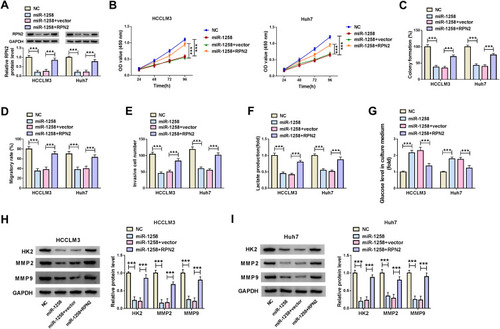
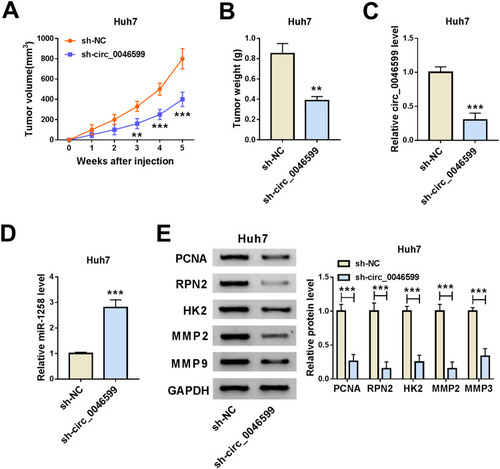
Knockdown of Circ_0046599 Inhibited the Growth of the Subcutaneous Transplanted HCC Tumor in vivo
At the same time, we also built the mice xenograft model to explore the effect of circ_0046599 silencing on HCC tumor growth in vivo. Through monitoring the tumor volume, we found that the tumor volume was significantly decreased after circ_0046599 knockdown (). Moreover, compared with the sh-NC group, the tumor weight in the sh-circ_0046599 group was remarkably reduced (). To confirm the success of circ_0046599 knockdown, we detected the expression of circ_0046599 in the tumors, and the results showed that circ_0046599 expression in the sh-circ_0046599 group was obviously lower than that in the sh-NC group (). Further, we also uncovered that miR-1258 expression was upregulated in the sh-circ_0046599 group compared to the sh-NC group (). In addition, WB analysis was used to detect the protein levels of proliferation-related marker (PCNA), RPN2, HK2, MMP2 and MMP9 in the tumors, and the results suggested that compared to the sh-NC group, the RPN2 expression was downregulated and the proliferation, metastasis and glycolysis process also were inhibited in the sh-circ_0046599 group (). Our data illuminated that circ_0046599 regulated the miR-1258/RPN2 axis to promote the tumor growth of HCC.
Figure 7 Circ_0046599 knockdown reduced the tumor growth of HCC tumor in vivo. The tumor volume (A) and tumor weight (B) were inhibited in the sh-circ_0046599 group compared with that in the sh-NC group. (C) QRT-PCR was used to evaluate the transfection efficiency of sh-circ_0046599. (D) The expression of miR-1258 in the tumors was detected by qRT-PCR. (E) The protein levels of PCNA, RPN2, HK2, MMP2 and MMP9 were measured by WB analysis in the tumors. **P < 0.01, ***P < 0.001.
Discussion
With the deepening of research, it has been implied that circRNAs act as an essential regulatory role in regulating gene transcription and expression, and the abnormal expression of circRNAs may be closely related to many cancer developments.Citation17 Many studies have demonstrated that circRNA has the potential to function as a key target for HCC treatment, prognosis and diagnosis, such as hsa-circ-0046600, hsa_circ_0000517 and hsa_circ_0027089.Citation18–Citation20 Although many circRNA functions have been demonstrated, a large number of circRNA functions in cancer have not yet been fully elucidated. Here, we focused on investigating the role of circ_0046599 in the progression of HCC. We uncovered that circ_0046599 was remarkably upregulated in HCC. Meanwhile, subsequent experiments indicated that downregulation of circ_0046599 could restrain the proliferation, migration, invasion and glycolysis process of HCC cells. Also, the in vivo experiments implied that circ_0046599 knockdown reduced the tumor growth of HCC through inhibiting the proliferation, metastasis and glycolysis process of HCC tumors. All these findings confirmed that circ_0046599 might function as an oncogene in HCC.
Subsequently, bioinformatics analysis was performed to elucidate the molecular mechanism of circ_0046599. Dual-luciferase reporter assay and RIP assay results revealed that circ_0046599 could sponge miR-1258 to regulate RPN2 expression. Previous studies had demonstrated that miR-1258 was poorly expressed in many cancers and might act as a tumor suppressor to involve in the regulation of cancer progression. Peng et al reported that miR-1258 could hinder the proliferation, metastasis and accelerate the apoptosis of cervical cancer.Citation21 Also, the results of Hwang et al confirmed that miR-1258 could suppress colorectal cancer cell proliferation and migration via targeting CKS1B.Citation22 More importantly, Zou et al showed that miR-1258 had decreased expression in HCC, and its overexpression could repress the proliferation and invasion of HCC.Citation23 Therefore, there was a reason to believe that miR-1258 might play a negative role in HCC progression. In our study, we discovered that miR-1258 was lowly expressed in HCC, which was consistent with the results of Zou et al.Citation23 In addition, miR-1258 expression was regulated by circ_0046599 in vitro and in vivo. The reversal effect of miR-1258 inhibitor on circ_0046599 silencing also suggested that miR-1258 could be sponged by circ_0046599 and could participate in the regulation of circ_0046599 on HCC progression.
RPN2 is an important membrane glycoprotein that mediates rough endoplasmic reticulum translocation and maintains endoplasmic reticulum specificity.Citation24 Moreover, many studies have found that the expression of RPN2 is associated with the malignant phenotype of many cancers.Citation25,Citation26 For example, Ding et al suggested that RPN2 expression was increased in glioma, and it could be involved in glioma progression regulated by the circNFIX/miR-378e axis.Citation27 Besides, Li et al found that esophageal cancer cell proliferation and metastasis could be suppressed by RPN2 knockdown.Citation28 In HCC, Huang et al confirmed that RPN2 was a vital regulator to promote the malignant progression of HCC.Citation29 Our study presented that RPN2 was highly expressed in HCC. Additionally, we also uncovered that miR-1258 regulated HCC cell proliferation, metastasis and glycolysis process by targeting RPN2. Our data confirmed that RPN2 could be targeted by miR-1258 to involve in the regulation of HCC progression. Furthermore, our results also indicated that circ_0046599 promoted RPN2 expression via targeting miR-1258. The introduction of the circ_0046599/miR-1258/RPN2 axis helped us to better understand the function of circ_0046599 in HCC.
In summary, our data concluded that circ_0046599 could serve as a ceRNA for miR-1258 to facilitate the proliferation, migration, invasion and glycolysis process of HCC through increasing RPN2 expression. Our results illustrated the role of circ_0046599 in the progression of HCC, suggesting that circ_0046599 might be a new potential therapeutic target of HCC.
Disclosure of Interest
The authors declare that they have no funding and no conflicts of interest in this work.
References
- Armengol C, Sarrias MR, Sala M. Hepatocellular carcinoma: present and future. Med Clin (Barc). 2018;150(10):390–397. doi:10.1016/j.medcli.2017.08.01029096967
- Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol. 2020;72(2):250–261. doi:10.1016/j.jhep.2019.08.02531954490
- Aspichueta P. Lipid-rich environment: a key role promoting carcinogenesis in obesity-related non-alcoholic fatty liver disease. Gut. 2018;67(8):1376–1377. doi:10.1136/gutjnl-2018-31604729540438
- Ruzic M, Pellicano R, Fabri M, et al. Hepatitis C virus-induced hepatocellular carcinoma: a narrative review. Panminerva Med. 2018;60(4):185–191. doi:10.23736/S0031-0808.18.03472-929856183
- Pinter M, Peck-Radosavljevic M. Review article: systemic treatment of hepatocellular carcinoma. Aliment Pharmacol Ther. 2018;48(6):598–609. doi:10.1111/apt.1491330039640
- Lurje I, Czigany Z, Bednarsch J, et al. Treatment strategies for hepatocellular carcinoma (-) a multidisciplinary approach. Int J Mol Sci. 2019;20(6):1465. doi:10.3390/ijms20061465
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.2144229313949
- Selcuk H. Prognostic factors and staging systems in hepatocellular carcinoma. Exp Clin Transplant. 2017;15(Suppl 2):45–49. doi:10.6002/ect.TOND16.L1128301998
- Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045. doi:10.1080/15476286.2016.127152427982727
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141. doi:10.1177/153537021770897828485684
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi:10.1080/15476286.2015.112216226649774
- Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. 2018;8:332. doi:10.3389/fonc.2018.0033230191143
- Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680. doi:10.1002/1878-0261.1246830719845
- Li M, Duan L, Li Y, Liu B. Long noncoding RNA/circular noncoding RNA-miRNA-mRNA axes in cardiovascular diseases. Life Sci. 2019;233:116440. doi:10.1016/j.lfs.2019.04.06631047893
- Fu Y, Cai L, Lei X, Wang D. Circular RNA ABCB10 promotes hepatocellular carcinoma progression by increasing HMG20A expression by sponging miR-670-3p. Cancer Cell Int. 2019;19(1):338. doi:10.1186/s12935-019-1055-z31889891
- Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi:10.1038/s41419-019-2176-y31822654
- Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10(8):258. doi:10.3390/cancers10080258
- Zhai Z, Fu Q, Liu C, et al. Emerging roles of hsa-circ-0046600 targeting the miR-640/HIF-1alpha signalling pathway in the progression of HCC. Onco Targets Ther. 2019;12:9291–9302. doi:10.2147/OTT.S22951431807009
- Wang X, Wang X, Li W, Zhang Q, Chen J, Chen T. Up-regulation of hsa_circ_0000517 predicts adverse prognosis of hepatocellular carcinoma. Front Oncol. 2019;9:1105. doi:10.3389/fonc.2019.0110531750237
- Zhu K, Zhan H, Peng Y, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 2020;41(3):296–302. doi:10.1093/carcin/bgz15431535687
- Peng X, Zhang Y, Gao J, Cai C. MiR-1258 promotes the apoptosis of cervical cancer cells by regulating the E2F1/P53 signaling pathway. Exp Mol Pathol. 2020;114:104368. doi:10.1016/j.yexmp.2020.10436831917289
- Hwang JS, Jeong EJ, Choi J, et al. MicroRNA-1258 inhibits the proliferation and migration of human colorectal cancer cells through suppressing CKS1B expression. Genes (Basel). 2019;10(11):912. doi:10.3390/genes10110912
- Zou H, Xu X, Luo L, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle. 2019;18(19):2398–2413. doi:10.1080/15384101.2019.161812031095447
- Crimaudo C, Hortsch M, Gausepohl H, Meyer DI. Human ribophorins I and II: the primary structure and membrane topology of two highly conserved rough endoplasmic reticulum-specific glycoproteins. EMBO J. 1987;6(1):75–82. doi:10.1002/j.1460-2075.1987.tb04721.x3034581
- Ono M, Tsuda H, Kobayashi T, et al. The expression and clinical significance of ribophorin II (RPN2) in human breast cancer. Pathol Int. 2015;65(6):301–308. doi:10.1111/pin.1229725881688
- Fujiwara T, Takahashi RU, Kosaka N, et al. RPN2 gene confers osteosarcoma cell malignant phenotypes and determines clinical prognosis. Mol Ther Nucleic Acids. 2014;3:e189. doi:10.1038/mtna.2014.3525181275
- Ding C, Wu Z, You H, et al. CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J Exp Clin Cancer Res. 2019;38(1):506. doi:10.1186/s13046-019-1483-631888753
- Li Y, Huang C, Bai Q, Yu J. Ribophorin II promotes cell proliferation, migration, and invasion in esophageal cancer cells in vitro and in vivo. Biosci Rep. 2019;39(5):BSR20182448. doi:10.1042/BSR2018244830940778
- Huang L, Jian Z, Gao Y, et al. RPN2 promotes metastasis of hepatocellular carcinoma cell and inhibits autophagy via STAT3 and NF-kappaB pathways. Aging (Albany NY). 2019;11(17):6674–6690. doi:10.18632/aging.10216731481647
