Abstract
Autophagy is an evolutionarily conserved lysosomal degradation pathway that eliminates cytosolic proteins, macromolecules, organelles, and protein aggregates. Activation of autophagy may function as a tumor suppressor by degrading defective organelles and other cellular components. However, this pathway may also be exploited by cancer cells to generate nutrients and energy during periods of starvation, hypoxia, and stress induced by chemotherapy. Therefore, induction of autophagy has emerged as a drug resistance mechanism that promotes cancer cell survival via self-digestion. Numerous preclinical studies have demonstrated that inhibition of autophagy enhances the activity of a broad array of anticancer agents. Thus, targeting autophagy may be a global anticancer strategy that may improve the efficacy of many standard of care agents. These results have led to multiple clinical trials to evaluate autophagy inhibition in combination with conventional chemotherapy. In this review, we summarize the anticancer agents that have been reported to modulate autophagy and discuss new developments in autophagy inhibition as an anticancer strategy.
Introduction
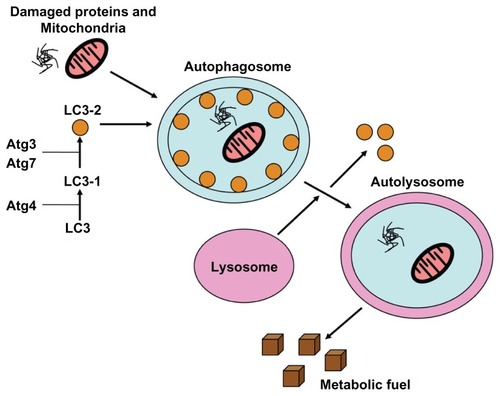
Autophagy is an evolutionarily conserved cellular catabolic degradation process that is characterized by the formation of double-membrane vesicles (autophagosomes) that engulf cellular components targeted for destruction. Autophagic degradation is an important regulator of cellular homeostasis as this process mediates the turnover of defective organelles, misfolded or aggregated proteins, and certain long-lived molecules.Citation1 Knockout animal studies demonstrated that some degree of basal autophagy is essential for viability as autophagy-deficient mice cannot survive the neonatal starvation period, and these mice die within one day of birth.Citation2 Ex vivo studies demonstrating that autophagy-deficient cells are significantly more susceptible to starvation-mediated cell death triggered by serum or growth factor deprivation than their autophagy-competent counterparts provided further support for a critical role for autophagy in the regulation of cellular homeostasis.Citation3 Autophagy is initiated by stress signals from the mammalian target of rapamycin complex 1 (mTORC1), which activates the kinase ULK1 (ATG1), and which then forms a complex with ATG13 and ATG17.Citation4 Autophagosome formation occurs after mTORC1 activity is inhibited and requires class 3 phosphoinositide 3-kinase (PI3K) activity as vacuolar sorting protein 34 forms a complex with Beclin-1.Citation5 Microtubule-associated protein light chain 3 (LC3) is incorporated into the membrane by ATG7 and ATG3. LC3 recruits adaptor proteins such as p62 and NIX that recognize protein aggregates and damaged organelles and recruits them into the autophagosome.Citation6,Citation7 Autophagosomes subsequently fuse with lysosomes to form autolysosomes, and their cargo is degraded by a number of different lysosomal proteases including the cathepsins (). In this review, we will summarize what is currently known regarding the regulation of autophagy, the role(s) of autophagy within the context of malignancy, and the therapeutic implications of autophagy activation versus autophagy inhibition in the treatment of cancer.
Figure 1 Autophagy produces metabolic fuel through the degradation of biomolecules.
Notes: Damaged proteins, organelles, and other biomolecules are sequestered into double-membrane vesicles called autophagosomes. LC3 is essential for autophagosome maturation. The mature autophagosomes fuse with the lysosome, and biomolecules are degraded by hydrolytic enzymes into metabolic fuel.
Abbreviation: LC3, lipidated cytosolic-associated protein light chain.
Established regulators of autophagy
Mammalian Target of Rapamycin (mTOR)
Although a number of signaling pathways have been implicated in the control of autophagy, the most well characterized autophagy regulator to date is mTOR. mTOR functions as two multiprotein complexes, mTORC1 and mTORC2, and each has unique binding partners and differential sensitivity to rapamycin and related compounds.Citation8 mTORC1 is able to form a complex with multiple binding partners, and its activity is inhibited by rapamycin and related drugs including temsirolimus/CCI-779, everolimus/RAD001, and ridaforolimus/AP23573.Citation9–Citation13 The PI3K/AKT/mTOR signaling cascade is an essential regulator of protein translation and cell proliferation. Its activity can be stimulated by growth factors and nutrients, although this pathway is constitutively active in many cancer types.
Constitutive PI3K/AKT signaling in malignant cells is frequently a consequence of mutations in PI3K or upstream growth factor receptors, AKT overexpression, or it can occur due to loss of the tumor suppressor phosphatase and tensin homolog (PTEN) on chromosome ten, which functions as a negative regulator of this pathway.Citation14,Citation15 Since activation of the PI3K/AKT cascade promotes mTOR activity, many tumor types exhibit high levels of mTOR activity due to constitutive upstream signaling events. In addition to its regulation by PI3K/AKT, mTOR activity is also controlled by AMP-activated kinase (AMPK), which functions as a sensor for cellular nutrient and energy levels. Upon its activation, mTORC1 stimulates protein synthesis via phosphorylation of the elongation factor 4E-BP1 and p70 S6 kinase. When nutrient and energy supplies are adequate, mTORC1 suppresses autophagy through the phosphorylation of the autophagy kinases ULK1 and ULK2.Citation16–Citation18
There are a number of different mechanisms through which the autophagy-related functions of the PI3K/AKT/mTOR pathway are regulated. For example, mTORC2 can inhibit autophagy through the phosphorylation of AKT.Citation19 Conversely, when levels of nutrients and metabolic fuel are diminished, the energy sensor AMPK can be activated upon its phosphorylation by the serine-threonine kinase LKB1. This subsequently leads to the activation of tuberous sclerosis protein (TSC2), a consequential reduction in mTORC1 activity, and the stimulation of autophagy. The activation of autophagy serves to maintain cell survival under nutrient-deficient conditions, as the breakdown of cellular components generates essential metabolic fuel. Similarly, increased autophagic activity has been frequently observed in malignant cells in response to treatment with therapeutic mTOR inhibitors, and this has been hypothesized to significantly reduce their clinical activity.Citation20
Beclin-1
Beclin-1 is a member of the BCL-2 homology domain 3 (BH3) containing proteins, which have been well characterized as critical regulators of apoptosis.Citation21 Beclin-1 is widely expressed in human tissues and is primarily localized to the endoplasmic reticulum, mitochondria, and the perinuclear membrane. Earlier studies demonstrated that the evolutionarily conserved domain of Beclin-1 is required for its proautophagic and tumor suppressive functions. The BH3 domain of Beclin-1 facilitates its self-association and its interactions with antiapoptotic members of the BCL-2 family. Recent studies have identified death-associated protein kinase (DAPK) as an important regulator of Beclin-1-mediated autophagy.Citation22 Phosphorylation of Beclin-1 by DAPK causes it to dissociate from BCL-2 family members and this event promotes the induction of autophagy.Citation22 Interestingly, DAPK has also been shown to have a Beclin-1 independent function in the stimulation of autophagy through its interactions with LC3.
A separate investigation defined an indirect role for c-Jun N-terminal kinase 1 (JNK1) in the control of Beclin-1-associated autophagy by demonstrating that JNK-mediated phosphorylation of BCL-2 disrupts its binding to Beclin-1, which ultimately leads to the induction of autophagy.Citation23 Ubiquitination also appears to contribute to the control of Beclin-1 function as its ubiquitination on lysine 63 was recently shown to facilitate its self-association and the formation of autophagosomes.Citation24 Additional studies are required to fully elucidate the finer mechanistic details regarding how different phosphorylation and ubiquitination events specifically control Beclin-1’s function within the context of autophagy. Finally, the generation of Beclin-1-deficient mice established the essential function of Beclin-1-mediated autophagy during embryonic development, as its loss yields an embryonic lethal phenotype characterized by death at embryonic day 7.5 or earlier due to impaired proamniotic canal closure.Citation25,Citation26
p53
The identification of the p53 transcriptional target damage-regulated autophagy modulator (DRAM) established a role for p53 in the regulation of autophagy. Initial studies demonstrated that DRAM1 expression was induced by DNA-damaging agents in a p53-dependent manner.Citation27,Citation28 Indications that DRAM1 may play a role in autophagy were initially revealed by its lysosomal localization. Mechanistic studies designed to elucidate DRAM1’s function(s) demonstrated that its expression promotes autophagosome formation.Citation27,Citation28 A subsequent study showed that DRAM could also be activated by the p53 related protein, p73. However, unlike p53, p73 retained its ability to induce autophagy in a manner that was independent of DRAM1 expression.Citation29 These findings indicated that other p53/p73 transcriptional targets may be able to compensate for the loss of DRAM1 with respect to the regulation of autophagy.
Several studies have identified a number of candidate transcriptional targets that may contribute to the control and execution of autophagy.Citation22,Citation30–Citation32 Additional research is required to define their individual roles and significance. Interestingly, more recent rigorous analyses of DRAM1 and its functions showed that DRAM1 encodes multiple isoforms and also belongs to a family of five related proteins, whose functions and transcriptional regulation remain to be fully elucidated.Citation33,Citation34 Ongoing studies will likely clarify these issues.
Autophagy in cancer
The roles of autophagy within the context of cancer remain somewhat controversial and appear to be quite divergent in the pre- and postmalignant states. Numerous studies have established that autophagy is an essential process in tumor suppression. The first evidence of this came from a study of mice with Beclin-1 haploinsufficiency (BECN1+/−), which displayed a significantly higher frequency of spontaneous malignancies (leukemias, lymphomas, and tumors of the lung and liver) as compared with their wild-type counterparts.Citation26 As mentioned earlier, deletion of both Beclin-1 alleles induces embryonic lethality.Citation25,Citation26 These phenotypic characteristics most likely result from the aberrant accumulation of organelles and specific proteins such as ubiquitinated keratins and the autophagy cargo adaptor p62 due to compromised autophagic degradation.Citation35 In particular, abrogation of p62 turnover due to the disruption of autophagy has been shown to lead to activation of the DNA damage response, disruption of cellular redox status, accumulation of defective mitochondria, increased susceptibility to degenerative and inflammatory diseases, and enhanced tumorigenicity.Citation35–Citation38 Specifically, several clinical correlative studies have provided additional support for a role for Beclin-1 in tumor suppression. The loss of Beclin-1 expression is linked to a poor prognosis in patients with solid malignancies. Low/deficient Beclin-1 expression is associated with shorter overall survival in patients with colon cancer and squamous-cell carcinoma of the esophagus.Citation39,Citation40 Conversely, elevated Beclin-1 levels appear to be associated with a favorable prognosis in patients with high-grade glioblastomas or hepatocellular tumors.Citation41,Citation42
However, a separate study of specimens from patients with nasopharyngeal tumors showed that high Beclin-1 expression was significantly correlated with poor outcomes.Citation43 Similarly, two independent studies investigated LC3 expression levels and their potential correlation with clinical outcomes in patients with cancer. A study of melanoma in situ (MIS) and melanoma specimens showed significantly higher levels of expression of LC3 and greater numbers of autophagosomes in both tissue types as compared with normal melanocytes and early stage melanoma in situ, indicating that the elevation of autophagic activity may be a later event in disease pathogenesis.Citation44 A larger assessment of LC3 levels in gastrointestinal tumors demonstrated overexpression of LC3 in more than 50% of all specimens analyzed. Interestingly, there was no clear link between LC3 levels and patient survival.Citation45 Collectively, these findings outline a potential role for autophagy as a contributor to the malignant phenotype, but suggest that caution should be taken regarding the potential use of any individual autophagy factor as a predictive biomarker.
The majority of pertinent studies conducted to date indicate that autophagy likely contributes to tumor suppression in healthy cells. However, it appears that autophagy may instead promote the pathogenesis of established malignancies by providing cancer cells with a mechanism to generate alternative sources of metabolic fuel via protein recycling, which facilitates their survival in nutrient/oxygen-deficient conditions.Citation46,Citation47 Considering that nutrient and oxygen deprivation are hallmark features of the tumor microenvironment, cancer cells may have an increased dependency upon autophagy for their survival. A number of studies have provided support for this hypothesis. For example, hypoxia-inducible transcription factor 1α (HIF-1α), which is constitutively active in many tumor types, was shown to promote BCL-2/adenovirus E1B 19kDa-interacting protein (BNIP3L)-dependent autophagy.Citation48,Citation49 Recent evidence suggests that hypoxic conditions may also trigger autophagy through a HIF-1α-independent mechanism that involves activation of the unfolded protein response.Citation50 Unfolded protein response-induced autophagy may serve as a mechanism to reduce endoplasmic reticular energy consumption while simultaneously generating metabolic fuel from the catabolism of endoplasmic reticulum membranes and other organelles such as mitochondria.Citation51
While most published studies support a prosurvival role for autophagy in cancer pathogenesis, there is also evidence for the induction of autophagy during the cell death process. In fact, some investigators in the field believe that autophagy can function as a unique caspase-independent mechanism of cell death that is distinct from apoptosis and necrosis as the minimum number of organelles or cellular components are degraded as required to support cell survival.Citation52 Others feel that autophagy associated with cell death likely reflects an attempt by cancer cells to survive the stress stimuli that initiated their death process and that it impedes rather than accelerates the efficiency of apoptotic/necrotic execution. The specific implications of this phenomenon are likely to be context-dependent and vary between in vitro and in vivo settings. Additional research is required in order to draw definitive conclusions. These reviews provide an excellent summary of what is currently known regarding autophagy as a potential mechanism of cell death.Citation53,Citation54
A number of studies have provided strong evidence implicating autophagy as a process that increases the survival capacity of both normal and malignant cells under stressful and metabolically challenging conditions both in culture and in the in vivo microenvironment.Citation2,Citation47,Citation55 Genetic impairment of autophagy significantly diminishes the ability of cells to withstand nutrient deprivation and confers an accelerated rate of cell death in response to these conditions as compared with experimental controls.Citation38 Although most evidence suggests that the survival advantage of autophagy-competent versus autophagy-deficient cells in the face of oxidative, genotoxic, or metabolic stress is derived from the autophagy-mediated catabolic generation of alternative energy sources, it is possible that other aspects of autophagy function could also promote cell survival and/or resistance to cell death. Ongoing research will likely define additional roles for autophagy in the regulation of cell survival.
Although low nutrient levels and hypoxia have been shown to trigger autophagy in cancer cells, numerous studies have shown that autophagy may also be stimulated in response to treatment with anticancer agents and radiation therapy. A wide array of therapeutic modalities with diverse mechanisms of action have been reported to induce autophagy including mTOR inhibitors, arsenic trioxide, etoposide, histone deacetylase (HDAC) inhibitors, tamoxifen, temozolomide, tyrosine kinase inhibitors, proteasome inhibitors, and ionizing radiation ().Citation56–Citation62 Based on earlier studies that showed that autophagy can represent both an alternative cell death pathway and a mechanism that promotes survival, autophagy is frequently viewed as a “double-edged sword.”
Table 1 Selected agents that modulate autophagy
The role that autophagy plays following chemotherapy remains highly controversial. It is possible that the induction of autophagy may contribute to the efficacy of some anticancer agents; conversely, it could also facilitate cell survival by maintaining bioenergetics following exposure to chemotherapeutic agents.Citation1 In order to better define the role of autophagy in the regulation of sensitivity to cancer therapeutics, investigators have quantified the impact of autophagy inhibition on the efficacy of numerous anticancer agents, including vorinostat, cyclophosphamide, and imatinib.Citation56,Citation57,Citation59,Citation60,Citation63 The overwhelming majority of relevant preclinical studies have demonstrated that disruption of autophagy significantly augments the efficacy of most classes of anticancer agents.Citation56–Citation59,Citation63 These studies suggest that autophagy inhibition may be an effective approach worthy of further investigation with broad applications in cancer therapy.
Therapeutic modulation of autophagy: new developments
As mentioned earlier, inhibition of autophagy in preclinical models increases the sensitivity of tumors to diverse classes of anticancer agents.Citation64 Chloroquine (CQ) and its analog, hydroxychloroquine (HCQ), are the only autophagic inhibitors currently being evaluated in clinical trials for cancer therapy. Both of these drugs disrupt lysosomal function and prevent the degradation of proteins within the autophagosome. HCQ is approved by the Food and Drug Administration for the treatment of malaria, rheumatoid arthritis, and lupus, and is currently being evaluated in numerous cancer clinical trials.Citation65 Most of these clinical studies are investigating HCQ in combination with standard of care agents (). Although these agents hold promise for their potential applications in cancer therapy due to their ability to inhibit autophagy and increase the efficacy of many standard of care treatments, CQ and HCQ induce ocular toxicities, such as retinopathy. It is also still unclear whether safely tolerated doses of HCQ or CQ can effectively and consistently inhibit autophagy in human tumors. These issues underscore the need for additional inhibitors of autophagy.
Table 2 Selected clinical trials with the autophagy inhibitor HCQ
Recent preclinical studies have identified two new autophagy inhibitors with potential clinical utility in cancer therapy, and presented with some advantages associated with CQ/HCQ. The first agent, lucanthone (Miracil D), is an existing drug that has been used for many years for the treatment of schistosomal parasites. Earlier investigations have shown that lucanthone inhibits topoisomerase 2 activity, as well as the activity of AP endonuclease, a critical regulator of DNA base excision repair.Citation66,Citation67 These findings prompted the evaluation of lucanthone as a potential sensitizer to chemotherapy and radiation. A more recent study defined a novel mechanism of action for lucanthone that includes the disruption of lysosomal function, inhibition of autophagy, and induction of apoptosis.Citation58 Lucanthone triggered apoptosis in cancer cells independently of p53 functional status. Moreover, knockdown studies demonstrated that the lysosomal protease cathepsin D was an important regulator of the proapoptotic effects of lucanthone. Notably, a direct comparison of the cellular sensitivity of a panel of human breast cancer cells to lucanthone and CQ showed that lucanthone was approximately tenfold more potent than CQ in this setting. The fact that earlier clinical studies of lucanthone did not demonstrate any evidence of drug-related ocular toxicity suggests that in addition to possessing increased anticancer activity, lucanthone may also have a better safety profile than CQ/HCQ.Citation68 These collective findings provide a strong rationale for the further investigation of lucanthone as a novel autophagy inhibitor with potential clinical applications in combination with anticancer agents that induce this survival pathway.
A second novel inhibitor of autophagy that warrants further investigation as an anticancer agent is Lys05. This new drug shares some structural homology with CQ and was specifically designed to inhibit autophagy more effectively through the inclusion of two aminoquinoline rings, a triamine linker, and C-7 chlorine.Citation69 Lys05 is a water-soluble compound that accumulates more readily within the lysosome and has improved deacidification effects compared with HCQ. Similar to lucanthone, Lys05 displayed significantly higher anticancer activity than CQ/HCQ in preclinical models without inducing significant observable toxicity. Additional studies are planned to further investigate the therapeutic anticancer potential of Lys05 as a novel inhibitor of autophagy.
Conclusion and future directions
Defining the role(s) of autophagy in malignant pathogenesis and in the regulation of therapeutic sensitivity is an important and challenging endeavor. A plethora of studies conducted to date have clearly shown that the induction of autophagy can be associated with both the execution of cell death as well as the protection from prodeath stimuli including metabolic stress and treatment with anticancer agents. These observations have generated many important scientific questions and opened up new fields of autophagy-related investigation within the context of cancer. Based on the currently available data in the literature, it is highly likely that the cellular consequences of autophagy induction are cell type-specific and treatment-dependent.
Several key issues that need to be better addressed experimentally include elucidating the individual roles of oncogenes in the regulation of cellular autophagy activity, defining the impact of the tumor microenvironment on autophagy function, and determining the most effective approaches to therapeutically target autophagy for cancer therapy. Of particular interest related to the latter issue regarding the modulation of autophagy as an anticancer strategy, numerous studies have demonstrated that the pharmacological and/or genetic disruption of autophagy in combination with an array of anticancer agents or ionizing radiation yields significant therapeutic benefit in preclinical cancer models. These findings have been clinically translated and more than 20 clinical trials investigating the established inhibitors of autophagy CQ and HCQ in combination with a variety of conventional and targeted cancer therapies are currently ongoing. These studies will likely yield important information regarding the safety and preliminary efficacy of these autophagy inhibitors as potential augmenters of cancer therapy.
Given that the ocular toxicities and minimal single agent anticancer efficacy of CQ and HCQ may limit their future applications in cancer therapy, the discovery of novel autophagy inhibitors is a high priority. Lucanthone and Lys05 are two new and exciting autophagy inhibitors that have demonstrated superior efficacy and tolerability compared with CQ/HCQ in preclinical studies. Ongoing preclinical studies and planned clinical trials with lucanthone and Lys05 will provide critical information regarding their potential applications in cancer therapy.
Acknowledgments
This work was supported by funding from The Institute for Drug Development, Cancer Therapy and Research Center at The University of Texas Health Science Center at San Antonio, USA.
Disclosure
The authors report no conflicts of interest in this work.
References
- CarewJSNawrockiSTClevelandJLModulating autophagy for therapeutic benefitAutophagy20073546446717495516
- KumaAHatanoMMatsuiMThe role of autophagy during the early neonatal starvation periodNature200443270201032103615525940
- KomatsuMWaguriSUenoTImpairment of starvation-induced and constitutive autophagy in Atg7-deficient miceJ Cell Biol2005169342543415866887
- JungCHJunCBRoSHULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machineryMol Biol Cell20092071992200319225151
- FunderburkSFWangQJYueZThe Beclin 1-VPS34 complex – at the crossroads of autophagy and beyondTrends Cell Biol201020635536220356743
- PankivSClausenTHLamarkTp62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagyJ Biol Chem200728233241312414517580304
- SchweersRLZhangJRandallMSNIX is required for programmed mitochondrial clearance during reticulocyte maturationProc Natl Acad Sci U S A200710449195001950518048346
- LoewithRJacintoEWullschlegerSTwo TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth controlMol Cell200210345746812408816
- HaraKMarukiYLongXRaptor, a binding partner of target of rapamycin (TOR), mediates TOR actionCell2002110217718912150926
- KimDHSarbassovDDAliSMmTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machineryCell2002110216317512150925
- PetersonTRLaplanteMThoreenCCDEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survivalCell2009137587388619446321
- SancakYThoreenCCPetersonTRPRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinaseMol Cell200725690391517386266
- Vander HaarELeeSIBandhakaviSGriffinTJKimDHInsulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40Nat Cell Biol20079331632317277771
- NeshatMSMellinghoffIKTranCEnhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTORProc Natl Acad Sci U S A20019818103141031911504908
- PodsypaninaKLeeRTPolitisCAn inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− miceProc Natl Acad Sci U S A20019818103201032511504907
- KamadaYFunakoshiTShintaniTNaganoKOhsumiMOhsumiYTor-mediated induction of autophagy via an Apg1 protein kinase complexJ Cell Biol200015061507151310995454
- YangZKlionskyDJMammalian autophagy: core molecular machinery and signaling regulationCurr Opin Cell Biol201022212413120034776
- HeCKlionskyDJRegulation mechanisms and signaling pathways of autophagyAnnu Rev Genet200943679319653858
- SunSYRosenbergLMWangXActivation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibitionCancer Res200565167052705816103051
- EastonJBHoughtonPJmTOR and cancer therapyOncogene200625486436644617041628
- ObersteinAJeffreyPDShiYCrystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only proteinJ Biol Chem200728217131231313217337444
- ZalckvarEBerissiHMizrachyLDAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagyEMBO Rep200910328529219180116
- WeiYPattingreSSinhaSBassikMLevineBJNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagyMol Cell200830667868818570871
- ShiCSKehrlJHTRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagySci Signal20103123ra4220501938
- YueZJinSYangCLevineAJHeintzNBeclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressorProc Natl Acad Sci U S A200310025150771508214657337
- QuXYuJBhagatGPromotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy geneJ Clin Invest2003112121809182014638851
- CrightonDWilkinsonSO’PreyJDRAM, a p53-induced modulator of autophagy, is critical for apoptosisCell2006126112113416839881
- CrightonDWilkinsonSRyanKMDRAM links autophagy to p53 and programmed cell deathAutophagy200731727417102582
- CrightonDO’PreyJBellHSRyanKMp73 regulates DRAM-independent autophagy that does not contribute to programmed cell deathCell Death Differ20071461071107917304243
- YeeKSWilkinsonSJamesJRyanKMVousdenKHPUMA- and Bax-induced autophagy contributes to apoptosisCell Death Differ20091681135114519300452
- FengZHuWde StanchinaEThe regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathwaysCancer Res20076773043305317409411
- BudanovAVKarinMp53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signalingCell2008134345146018692468
- O’PreyJSkommerJWilkinsonSRyanKMAnalysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagyCell Cycle20098142260226519556885
- MahLYO’PreyJBaudotADHoekstraARyanKMDRAM-1 encodes multiple isoforms that regulate autophagyAutophagy201281182822082963
- MathewRKarpCMBeaudoinBAutophagy suppresses tumorigenesis through elimination of p62Cell200913761062107519524509
- CadwellKLiuJYBrownSLA key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cellsNature2008456721925926318849966
- HaraTNakamuraKMatsuiMSuppression of basal autophagy in neural cells causes neurodegenerative disease in miceNature2006441709588588916625204
- MathewRKarantza-WadsworthVWhiteERole of autophagy in cancerNat Rev Cancer200771296196717972889
- LiBXLiCYPengRQThe expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancersAutophagy20095330330619066461
- ChenYLuYLuCZhangLBeclin-1 expression is a predictor of clinical outcome in patients with esophageal squamous cell carcinoma and correlated to hypoxia-inducible factor (HIF)-1alpha expressionPathol Oncol Res200915348749319130303
- PirtoliLCeveniniGTiniPThe prognostic role of Beclin 1 protein expression in high-grade gliomasAutophagy20095793093619556884
- DingZBShiYHZhouJAssociation of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinomaCancer Res200868229167917519010888
- WanXBFanXJChenMYElevated Beclin 1 expression is correlated with HIF-1alpha in predicting poor prognosis of nasopharyngeal carcinomaAutophagy20106339540420150769
- LazovaRKlumpVPawelekJAutophagy in cutaneous malignant melanomaJ Cutan Pathol201037225626819615007
- YoshiokaAMiyataHDokiYLC3, an autophagosome marker, is highly expressed in gastrointestinal cancersInt J Oncol200833346146818695874
- KondoYKanzawaTSawayaRKondoSThe role of autophagy in cancer development and response to therapyNat Rev Cancer20055972673416148885
- LumJJBauerDEKongMGrowth factor regulation of autophagy and cell survival in the absence of apoptosisCell2005120223724815680329
- BellotGGarcia-MedinaRGounonPHypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domainsMol Cell Biol200929102570258119273585
- BandMJoelAHernandezAAviviAHypoxia-induced BNIP3 expression and mitophagy: in vivo comparison of the rat and the hypoxia-tolerant mole rat, Spalax ehrenbergiFASEB J20092372327233519255257
- DeYoungMPHorakPSoferASgroiDEllisenLWHypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttlingGenes Dev200822223925118198340
- GlickDBarthSMacleodKFAutophagy: cellular and molecular mechanismsJ Pathol2010221131220225336
- HippertMMO’ToolePSThorburnAAutophagy in cancer: good, bad, or both?Cancer Res200666199349935117018585
- KreuzalerPWatsonCJKilling a cancer: what are the alternatives?Nat Rev Cancer201212641142422576162
- KroemerGLevineBAutophagic cell death: the story of a misnomerNat Rev Mol Cell Biol20089121004101018971948
- DegenhardtKMathewRBeaudoinBAutophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesisCancer Cell2006101516416843265
- AmaravadiRKYuDLumJJAutophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphomaJ Clin Invest2007117232633617235397
- BellodiCLidonniciMRHamiltonATargeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cellsJ Clin Invest200911951109112319363292
- CarewJSEspitiaCMEsquivelJA2ndLucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosisJ Biol Chem201128686602661321148553
- CarewJSMedinaECEsquivelJA2ndAutophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulationJ Cell Mol Med201014102448245919583815
- CarewJSNawrockiSTGilesFJClevelandJLTargeting autophagy: a novel anticancer strategy with therapeutic implications for imatinib resistanceBiologics20082220120419707354
- TakeuchiHKondoYFujiwaraKSynergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitorsCancer Res20056583336334615833867
- ZhuKDunnerKJrMcConkeyDJProteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cellsOncogene201029345146219881538
- CarewJSNawrockiSTKahueCNTargeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistanceBlood2007110131332217363733
- AmaravadiRKLippincott-SchwartzJYinXMPrinciples and current strategies for targeting autophagy for cancer treatmentClin Cancer Res201117465466621325294
- GlaumannHAhlbergJComparison of different autophagic vacuoles with regard to ultrastructure, enzymatic composition, and degradation capacity – formation of crinosomesExp Mol Pathol19874733463623678466
- DassonnevilleLBaillyCStimulation of topoisomerase II-mediated DNA cleavage by an indazole analogue of lucanthoneBiochem Pharmacol19995881307131210487533
- LuoMKelleyMRInhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthoneAnticancer Res20042442127213415330152
- MilliganAJKatzHRLeeperDBEffect of lucanthone hydrochloride on the radiation response of intestine and bone marrow of the Chinese hamsterJ Natl Cancer Inst197860510231028642024
- McAfeeQZhangZSamantaAAutophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiencyProc Natl Acad Sci U S A2012109218253825822566612
- YazbeckVYBuglioDGeorgakisGVTemsirolimus downregulates p21 without altering cyclin D1 expression and induces autophagy and synergizes with vorinostat in mantle cell lymphomaExp Hematol200836444345018343280
- CaoCSubhawongTAlbertJMInhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cellsCancer Res20066620100401004717047067
- AlonsoMMJiangHYokoyamaTDelta-24-RGD in combination with RAD001 induces enhanced anti-glioma effect via autophagic cell deathMol Ther200816348749318253154
- ShaoYGaoZMarksPAJiangXApoptotic and autophagic cell death induced by histone deacetylase inhibitorsProc Natl Acad Sci U S A200410152180301803515596714
- EllisLBotsMLindemannRKThe histone deacetylase inhibitors LAQ824 and LBH589 do not require death receptor signaling or a functional apoptosome to mediate tumor cell death or therapeutic efficacyBlood2009114238039319383971
- DingWXNiHMGaoWOncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagyMol Cancer Ther2009872036204519584239
- BascianiSVonaRMatarresePImatinib interferes with survival of multi drug resistant Kaposi’s sarcoma cellsFEBS Lett2007581305897590318061581
- BilirAErguvenMOktemGPotentiation of cytotoxicity by combination of imatinib and chlorimipramine in gliomaInt J Oncol200832482983918360710
- ErtmerAHuberVGilchSThe anticancer drug imatinib induces cellular autophagyLeukemia200721593694217330103
- MilanoVPiaoYLaFortuneTde GrootJDasatinib-induced autophagy is enhanced in combination with temozolomide in gliomaMol Cancer Ther20098239440619190119
- MartinAPParkMAMitchellCBCL-2 family inhibitors enhance histone deacetylase inhibitor and sorafenib lethality via autophagy and overcome blockade of the extrinsic pathway to facilitate killingMol Pharmacol200976232734119483105
- HuangHLChenYCHuangYCLapatinib induces autophagy, apoptosis and megakaryocytic differentiation in chronic myelogenous leukemia K562 cellsPLoS One2011612e2901422216158
- BurschWEllingerAKienzlHActive cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagyCarcinogenesis1996178159516078761415
- TurzanskiJDanielsIHaynesAPInvolvement of macroautophagy in the caspase-independent killing of Burkitt lymphoma cell lines by rituximabBr J Haematol2009145113714019183195
- GiannopoulouEAntonacopoulouAMatsoukaPKalofonosHPAutophagy: novel action of panitumumab in colon cancerAnticancer Res200929125077508220044619
- LiXFanZThe epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complexCancer Res201070145942595220634405
- VossVSenftCLangVThe pan-Bcl-2 inhibitor (−)-gossypol triggers autophagic cell death in malignant gliomaMol Cancer Res2010871002101620587533
- MaiuriMCCriolloATasdemirEBH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L)Autophagy20073437437617438366