Abstract
Background
Circular RNAs (circRNAs) have increasingly been investigated in different cancers due to their regulatory roles. In this study, hsa_circ_0046263 will be detailedly researched in non-small cell lung cancer (NSCLC).
Methods
The analyses of hsa_circ_0046263, microRNA-940 (miR-940), and neuro-oncological ventral antigen 2 (NOVA2) levels were administrated by quantitative real-time polymerase chain reaction (qRT-PCR). The proliferation detection was conducted using Cell Counting Kit-8 (CCK-8) and colony formation assays. Cell cycle and apoptosis were evaluated by flow cytometry. Transwell assay for migration and invasion was used to determine cell metastatic capacity. Overall protein levels were examined adopting Western blot. Target binding analysis was completed via dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. The effect of hsa_circ_0046263 on NSCLC in vivo was studied by xenograft model in mice.
Results
Hsa_circ_0046263 was overtly upregulated in NSCLC with important prognostic value. In vitro experiments indicated that hsa_circ_0046263 knockdown caused inhibitory effects on NSCLC cell proliferation, cell cycle, and metastasis but stimulative effect on apoptosis. Molecular mechanism analysis demonstrated that hsa_circ_0046263 served as a miR-940 sponge to act in the development of NSCLC. Moreover, miR-940 targeted NOVA2 and NOVA2 was regulated by hsa_circ_0046263/miR-940 axis. NOVA2 overexpression also neutralized the miR-940-mediated progression inhibition of NSCLC cells. In vivo assays suggested that hsa_circ_0046263 enhanced NSCLC tumorigenesis by targeting miR-940/NOVA2 axis.
Conclusion
Hsa_circ_0046263 was identified as a cancer-promoting factor in NSCLC via sponging miR-940 and upregulating NOVA2, which presented a clear mechanism of NSCLC occurrence and progression.
Introduction
Lung cancer has taken the top spot of cancer mortality for consecutive years.Citation1,Citation2 Approximate 85% of lung cancer patients are diagnosed as non-small cell lung cancer (NSCLC),Citation3 and local or distant metastasis is the primary cause of death in NSCLC patients.Citation4,Citation5 Mounting medical research has been performed for developing effective therapies against the metastatic and advanced NSCLC, especially the novel targeted therapy at the molecular level.Citation6,Citation7
Circular RNAs (circRNAs) are a widespread class of RNA transcripts in eucaryotes with tissue/cell-specific expression patterns and possess high stabilization with benefit from their closed-loop structures.Citation8 CircRNAs have been identified as crucial molecular regulators in the diverse biological processes of NSCLC, like cell proliferation, cell cycle, migration, and invasion.Citation9 Different regulatory mechanisms of circRNAs have been found in cancer progression, such as “miRNA sponge” and RNA-binding protein mechanisms.Citation10 CircRNAs have largely been reported as cancer-regulatory molecules by playing their sponge effect on microRNAs (miRNAs) and affecting the transcription-translation of genes.Citation11–Citation13 Wang et al claimed that hsa_circ_0046263 (circP4HB) was a metastatic biomarker in NSCLC via being a miR-133-5p sponge.Citation14 The further functional mechanism of hsa_circ_0046263 in NSCLC is needed to be explored.
MiRNAs are small RNAs without the ability to code proteins like circRNAs but still can modulate tumor process via acting on its targets at the position of 3ʹuntranlasted regions (3ʹUTRs).Citation15 For instance, miR-223 promoted cell proliferation of breast cancer by suppressing FOXO1Citation16 and miRNA-148a inhibited NSCLC cell migration and invasion by targeting Wnt1.Citation17 Previous studies have confirmed microRNA-940 (miR-940) as a tumor inhibitor in NSCLCCitation18,Citation19 and neuro-oncological ventral antigen 2 (NOVA2) as an oncogene to abolish the miR-7-5p-mediated cancer repression in NSCLC.Citation20 It has been unexplored about the potential target relation between miR-940 and NOVA2.
Here it is the first investigation of the interaction between hsa_circ_0046263 and miR-940, as well as miR-940 and NOVA2. The novel circRNA/miRNA/mRNA signal mechanism based on hsa_circ_0046263 is also disclosed in NSCLC during this current study.
Materials and Methods
Tissue Samples from Patients
A sum of 45 paired NSCLC tissues and the corresponding adjacent tissues were collected from 45 NSCLC patients after the surgical resection at Dongying People’s Hospital, then frozen in liquid nitrogen for standby application. No preoperative treatment was performed for these patients, and they have provided the written informed consent to support this medical research. All procedures were approved by the Ethical Committee of Dongying People’s Hospital (IRB No. 2019DY536).
Cell Culture
Two NSCLC cell lines (NCI-H1299 and A-549) and the bronchus epithelial cell line (BEAS-2B) acquired from American Type Culture Collection (ATCC, Manassas, VA, USA) were all cultured using Roswell Park Memorial Institute-1640 (RPMI1640; Gibco, Carlsbad, CA, USA) mixed with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin solution (Gibco) in a humidified CO2 (5%) incubator controlled temperature at 37°C.
Cell Transfection
Cells at the logarithmic phase were transplanted into the new 96-well plates to cultivate for 70% adherence, and then cell transient transfection was implemented via Lipofectamine™ 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the detailed instruction of the manufacturer. The used oligonucleotides were purchased from GenePharma (Shanghai, China): small interfering RNA against hsa_circ_0046263 (si-hsa_circ_0046263#1 and si-hsa_circ_0046263#2), miR-940 mimic and inhibitor (miR-940 and anti-miR-940) and the control groups (si-NC, NC, and anti-NC). All vectors were from RIBOBIO (Guangzhou, China): pCE-RB-Mam-hsa_circ_0046263 (hsa_circ_0046263)/pCE-RB-Mam (circ-NC), pEXP-RB-Mam-NOVA2 (NOVA2)/pEXP-RB-Mam (vector). For stable transfection, lentiviral vectors (RIBOBIO) of short hairpin RNA against hsa_circ_0046263 (sh-hsa_circ_0046263) and negative control (sh-NC) were, respectively, transfected into A-549 cells for 72 h. Then, cells with stable expression were screened for 7 days using antibiotic puromycin (Gibco).
Extraction of Total RNA and the Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA Extractor (Trizol; Sangon, Shanghai) was added to tissues or cells for extracting total RNA, followed by the detection of NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, 1 μg RNA was reversely transcribed into complementary DNA (cDNA) using First Strand cDNA Synthesis Kit ReverTra Ace -α- (Toyobo, Kita-Ku, Osaka, Japan). SYBR® Green Realtime PCR Master Mix (Toyobo) was applied for the administration of qRT-PCR with the obtained cDNA (2 μL) as the amplified template. The collected data were analyzed by using the 2−∆∆Ct method to calculate the relative levels of genes. Glyceraldehyde-phosphate dehydrogenase (GAPDH) and U6 were chosen as the endogenous controls, respectively, for hsa_circ_0046263/NOVA2 and miR-940. Primers for hsa_circ_0046263 included forward (F): 5ʹ-ACCATTTGGGATCACTTCCA-3ʹ and reverse (R): 5ʹ-CTTCTTCAGCCAGTTCACGA-3ʹ; for miR-940 included F: 5ʹ-GCATCGTTCCTTCAAGCCGATCT-3ʹ and R: 5ʹ-TGGGTGAGTCGTTCGG-3ʹ; for NOVA2 included F: 5ʹ-TGATTCAGCCAAGAGCTCGC-3ʹ and R: 5ʹ-AGGAGTCCCACCATCTGACA-3ʹ; for GAPDH included F: 5ʹ-GTGAACCATGAGAAGTATG-3ʹ and R: 5ʹ-CGGCCATCACGCCACAGTTTC-3ʹ; for U6 included F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Ribonuclease R (RNase R) and Actinomycin D Treatments
RNase R (Epicentre Technologies, Madison, WI, USA) was exposed to 4 μg total RNA with 3 U/μg for 60 min at 37°C, and culture medium was added with 2 mg/mL Actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) for 0 h, 4 h, 8 h, 12 h, and 24 h. After the acquisition of cDNA, the examination of hsa_circ_0046263 and GAPDH was carried out using qRT-PCR.
Cell Proliferation Detection
Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China) was applied for evaluating cell proliferative ability. NCI-H1299 and A-549 cells were, respectively, seeded into the 96-well plates to be cultured overnight. At 24 h, 48 h, and 72 h post-transfection, CCK-8 solution was added with 10 µL/well followed by reading the absorbance (450 nm) by a microplate reader after 4-h incubation at 37°C.
Cell proliferation was also assessed by colony formation assay. Two hundred transfected cells were seeded into each well of a 6-well plate and cultivated at 37°C with 5% CO2. About 12 d later, the macroscopic colonies could be observed in the wells. At room temperature, 4% Paraformaldehyde Fix Solution (Sangon, Shanghai) and crystal violet (Sangon) were employed to fix and stain colonies, respectively. After obtaining the images, the colony counting was manually performed.
Cell Cycle and Apoptosis Measurement
Flow cytometry was used for the analysis of cell cycle and apoptosis. For cell cycle, Propidium Iodide (PI) was used as a DNA fluorescent dye for cell staining by using Cell Cycle Assay Kit (Dojindo Molecular Technologies, Kumamoto, Japan) as per user’s guidelines. Through the detection of a flow cytometer (BD Biosciences, San Diego, CA, USA), the distributed proportions of cell G1, S, and G2 phases were obtained. For cell apoptosis, Annexin V-FITC/PI double-staining was performed by Apoptosis Detection Kit (BD Biosciences) according to the manufacture’s specification. The apoptotic cells were discerned following the analysis by the flow cytometer (BD Biosciences). Generally, the early apoptotic cells were stained by positive Annexin V and negative PI while those late apoptotic cells were stained by doubly positive Annexin V and PI.
Cell Migration and Invasion Evaluation
Transwell assay was performed using a 24-well transwell chamber (8 µm; Corning Inc., Corning, NY, USA). The top surface of the membranes in the upper chamber required to be pre-coated with matrigel (Corning Inc.) in the detection for invasion. Cell suspension containing a total of 1 × 104 cells in serum-free medium was added into the upper chamber. After the addition of culture medium with 10% FBS in the lower chamber for 24 h, cells on the top surface of the membranes were carefully erased by a wet cotton swab. Migratory and invasive cells on the low surface of the membranes were fixated in methanol (Sangon) and dyed in crystal violet (Sangon). Ultimately, an inverted microscope (Olympus, Tokyo, Japan) was used for photo shooting (100 × magnification) followed by the calculation of cell number.
Protein Preparation and Western Blot
Fresh protein lysis solution was produced by mixing RIPA Buffer (Sigma-Aldrich), protease inhibitor PMSF (Sangon), and phosphatase inhibitor Sodium orthovanadate (V) dodecahydrate (Sangon) with the ratio of 98:1:1 (v/v/v). Total proteins were extracted by adding the protein lysis solution for incubation on ice for 30 min and collecting the supernatant through centrifugation at 7000 rpm/min for 15 min. The operating steps for Western blot followed the detailed descriptions published previously.Citation21 The antibodies were all bought from Abcam (Cambridge, UK): anti-proliferating cell nuclear antigen (anti-PCNA; ab18197, 1:1000), anti-Cyclin D1 (ab226977, 1:1000), anti-total-caspse3 (anti-t-caspase 3; ab4051, 1:500), anti-cleaved caspase 3 (anti-C-caspase 3; ab2302, 1:1000), anti-matrix metalloproteinase 9 (anti-MMP9, ab38898, 1:1000), anti-NOVA2 (Sigma-Aldrich, AV40399, 1:1000), anti-GAPDH (ab9485, 1:1000) and Goat Anti-Rabbit IgG H&L (HRP) second antibody (ab205718, 1:3000). GAPDH acted as the internal control in this assay. ImageLab software version 4.1 (Bio-Rad, Hercules, CA, USA) was exploited to perform the grey level analysis.
Dual-Luciferase Reporter Assay
Luciferase plasmids for hsa_circ_0046263 and NOVA2 3ʹUTR were constructed by cloning their wild-type (wt) or mutant-type (mut) sequences that aimed at the miR-940-binding sites into the psiCHECK-2 vector (Promega, Madison, WI, USA). The recombinant luciferase plasmids were expressed as hsa_circ_0046263-wt, hsa_circ_0046263-mut, NOVA2-wt, and NOVA2-mut. Then, NCI-H1299 and A-549 cells were digested with trypsin (Gibco) and plated into the 24-well plate for 24 h, followed by the following transfection: hsa_circ_0046263-wt+NC/miR-940, hsa_circ_0046263-mut+NC/miR-940, NOVA2-wt+NC/miR-940 and NOVA2-mut+NC/miR-940. Forty-eight hours later, the relative luciferase activity (firefly/renilla) of each transfection group was determined via the dual-luciferase reporter assay system (Promega).
RNA Immunoprecipitation (RIP) Assay
NCI-H1299 and A-549 cells were harvested and RIP assay was implemented via Imprint® RNA Immunoprecipitation Kit (Sigma-Aldrich). Firstly, cells were dissolved in RIP lysis buffer and incubated with protein A magnetic beads conjugated with Anti-Argonaute-2 (Anti-Ago2) and Anti-immunoglobulin G (Anti-IgG) at 4°C. On the next day, total RNA was collected and the genic level analysis was completed by qRT-PCR.
Xenograft Models in Mice
An amount of 10 Male BALB/c nude mice (about 6-weeks old, 25 g) were purchased from Shanghai Animal Experimental Center (Shanghai, China). A-549 cells transfected with sh-hsa_circ_0046263 or sh-NC vector were injected subcutaneously into the flank of mice (5 mice/group) to establish the xenograft models in vivo. Every 5 d after injection, tumor volume (length × width2 × 0.5) was obtained by using tumor length and width as the determined indicators. After the euthanasia for all nude mice at 35 d, tumor weight was measured and the related expression levels were analyzed by qRT-PCR and Western blot. The present animal experiment got empowerment from the Animal Ethical Committee of Dongying People’s Hospital and the Management and Use Guidelines of Laboratory Animals of NIH were strictly followed.
Statistical Analysis
Statistical analyses for data that were expressed the mean ± standard deviation (SD) were conducted by SPSS 24.0 software (IBM Corp., Armonk, NY, USA). The survival curve was generated by Kaplan-Meier plot and analyzed by Log-rank test. All genic linear analyses in NSCLC tissues were performed using Spearman correlation coefficient. The comparison of difference was analyzed via Student’s t-test for two groups and one-way analysis of variance (ANOVA) followed by Tukey’s test for more than three groups.
Results
High hsa_circ_0046263 Was Found in NSCLC and Could Predict Poor Prognosis
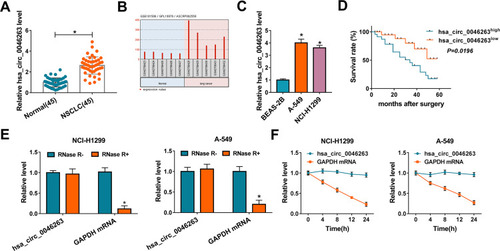
The expression detection of hsa_circ_0046263 was first performed in NSCLC tissues. As illustrated in , the relative hsa_circ_0046263 level was signally increased in NSCLC tissues via the comparison with Normal samples, in conformity with the analysis of GSE101586 dataset (). Also, there was the upregulated hsa_circ_0046263 in NCI-H1299 and A-549 cells contrasted to control BEAS-2B cells (). Through the analysis of 5-year survival rate after surgery, we found that survival rate of hsa_circ_0046263high group was lower than that of hsa_circ_0046263low group (), indicating the potence of high hsa_circ_0046263 as predictive marker for poor prognosis of NSCLC patients. To identify that hsa_circ_0046263 was more stable than linear RNA, external RNase R and Actinomycin D treatments were performed. Compared to linear GAPDH mRNA, hsa_circ_0046263 was not easy to be degraded by RNase R () and Actinomycin D () both in NCI-H1299 and A-549 cells. The upregulation of hsa_circ_0046263 and its prognostic value implied the regulatory potential of hsa_circ_0046263 in NSCLC.
Figure 1 High hsa_circ_0046263 was found in NSCLC and could predict poor prognosis. (A) The qRT-PCR was used to measure hsa_circ_0046263 in NSCLC and normal tissues. (B) The dataset analysis of hsa_circ_0046263 in lung cancer. (C) Hsa_circ_0046263 detection in BEAS-2B, A-549 and NCI-H1299 cells was performed by qRT-PCR. (D) Survival curves of NSCLC patients were analyzed using Log-rank test. (E and F) Hsa_circ_0046263 and GAPDH mRNA levels were examined via qRT-PCR following RNase R digestion in isolated RNA (E) or Actinomycin D treatment for cells (F). *P < 0.05.
Downregulated hsa_circ_0046263 Exerted the Inhibition of Cell Proliferation, Cell Cycle, Metastasis and Motivation of Cell Apoptosis in NSCLC Cells
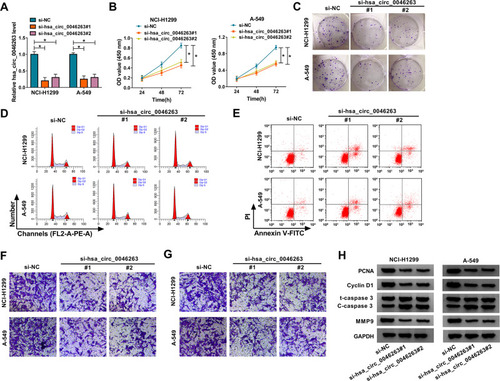
To perform the functional analysis of hsa_circ_0046263, siRNA transfection was first used for obstructing the normal expression of hsa_circ_0046263. After the analysis of qRT-PCR, hsa_circ_0046263 level was found to be mostly reduced by si-hsa_circ_0046263#1 and si-hsa_circ_0046263#2 in NCI-H1299 and A-549 cells, relative to si-NC group (). CCK-8 () and colony formation assay ( and Supplementary Figure 1A) showed that the deficiency of hsa_circ_0046263 affected cell proliferation with an inhibitory influence. In terms of cell cycle, cell distribution at S phase was significantly declined while that at G1 phase was increased in si-hsa_circ_0046263#1 or si-hsa_circ_0046263#2 group ( and Supplementary Figure 1B), displaying that cell cycle progression was impeded by hsa_circ_0046263 downregulation. Apoptotic cell percentage by flow cytometry was largely upregulated in NCI-H1299 and A-549 cells downregulated hsa_circ_0046263 ( and Supplementary Figure 1C). With the employment of transwell assay, the overt repression was observed in cell migratory ( and Supplementary Figure 1D) and invasive ( and Supplementary Figure 1E) capacities. Moreover, hsa_circ_0046263 knockdown led to the protein expressed downregulation of PCNA (proliferation maker), Cyclin D1 (cell cycle marker), MMP9 (invasion marker), and upregulation of C-caspase 3/total caspase 3 (apoptosis indication) ( and Supplementary Figure 1F-G). Doubtlessly, the downregulated hsa_circ_0046263 repressed the NSCLC process.
Figure 2 Downregulated hsa_circ_0046263 exerted the inhibition of cell proliferation, cell cycle, metastasis and motivation of cell apoptosis in NSCLC cells. (A) After transfection of si-NC, si-hsa_circ_0046263#1 or si-hsa_circ_0046263#2, the hsa_circ_0046263 expression determination was conducted using qRT-PCR. (B and C) Cell proliferation assessment was completed via CCK-8 (B) and colony formation assay (C). (D and E) Flow cytometry was administrated for measuring cell cycle (D) and apoptosis (E). (F and G) The effects of the above transfection on cellular migration (F) and invasion (G) were analyzed by transwell assay. (H) Western blot was applied for detecting the proteins associated with these cellular behaviors. *P < 0.05.
Hsa_circ_0046263 Sponged miR-940 in NSCLC Cells
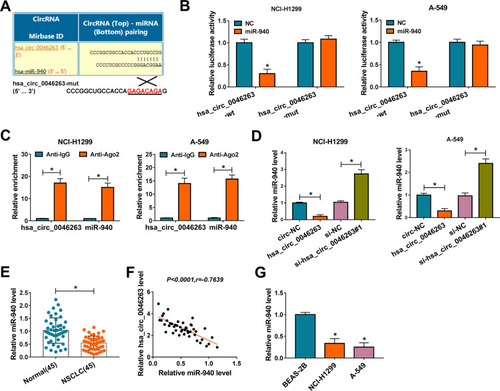
Emerging studies have demonstrated that circRNAs acted in cancer development via sponging miRNAs.Citation22,Citation23 To explore the potential of hsa_circ_0046263 as a specific miRNA sponge, we used circinteractome as a prediction software and found the binding sites between the sequences of hsa_circ_0046263 and miR-940 (). Later, dual-luciferase reporter assay revealed the suppressive effect of miR-940 overexpression on the relative luciferase activity of hsa_circ_0046263-wt vector, with no obvious effect on hsa_circ_0046263-mut vector (). The binding of hsa_circ_0046263 and miR-940 was also affirmed by RIP assay, in which they were abundantly captured by Ago2 antibody compared with IgG antibody (). Contraposed to transfection of circ-NC and si-NC, miR-940 was down-regulated in hsa_circ_0046263 group and its expressed promotion was found in si-hsa_circ_0046263#1 group (), showing the negative regulation of hsa_circ_0046263 on miR-940 level. The expression of miR-940 was notably repressed in NSCLC samples relative to Normal samples () and hsa_circ_0046263 was negatively correlated with miR-940 level (P<0.0001, r=−0.7639) in NSCLC tissues (). NCI-H1299 and A-549 cells also expressed miR-940 lowly by comparison with BEAS-2B cells (). Altogether, the sponge influence of hsa_circ_0046263 on miR-940 was validated.
Figure 3 Hsa_circ_0046263 sponged miR-940 in NSCLC cells. (A) The binding sites between hsa_circ_0046263 and miR-940 were presented by circinteractome. (B and C) Dual-luciferase reporter (B) and RIP (C) assays were implemented for testifying the interactive capacity between hsa_circ_0046263 and miR-940. (D) The qRT-PCR was exploited to evaluate the influences of hsa_circ_0046263 overexpression and inhibition on miR-940. (E) The analysis of miR-940 level in NSCLC and Normal samples was implemented via qRT-PCR. (F) The linear relation between hsa_circ_0046263 and miR-940 in NSCLC tissues was analyzed using Spearman correlation coefficient. (G) The miR-940 expression in NSCLC and normal BEAS-2B cells was carried out by qRT-PCR. *P < 0.05.
Silence of Hsa_circ_0046263 Decelerated NSCLC Progression by Promoting miR-940
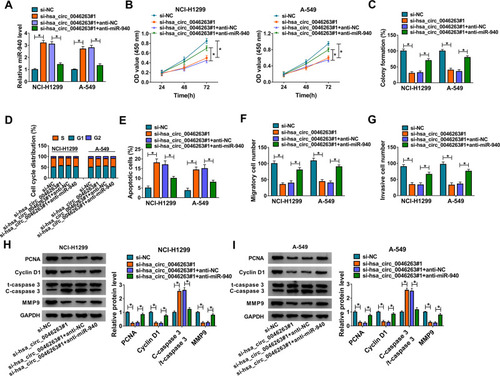
To investigate whether the function of hsa_circ_0046263 in NSCLC was associated with its sponge effect on miR-940, the transfection of si-hsa_circ_0046263 #1, si-hsa_circ_0046263#1+anti-miR-940 or the matched control groups was administrated in NCI-H1299 and A-549 cells. The miR-940 expression in si-hsa_circ_0046263#1+anti-miR-940 was markedly repressed by contrast to si-hsa_circ_0046263#1+anti-NC group (), which suggested the excellent inhibition of miR-940 by anti-miR-940. Importantly, the si-hsa_circ_0046263#1-induced proliferation () and cell cycle () retardment, apoptosis acceleration () and metastasis inhibition () were partly assuaged by anti-miR-940 transfection. The relaxative trends of miR-940 inhibitor to the downregulated hsa_circ_0046263 on these cellular behaviors were also verified by the examination of markers in Western blot (). Expectedly, the action of hsa_circ_0046263 in NSCLC was achieved by the negative modulation of miR-940.
Figure 4 Silence of hsa_circ_0046263 decelerated NSCLC progression by promoting miR-940. (A) In the following transfection groups: si-NC, si-hsa_circ_0046263#1, si-hsa_circ_0046263#1+anti-NC and si-hsa_circ_0046263#1+anti-miR-940, qRT-PCR was used for miR-940 determination. (B and C) The evaluation of cell proliferation was performed by CCK-8 (B) and colony formation assay (C). (D–G) Flow cytometry and transwell assay were respectively employed to assess cell cycle or apoptosis (D and E) and cell migration or invasion (F and G). (Hand I) The related protein markers were examined via Western blot. *P < 0.05.
Hsa_circ_0046263 Facilitated the Level of NOVA2 by Binding to miR-940
FAM83F and Snail have been identified as the targets for miR-940 in NSCLC.Citation18,Citation19 Targetscan prediction manifested that NOVA2 3ʹUTR sequence harbored the miR-940-binding sites (). The result of dual-luciferase reporter assay revealed the combination between NOVA2-wt and miR-940 not NOVA2-mut and miR-940 (). The high enrichment in Ago2 protein further proved the interplay of NOVA2 and miR-940 (). Interestingly, there was an inhibitory influence of si-hsa_circ_0046263#1 on NOVA2 protein level but this inhibition was abolished through the introduction of anti-miR-940 (), presenting that the positive regulation of hsa_circ_0046263 on NOVA2 by targeting miR-940. NOVA2 was an aberrantly upregulated molecule in NSCLC tissues as the qRT-PCR and Western blot results in . Spearman correlation coefficient showed a positive relation (P<0.0001, r=0.6323) between hsa_circ_0046263 and NOVA2 () but a negative association (P<0.0001, r=−0.7404) between miR-940 and NOVA2 () in NSCLC samples. Keeping pace with the expression in NSCLC tissues, the protein level of NOVA2 was elevated in NSCLC cells (NCI-H1299 and A-549) in contrast with BEAS-2B cells (). These analyses explained the target relation of miR-940 for NOVA2 and the regulatory effect of hsa_circ_0046263 on NOVA2 via mediating miR-940 level.
Figure 5 Hsa_circ_0046263 facilitated the level of NOVA2 by binding to miR-940. (A) Targetscan software was applied to perform the binding site analysis between miR-940 and NOVA2. (B and C) The combination between miR-940 and NOVA2 was attested by dual-luciferase reporter (B) and RIP (C) assays. (D) NOVA2 protein expression was assayed by Western blot in NCI-H1299 and A-549 cells with transfection of si-NC, si-hsa_circ_0046263#1, si-hsa_circ_0046263#1+anti-NC and si-hsa_circ_0046263#1+anti-miR-940. (E and F) The qRT-PCR and Western blot were conducted for the detection of NOVA2 mRNA (E) and protein (F) levels. (G and H) Spearman correlation coefficient was used to analyze the correlation between hsa_circ_0046263 and NOVA2 (G), miR-940 and NOVA2 (H). (I) Western blot was carried out to examine the protein expression of NOVA2 in NSCLC cells and control BEAS-2B cells. *P < 0.05.
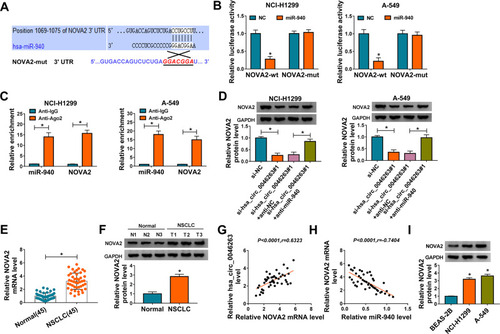
MiR-940 Functioned as a Tumor-Inhibitory Molecule in NSCLC via Targeting NOVA2
The rescued assays were carried out to research the regulatory mechanism of miR-940 and NOVA2 in NSCLC. Firstly, NOVA2 protein level was presented to be enhanced in miR-940+NOVA2 group by comparison to miR-940+vector group (), indicating that NOVA2 vector greatly mitigated the miR-940-mediated NOVA2 downregulation. Then, we found that miR-940 significantly suppressed cell proliferation by performing CCK-8 and colony formation assays, whereas the supplement of NOVA2 limited this effect (). Flow cytometry revealed that NOVA2 overexpression counterbalanced the transition inhibition from cell G1 phase to S phase () and the promotive cell apoptosis () evoked by miR-940 in NCI-H1299 and A-549 cells. In addition, the miR-940-induced decline of migratory () and invasive () cell number was relieved with the increase of NOVA2 expression. After the protein detection of cellular markers associated with the above cellular processes by Western blot in , it was again affirmed that miR-940 worked as an inhibitor in NSCLC development by downregulating NOVA2.
Figure 6 MiR-940 functioned as a tumor-inhibitory molecule in NSCLC via targeting NOVA2. (A) The measurement of NOVA2 was performed through Western blot after NCI-H1299 and A-549 cells were transfected with NC, miR-940, miR-940+vector and miR-940+NOVA2. (B and C) CCK-8 (B) and colony formation (C) assays were administrated for analyzing the proliferative ability of the above transfected cells. (D–G) The assessment of cell cycle/apoptosis (D and E) and migration/invasion (F and G) were achieved by flow cytometry and transwell assay. (H and I) The analyses of proteins regarding cellular processes were executed by Western blot. *P < 0.05.
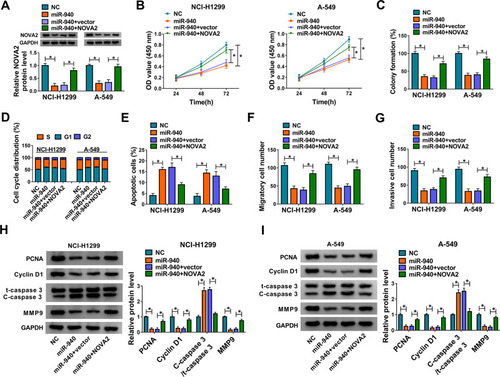
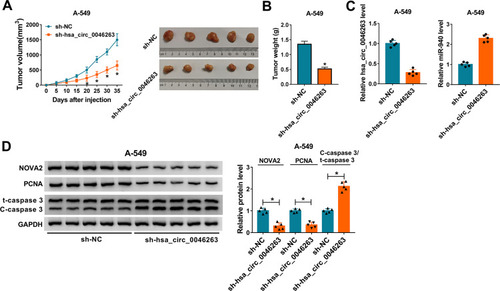
NSCLC Progression in vivo Was Promoted by Hsa_circ_0046263 That Modulated NOVA2 via miR-940
Animal experiment was conducted to prove the role of hsa_circ_0046263 in vivo. By monitoring the changes of tumor volume after cell injection, we noticed that there was prominent inhibition of tumor volume in sh-hsa_circ_0046263 group contrasted with sh-NC group at the observational late period of 20–35 d (). The dissected tumors of sh-hsa_circ_0046263 group were also lighter than that of sh-NC group (). The injection of cells transfected with sh-hsa_circ_0046263 induced the expression downregulation of hsa_circ_0046263 and upregulation of miR-940, by comparison with the injection of cells transfected with sh-NC (). Western blot suggested that knockdown of hsa_circ_0046263 restrained NOVA2 and PCNA levels but elevated C-caspase 3/t-caspase 3 level (). Thus, hsa_circ_0046263 contributed to the progression of NSCLC in vivo through the dependence of miR-940/NOVA2 axis.
Figure 7 NSCLC progression in vivo was promoted by hsa_circ_0046263 that modulated NOVA2 via miR-940. (A and B) After cell injection of sh-NC or sh-hsa_circ_0046263 group in mice (5 mice/group), tumor volume was measured every 5 d (A) and tumors were weighed after 35 d. (C) The qRT-PCR was adopted to determine hsa_circ_0046263 and miR-940 expression levels of two groups in 10 mice. (D) The protein levels of NOVA2, PCNA and C-caspase 3/t-caspase 3 in tumor tissues of 10 mice were all tested via Western blot. *P < 0.05.
Discussion
In different types of cancers, numerous circRNAs with abnormal expression have pivotal significance on cancer diagnosis, treatment, and prognosis.Citation24–Citation26 In the present exploration, we selected hsa_circ_0046263 that was reported to be upregulated in NSCLCCitation14 as a research subject. Consistently, distinct upregulation of hsa_circ_0046263 was also found in our collected NSCLC tissues and cultured NCI-H1299 or A-549 cells. Moreover, hsa_circ_0046263 could be a prognostic marker for predicting a poor survival rate after our survival analysis for patients. And hsa_circ_0046263 had the characteristic of high stability to resist the interference of RNase R and Actinomycin D. Functionally, the differentially upregulated circRNAs can generate the promoting influence on the progression and malignance of cancers. For example, circRNA_0000285 expression was apparently boosted in cervical cancer and it contributed to cell proliferation and metastasis.Citation27 Tang et al have asserted the overexpression of hsa_circ_0001982 in breast cancer tissues and cells, as well as its promotion on oncogenesis.Citation28 Also, the upregulated circERBB2 in gallbladder cancerCitation29 and circFARSA in NSCLCCitation30 were, respectively, testified as a pro-cancer factor. Herein, the functional investigation of hsa_circ_0046263 was conducted on various cellular behaviors of NSCLC. After hsa_circ_0046263 was knocked down in NSCLC cells by siRNA transfection, we found that cell proliferation, cell cycle progression, cell migration, and invasion were all blocked while cell apoptosis was enhanced. Through the detection of marker genes using Western blot, the above effects were again affirmed. Thus, hsa_circ_0046263 was indeed able to accelerate the tumorigenesis and metastasis of NSCLC.
CircRNAs have previously considered as natural sponges for miRNAs, which then led to the regulation of cellular processes in cancers. CircRNA cTFRC motivated the evolution of bladder cancer by serving as a miR-107 spongeCitation31 and hsa_circ_0007059 sponged miR-378 to restrain cell proliferation and epithelial–mesenchymal transition of lung cancer.Citation32 In this chapter, miR-940 was shown to have a direct combination with hsa_circ_0046263 and there was a negative expression regulation of miR-940 by hsa_circ_0046263. According to the issued reports, miR-940 played a repressive role in the progression of many common cancers, such as hepatocellular carcinoma,Citation33 esophageal squamous cell carcinoma,Citation34 and ovarian cancer.Citation35 Also, miR-940 inhibitor promoted NSCLC progression to restore the tumor inhibition induced by hsa_circ_0046263 knockdown. In other words, hsa_circ_0046263 expedited NSCLC development through sponging miR-940.
By functioning as the sponges for miRNAs, circRNAs can affect gene regulation to control the advancement of cancer process.Citation36 As reported, silencing hsa_circRNA_002178 hampered the evolution of breast cancer depending on the inhibition of COL1A1 mediated by miR-328-3pCitation37 and circZFR sponged miR-101-3p to promote cell metastasis of NSCLC by regulating CUL4B.Citation38 Here, target analysis revealed that miR-940 interplayed with NOVA2 in NSCLC cells, and NOVA2 was positively modulated by hsa_circ_0046263 via miR-940. Our qRT-PCR manifested that NOVA2 expression was significantly upregulated in NSCLC tissue samples and cell lines compared with normal tissues and cells. NOVA2 has been announced to serve for an oncogenic gene in colorectal cancerCitation39 and glioma.Citation40 Also, the reverted experiment here suggested that the miR-940 triggered cell proliferation and metastasis inhibition, cell cycle arrest, and apoptosis enhancement by downregulating the NOVA2. These results again implied the tumorigenic role of NOVA2 in the progression of NSCLC. Furthermore, we found that hsa_circ_0046263 could facilitate NSCLC progression in vivo by regulating miR-940 and NOVA2.
Conclusion
In consequence, hsa_circ_0046263 was demonstrated to heighten the carcinogenic and metastatic abilities of NSCLC via motivating the miR-940-targeted NOVA2 expression. All results in this report have proposed a hsa_circ_0046263/miR-940/NOVA2 regulatory axis in NSCLC, unraveling the pathological mechanism of NSCLC by hsa_circ_0046263 in another molecular way.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.2149230207593
- Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85(1). doi:10.5334/aogh.2419
- Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, Version 6.2015. J Natl Compr Canc Netw. 2015;13(5):515–524. doi:10.6004/jnccn.2015.007125964637
- Mirrielees JA, Kapur JH, Szalkucki LM, et al. Metastasis of primary lung carcinoma to the breast: a systematic review of the literature. J Surg Res. 2014;188(2):419–431. doi:10.1016/j.jss.2014.01.02424560348
- Page S, Milner-Watts C, Perna M, et al. Systemic treatment of brain metastases in non-small cell lung cancer. Eur J Cancer. 2020;132:187–198. doi:10.1016/j.ejca.2020.03.00632380429
- Prabhu VV, Devaraj SN. KAI1/CD82, metastasis suppressor gene as a therapeutic target for non-small-cell lung carcinoma. J Environ Pathol Toxicol Oncol. 2017;36(3):269–275. doi:10.1615/JEnvironPatholToxicolOncol.201702461929283339
- Xia L, Liu Y, Wang Y. PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: current status and future directions. Oncologist. 2019;24(Suppl1):S31–S41. doi:10.1634/theoncologist.2019-IO-S1-s0530819829
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-731395983
- Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889. doi:10.1007/s00432-019-03045-431630262
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.00326052092
- Tian Y, Xu Z, Fu J. CircularRNA-9119 promotes the proliferation of cervical cancer cells by sponging miR-126/MDM4. Mol Cell Biochem. 2020;470(1–2):53–62. doi:10.1007/s11010-020-03745-332385717
- Lin S, Song S, Sun R, et al. Oncogenic circular RNA Hsa-circ-000684 interacts with microRNA-186 to upregulate ZEB1 in gastric cancer. FASEB J. 2020;34(6):8187–8203. doi:10.1096/fj.201903246R32388910
- Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099. doi:10.1016/j.gene.2019.14409931479715
- Wang T, Wang X, Du Q, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513(4):904–911. doi:10.1016/j.bbrc.2019.04.10831005252
- Li M, Huo X, Davuljigari CB, Dai Q, Xu X. MicroRNAs and their role in environmental chemical carcinogenesis. Environ Geochem Health. 2019;41(1):225–247. doi:10.1007/s10653-018-0179-830171477
- Wei YT, Guo DW, Hou XZ, Jiang DQ. miRNA-223 suppresses FOXO1 and functions as a potential tumor marker in breast cancer. Cell Mol Biol (Noisy-Le-Grand). 2017;63(5):113–118. doi:10.14715/cmb/2017.63.5.2128719355
- Chen Y, Min L, Ren C, et al. miRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS One. 2017;12(2):e0171751. doi:10.1371/journal.pone.017175128199399
- Jiang K, Zhao T, Shen M, et al. MiR-940 inhibits TGF-β-induced epithelial-mesenchymal transition and cell invasion by targeting Snail in non-small cell lung cancer. J Cancer. 2019;10(12):2735–2744. doi:10.7150/jca.3180031258781
- Gu GM, Zhan YY, Abuduwaili K, et al. MiR-940 inhibits the progression of NSCLC by targeting FAM83F. Eur Rev Med Pharmacol Sci. 2018;22(18):5964–5971. doi:10.26355/eurrev_201809_1592730280778
- Xiao H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24:60. doi:10.1186/s11658-019-0188-331832068
- Zheng H, Wang JJ, Zhao LJ, Yang XR, Yu YL. Exosomal miR-182 regulates the effect of RECK on gallbladder cancer. World J Gastroenterol. 2020;26(9):933–946. doi:10.3748/wjg.v26.i9.93332206004
- Xu G, Ye D, Zhao Q, et al. circNFIC suppresses breast cancer progression by sponging miR-658. J Cancer. 2020;11(14):4222–4229. doi:10.7150/jca.3883032368305
- Chu YL. Circ_0067934 correlates with poor prognosis and promotes laryngeal squamous cell cancer progression by sponging miR-1324. Eur Rev Med Pharmacol Sci. 2020;24(8):4320–4327. doi:10.26355/eurrev_202004_2101332373969
- Fu B, Zhang A, Li M, et al. Circular RNA profile of breast cancer brain metastasis: identification of potential biomarkers and therapeutic targets. Epigenomics. 2018;10(12):1619–1630. doi:10.2217/epi-2018-009030810051
- Gao Y, Zhang C, Liu Y, Wang M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci Trends. 2019;13(2):204–211. doi:10.5582/bst.2019.0102131019161
- Wei J, Wei W, Xu H, et al. Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 2020;27(2):139–145. doi:10.3233/CBM-18202930689557
- Chen RX, Liu HL, Yang LL, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci. 2019;23(20):8771–8778. doi:10.26355/eurrev_201910_1927131696463
- Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36(11):901–908. doi:10.1089/dna.2017.386228933584
- Huang X, He M, Huang S, et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4-dependent rDNA transcription. Mol Cancer. 2019;18(1):166. doi:10.1186/s12943-019-1098-831752867
- Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–2791. doi:10.1002/cam4.151429722168
- Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18(1):27. doi:10.1186/s12943-019-0951-030782157
- Gao S, Yu Y, Liu L, Meng J, Li G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019;233:116692. doi:10.1016/j.lfs.2019.11669231351967
- Li P, Xiao Z, Luo J, Zhang Y, Lin L. MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J Cell Mol Med. 2019;23(4):2475–2488. doi:10.1111/jcmm.1412130710422
- Wang H, Song T, Qiao Y, Sun J. miR-940 inhibits cell proliferation and promotes apoptosis in esophageal squamous cell carcinoma cells and is associated with post-operative prognosis. Exp Ther Med. 2020;19(2):833–840. doi:10.3892/etm.2019.827932010243
- Wang F, Wang Z, Gu X, Cui J. miR-940 upregulation suppresses cell proliferation and induces apoptosis by targeting pKC-delta in ovarian cancer OVCAR3 cells. Oncol Res. 2017;25(1):107–114. doi:10.3727/096504016X1473277215014528081739
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi:10.1080/15476286.2015.112216226649774
- Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. 2020;24(3):2189–2201. doi:10.1111/jcmm.1487531957232
- Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif Cells Nanomed Biotechnol. 2019;47(1):3410–3416. doi:10.1080/21691401.2019.165262331407591
- Gallo S, Arcidiacono MV, Tisato V, et al. Upregulation of the alternative splicing factor NOVA2 in colorectal cancer vasculature. Onco Targets Ther. 2018;11:6049–6056. doi:10.2147/OTT.S17167830275709
- Li G, Huang M, Cai Y, Yang Y, Sun X, Ke Y. Circ-U2AF1 promotes human glioma via derepressing neuro-oncological ventral antigen 2 by sponging hsa-miR-7-5p. J Cell Physiol. 2019;234(6):9144–9155. doi:10.1002/jcp.2759130341906
