Abstract
Background
Circular RNA circSKA3 plays an oncogenic role in breast cancer, while its role in glioblastoma (GBM) is unknown. This study aimed to explore the role of circSKA3 in GBM.
Methods
Differential expression of circSKA3 and miR-1 in GBM and adjacent non-cancer tissue samples were analyzed by RT-qPCR. GBM cells were transfected with circSKA3 expression vector or miR-1 mimic, followed by RT-qPCR to explore the potential crosstalk between them. Methylation-specific PCR (MSP) was carried out to assess the role of circSKA3 in regulating the methylation of miR-1 gene. The role of circSKA3 and miR-1 in regulating GBM cell proliferation was analyzed by CCK-8 assay.
Results
We found that circSKA3 was upregulated in GBM and inversely correlated with miR-1 across GBM tissues. High expression levels of circSKA3 and low expression levels of miR-1 were significantly correlated with the poor survival of GBM patients. In GBM cells, overexpression of circSKA3 increased the methylation of miR-1 gene and decreased the expression of miR-1. CCK-8 assay showed that overexpression of circSKA3 reduced the inhibitory effects of miR-1 on cell proliferation.
Conclusion
Therefore, circSKA3 may downregulate miR-1 through methylation in GBM to promote cancer cell proliferation.
Keywords:
Introduction
Glioblastoma (GBM), also known as grade IV astrocytoma, is a rare type of malignancy that only affects about 2 out of 100,000 people per year.Citation1,Citation2 Despite the low incidence rate, GBM is considered as a common cause of cancer-related mortality, mainly due to its highly aggressive and infiltrative nature.Citation3 Although distant metastasis is rare among GBM patients (about 2%) the widespread infiltration of GBM throughout the brain causes unacceptable high mortality rate.Citation4,Citation5 It is estimated that the median survival time of GBM is only 10 to 16 months, and only about 8–12% of patients can survive more than 2 years after diagnosis.Citation5 Although GBM patients can usually be treated with surgeries, radiotherapy and chemotherapy, these treatments can only elongate patients’ survival.Citation6 It is estimated that only less than 25% of GBM patients can survive more than a year after active treatment.Citation6 At present, there is no cure for GBM.Citation7
More effective treatment strategies are needed to improve the survival of GBM patients. Studies on the molecular pathogenesis of GBM have revealed a considerable number of molecular players in the growth and spread of GBM.Citation8,Citation9 In effect, certain molecular factors, such as STAT3, have been characterized as potential targets to develop novel anti-GBM approaches, such as targeted therapy that can be performed to suppress cancer growth by affecting related gene expression.Citation10 However, to the best of our knowledge, effective and safe targets for GBM targeted therapy remain lacking. Circular RNAs (circRNAs) are covalently closed RNA (single-strand) transcripts that regulate gene expression rather than coding proteins to participate in human cancers.Citation11,Citation12 Therefore, circRNAs may serve as targets for targeted therapy. However, the functions of most circRNAs in GBM remain unknown. CircRNA circSKA3 has been characterized with an oncogenic role in breast cancer,Citation13 while its role in GBM is unknown. Our preliminary microarray analysis revealed the inverse correlation between circSKA3 and miR-1, a tumor suppressor in GBM.Citation14 This study was performed to explore the interaction between circSKA3 and miR-1 in GBM.
Patients and Methods
Patients and Tissue Acquisition
This study enrolled 56 GBM patients admitted at the First Affiliated Hospital of Jinan University between May 2016 and June 2018. Age range of the patients was 55 to 70 years old, with a median age of 63 years old. The 56 patients excluded those with initiated therapy or diagnosed with multiple clinical disorders. This study also excluded recurrent GBM cases and only newly diagnosed cases were included. The Ethics Committee of the aforementioned hospital approved this study (No. JNU353323). This study was performed following the Declaration of Helsinki. Informed consent was provided by all patients. Fine needle aspiration was carried out to collect GBM tissue and adjacent (within 3 cm around tumors) non-cancer tissue samples from the patients. All tissue samples were kept in liquid nitrogen prior to the subsequent assays.
Treatment and Follow-Up
All patients were treated with radiation therapy. From admission, a 2-year follow-up was performed on all patients. Follow-up was performed every month through telephone to record patients’ survival. Patients who were lost during follow-up and the died of causes unrelated to GBM were excluded.
GBM Cells and Transient Transfections
Human GBM cell line U87 obtained from ATCC (USA) was used to serve as the cell model of GBM. Cells were cultivated in an incubator at 37 °C with 95% humidity and 5% CO2. Cell culture medium was DMEM containing 5% FBS. Cells were cultivated until about 85% confluence prior to subsequent assays. To overexpress circSKA3 and miR-1, mimic of miR-1 and miRNA negative control (NC) were synthesized by Sangon (Shanghai, China), and pcDNA3.1(+) CircRNA Mini Vector (Addgene) was used to prepare the expression vector of circSKA3, followed by transfecting 108 cells with either 40 nM miR-1 mimic of 1 μg circSKA3 expression vector using Lipofectamine 2000 (Invitrogen). In each experiment, NC (NC miRNA- or empty vector-transfected cells) and C (untransfected control cells) were included. Cells with transfections were cultivated for further 48 h prior to the subsequent assays.
RNA Isolation and Removal of Genomic DNA
Total RNAs were isolated from tissues and U87 cells using RNAstorm™ RNA Isolation Kit (Biotium). RNA samples were digested with DNase I (Invitrogen) at 37 °C for 80 min to remove genomic DNA. RNA integrity was checked by performing 5% gel (urea-PAGE) electrophoresis. RNA purity was reflected by OD260/280 ratios.
RT-qPCR
RNA samples with satisfactory integrity and an OD 260/280 ratio around 2.0 (pure RNA) were reverse transcribed into cDNA using a Reverse Transcription System (A5001, Promega Corporation). The qPCR reactions were prepared using SYBR Green Master Mix (Bio-Rad). The internal control for circSKA3 was 18S rRNA. The expression levels of mature miR-1 were measured using All-in-OneTM miRNA qRT-PCR Detection Kit (GeneCopoeia). Poly (A) was first added, followed by using poly (T) as reverse primer to perform reverse transcription. Finally, qPCRs were performed using sequence-specific forward and pol (T) as reverse primer. Ct values of target genes were normalized to corresponding controls using of 2−ΔΔCq method.
Methylation-Specific PCR (MSP)
Genomic DNA was isolated from U87 cells with transfections using Sera-Xtracta Genomic DNA Kit (Cytiva). After that, genomic DNA was converted suing DNA Methylation-GoldTM kit (ZYMO RESEARCH). Taq 2X Master Mix (NEB) was used to perform both routine PCRs and MSPs to analyze the methylation of miR-1 gene.
Cell Counting Kit 8 (CCK-8) Assay
A CCK-8 kit (Dojindo) was used to analyze the proliferation of U87 cells at 48 h post-transfection. In brief, 3000 cells in 0.1 mL medium was added into each well of a 96-well cell culture plate. At 37 °C, cells were cultivated and CCK-8 solution was added to reach 10% at 2 h prior to the measurement of OD values at 450 nm. OD values were measured every 24 h until 96 h.
Statistical Analyses
ANOVA Tukey’s test was used to compare differences among multiple groups. Paired t test was used to compare differences between paired tissue samples. Pearson’s correlation coefficient was used to analyze correlations. To analyze the prognostic values of circSKA3 and miR-1 for GBM, the 56 GBM patients were divided into high and low circSKA3 or miR-1 groups (n = 28, cutoff value = the median expression level of circSKA3 or miR-1 in GBM tissues). Survival curves were plotted based on the 2-year follow-up study. Log rank test was used to compare survival curves. P < 0.05 was considered as statistically significant.
Results
CircSKA3 Was Upregulated in GBM and Inversely Correlated with miR-1
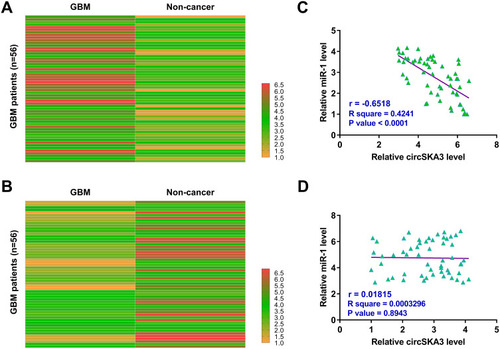
The differential expression of circSKA3 and miR-1 in GBM tissues and paired non-cancer tissues determined by RT-qPCRs. Heatmaps were plotted using Heml 1.0 software to reflect differential expression. It was observed that circSKA3 was upregulated () and miR-1 was downregulated () in GBM tissues compared to that in non-cancer tissues. Pearson’s correlation coefficient analysis showed that the expression of circSKA3 and miR-1 were inversely and significantly correlated across GBM tissues (), but not non-cancer tissues ().
Figure 1 CircSKA3 was upregulated in GBM and inversely correlated with miR-1. GBM tissues and paired non-cancer tissues were collected from the 56 GBM patients, followed by performing RNA isolations and RT-qPCRs to analyze the differential expression of circSKA3 (A) and miR-1 (B) in GBM. Heatmaps were plotted using Heml 1.0 software to reflect differential expression. Pearson’s correlation coefficient was performed to analyze the correlations between circSKA3 and miR-1 across GBM tissues (C) and not non-cancer tissues (D).
Altered Expression of circSKA3 and miR-1 Showed Prognostic Values for GBM
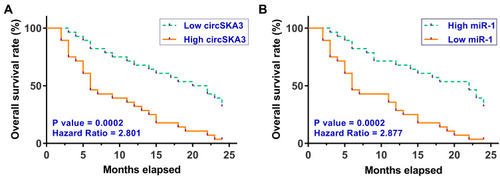
Survival analysis was performed to explore the prognostic value of circSKA3 or miR-1 for GBM. It was observed that patients in the high circSKA3 level group exhibited lower overall survival rate compared to that in the low circSKA3 level group (). In contrast, patients in the low miR-1 level group showed lower overall survival rate compared to that in the high miR-1 level group ().
Figure 2 Altered expression of circSKA3 and miR-1 showed prognostic values for GBM. To analyze the prognostic values of circSKA3 (A) and miR-1 (B) for GBM, the 56 GBM patients were divided into high and low circSKA3 or miR-1 groups (n = 28, cutoff value = the median expression level of circSKA3 or miR-1 in GBM tissues). Based on the 2-year follow-up study, survival curves were plotted. Log rank test was used to compare survival curves.
Overexpression of circSKA3 Decreased the Expression Levels of miR-1 in U87 Cells Through Methylation
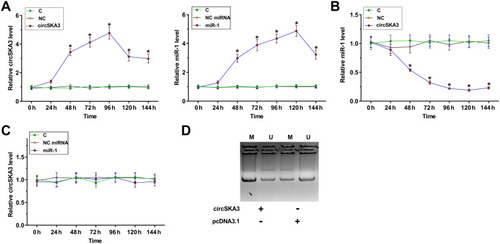
To explore the interaction between circSKA3 and miR-1 in GBM, U87 cells were transfected with either circSKA3 expression vector, or miR-1 mimic. Overexpression of circSKA3 and miR-1 were checked every 24 h until 144 h. It was observed that circSKA3 and miR-1 was significantly overexpressed (>2-fold) from 48 h to 144 h (, p < 0.05). Overexpression of circSKA3 significantly decreased the expression levels of miR-1 between 48 h and 144 h (, p < 0.05), while overexpression of miR-1 did not affect the expression of circSKA3 at each time point (). The methylation of miR-1 gene in cells transfection with either empty vector or circSKA3 expression vector was analyzed by MSP. Compared to empty vector-transfected cells, circSKA3 expression vector-transfected cells showed significantly increased methylation of the miR-1 gene (). It is well established that methylation plays a critical role in repressing gene expression. Therefore, circSKA3 may downregulate miR-1 through methylation in U87 cells.
Figure 3 Overexpression of circSKA3 decreased the expression levels of miR-1 in U87 cells through methylation. To explore the interaction between circSKA3 and miR-1 in GBM, U87 cells were transfected with either circSKA3 expression vector, or miR-1 mimic. Overexpression of circSKA3 and miR-1 was checked every 24 h until 144 h (A). The effects of overexpression of circSKA3 on the expression of miR-1 (B) and the effects of overexpression of miR-1 on the expression of circSKA3 (C) were analyzed by RT-qPCR. *p < 0.05. (D) The methylation of miR-1 gene in cells transfection with either empty vector or circSKA3 expression vector was analyzed by MSP. M, methylation PCR product; U, un-methylation PCR product.
Overexpression of circSKA3 Increased U87 Cell Proliferation Through miR-1
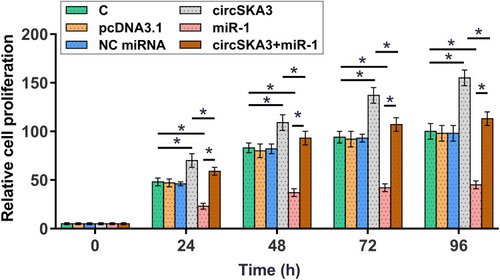
CCK-8 assay was performed to investigate the role of circSKA3 and miR-1 in regulating U87 cell proliferation. It was observed that overexpression of circSKA3 increased cell proliferation, and overexpression of miR-1 decreased cell proliferation. In addition, co-transfection of circSKA3 expression vector and miR-1 mimic showed higher cell proliferation rate than miR-1 mimic transfection alone, and lower cell proliferation rate than circSKA3 expression vector transfection alone (, p < 0.05).
Figure 4 Overexpression of circSKA3 increased U87 cell proliferation through miR-1. CCK-8 assay was performed to analyze the role of circSKA3 and miR-1 in regulating U87 cell proliferation. Cells were transected with circSKA3 expression vector or miR-1 mimic, or co-transfected with circSKA3 expression vector and miR-1 mimic. Cell proliferation was monitored by determining OD values at 450 nm every 24 h until 96 h.*p < 0.05.
Discussion
In this study, we analyzed the differential expression of circSKA3 in GBM and explored its potential interaction with miR-1. Our data showed that circSKA3 was upregulated in GBM and it may downregulate miR-1 through methylation to promote GBM cell proliferation.
The function of circSKA3 has only been explored in breast cancer.Citation13 It was observed that circSKA3 was upregulated in breast cancer and it can bind integrin β1 to induce the formation of invadopodium, thereby promoting the invasion of cancer cells.Citation13 To the best of our knowledge, the involvement of circSKA3 in other malignancies is unknown. We showed that circSKA3 was upregulated in GBM and overexpression of circSKA3 increased the proliferation of GBM cells. Therefore, circSKA3 may function as an oncogene in GBM by increasing cell proliferation.
GBM patients are suffering from extremely poor prognosis. It is well established that the accurate prognosis of GBM will provide guidance for the determination of treatment strategies, thereby improving patients’ survival. In this study we showed that high expression levels of circSKA3 were closely correlated with poor survival of GBM patients, suggesting that circSKA3 may serve as a prognostic biomarker for GBM. However, the reliability remains to be further tested by more clinical trials with bigger sample size.
MiR-1 is a well characterized tumor suppressor in different types of cancer including GBM.Citation14 It has been reported that miR-1 is downregulated in GBM and it can target fibronectin to suppress GBM.Citation14 Consistently, our study confirmed the downregulation of miR-1 in GBM and its inhibitory effects on GBM cell proliferation. Based on our knowledge, the upstream regulators of miR-1 in GBM are still unknown. Besides regulating gene expression at transcription and translational levels, circRNAs may also serve as the competing endogenous RNA of miRNAs to suppress their functions.Citation15 However, no promising bind site of miR-1 was found in the sequence of circSKA3. Interestingly, our data showed that overexpression of circSKA3 decreased the expression levels of miR-1 by increasing methylation of the miR-1 gene. However, the underlying mechanism is unknown. We showed that circSKA3 and miR-1 were only closely correlated in GBM tissues but not in non-cancer tissues, suggesting the involvement of certain pathological factors in the interaction between them.
Conclusion
In conclusion, circSKA3 is upregulated in GBM and predicts the poor survival of GBM patients. In addition, circSKA3 may downregulate miR-1 through methylation to promote GBM cell proliferation.
Disclosure
The authors declare that they have no competing interests.
References
- Alexander BM, Cloughesy TF. Adult glioblastoma. J Clin Oncol. 2017;35(21):2402–2409. doi:10.1200/JCO.2017.73.011928640706
- Davis ME. Glioblastoma: overview of disease and treatment. Clin J Oncol Nurs. 2016;20(5 Suppl):S2–S8. doi:10.1188/16.CJON.S1.2-8
- Babu R, Komisarow JM, Agarwal VJ, et al. Glioblastoma in the elderly: the effect of aggressive and modern therapies on survival. J Neurosurg. 2016;124(4):998–1007. doi:10.3171/2015.4.JNS14220026452121
- Smoll NR, Schaller K, Gautschi OP. Long-term survival of patients with glioblastoma multiforme (GBM). J Clin Neurosci. 2013;20(5):670–675. doi:10.1016/j.jocn.2012.05.04023352352
- Wick W, Osswald M, Wick A, Winkler F. Treatment of glioblastoma in adults. Ther Adv Neurol Disord. 2018;11:1756286418790452. doi:10.1177/175628641879045230083233
- Nam JY, de Groot JF. Treatment of glioblastoma. J Oncol Pract. 2017;13(10):629–638. doi:10.1200/JOP.2017.02553629020535
- Bianco J, Bastiancich C, Jankovski A, des Rieux A, Préat V, Danhier F. On glioblastoma and the search for a cure: where do we stand? Cell Mol Life Sci. 2017;74(13):2451–2466. doi:10.1007/s00018-017-2483-328210785
- Obacz J, Avril T, Le Reste PJ, et al. Endoplasmic reticulum proteostasis in glioblastoma-From molecular mechanisms to therapeutic perspectives. Sci Signal. 2017;10:470. doi:10.1126/scisignal.aal2323
- Lieberman F. Glioblastoma update: molecular biology, diagnosis, treatment, response assessment, and translational clinical trials. F1000Res. 2017;6:1892. doi:10.12688/f1000research.11493.129263783
- Esposito CL, Nuzzo S, Catuogno S, Romano S, de Nigris F, de Franciscis V. STAT3 gene silencing by Aptamer-siRNA Chimera as selective therapeutic for glioblastoma. Mol Ther Nucleic Acids. 2018;10:398–411. doi:10.1016/j.omtn.2017.12.02129499951
- Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi:10.1016/j.gde.2017.11.00729245064
- Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi:10.1016/j.canlet.2018.03.03529625140
- Du WW, Yang W, Li X, et al. The Circular RNA circSKA3 binds integrin β1 to induce invadopodium formation enhancing breast cancer invasion. Mol Ther. 2020;28(5):1287–1298. doi:10.1016/j.ymthe.2020.03.00232229309
- Yang CH, Wang Y, Sims M, Cai C, Pfeffer LM. MicroRNA-1 suppresses glioblastoma in preclinical models by targeting fibronectin. Cancer Lett. 2019;465:59–67. doi:10.1016/j.canlet.2019.08.02131491450
- Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi:10.18632/oncotarget.1915429069868
