Abstract
Purpose
CircRNA CircRIMS has been characterized as an oncogenic circRNA in gastric cancer, while its role in other cancers is unknown. This study aimed to explore the role of CircRIMS in esophageal squamous cell carcinoma (ESCC).
Patients and Methods
Tissues collected from 60 ESCC patients were subjected to extractions of total RNA and RT-qPCRs to analyze the differential expression of CircRIMS and miR-613. The 60 ESCC patients were followed up for 5 years to analyze the prognostic value of CircRIMS for ESCC. The interaction between CircRIMS and miR-613 was showed by luciferase activity assay and fluorescence in situ hybridization. The role of CircRIMS in regulating miR-613 expression and methylation was analyzed by overexpression experiments, RT-qPCRs and Western blot assay. The role of CircRIMS and miR-613 in regulating cell proliferation was analyzed using the BrdU assay. ESCC xenograft model was used to demonstrate the role of CircRIMS and miR-613 in vivo.
Results
We found that CircRIMS was overexpressed in ESCC and predicted poor survival. In addition, miR-613 was under expressed in ESCC and inversely correlated with CircRIMS. In ESCC cells, CircRIMS overexpression decreased the expression of miR-613 and increased the methylation of miR-613 gene. Cell proliferation assay showed that CircRIMS overexpression reduced the inhibitory effects of miR-613 overexpression on cell proliferation. Animal experience finally illustrated that CircRNA CircRIMS downregulated miR-613 through methylation to promote tumor growth.
Conclusion
Therefore, CircRIMS may downregulate miR-613 through methylation to increase cell proliferation in ESCC.
Introduction
Esophageal cancer is one of the most common types of cancer among males.Citation1 Esophageal squamous cell carcinoma (ESCC) as the most common subtype of esophageal cancer accounts for about 95% of all cases.Citation1,Citation2 With the development of anti-ESCC approaches, the survival of patients with resectable tumors, such as localized tumors, has been significantly improved by neoadjuvant chemoradiotherapy in combination with surgery.Citation3,Citation4 Nevertheless, the 5-year survival rate for ESCC patients was less than 20%.Citation5,Citation6 Therefore, it is necessary to explore new biomarkers of ESCC to develop the effective treatment of ESCC.
With the increased understanding of cancer biology, molecularly targeted therapy, which can be applied to treat cancers by regulating cancer-related gene expression, has been developed.Citation7–Citation9 In effect, certain molecular players, such as NRF2 signaling, HER2 gene and FGFR2 gene, have been proven to be potential targets for targeted ESCC therapy.Citation10,Citation11 However, the molecularly targeted therapy for ESCC is still under research and more targets are needed. Circular RNAs (circRNAs) are covalently closed single-strand RNA transcripts that participate in cancer biology by regulating gene expression rather than coding proteins, suggesting their potential role as targets for cancer treatment.Citation12,Citation13 However, the function of most circRAs in cancer biology remains to be analyzed. Evidence has been approved that the critical role of circular RNAs in ovarian cancer and CircRNA CircRIMS has been characterized as an oncogenic circRNA in gastric cancer,Citation14 while its role in other cancers is unknown. MicroRNAs (miRNAs) are a class of small non-coding RNAs with about 21–25 nucleotides that can regulate the expression of post-transcriptional target genes (b, c, f). A number of miRNAs play important roles in biological and pathological processes in ESCC. In ESCC, miR-613 has been identified as a diagnostic and prognostic biomarker for patients (h) and could suppress cell migration and invasion.
This study was performed to analyze the relationships between CircRIMS andmiR-613 in ESCC.
Patients and Methods
Patients and Follow-Up
A total of 60 ESCC patients (37 males and 23 females; 62.4 ± 5.6 years) who were diagnosed as ESCC through histopathological exam between May 2015 and August 2015 were enrolled in this study. This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of General Hospital of Eastern Theater Command. All patients were diagnosed for the first time. This study excluded the factors that can potentially affect the expression of the target gene of the present study, such as initiated therapy within 3 months prior to admission, other clinical disorders and history of malignancies, from the 60 ESCC patients. All patients signed informed consent. The 60 patients were grouped into stage I or II (n=32), and III or IV (n=28). All patients were followed up for 5 years from the day of admission to record survival. Follow-up was performed through a monthly manner and all patients completed the follow-up or died of ESCC during follow-up.
ESCC Tissue Acquisition and ESCC Cells
The 60 ESCC patients were subjected to a collection of ESCC and adjacent (within an area of 3 cm around the tumors) paired non-tumor tissue samples through fine needle aspiration. All tissue samples were confirmed by a histopathological exam. Tissue samples were kept in liquid nitrogen prior to the subsequent assays. ESCC cell model used in this study was KYSE450 human ESCC cell line (Laboratory of Birth Defects and Reproductive Health, China) and approved by the Ethics Committee of General Hospital of Eastern Theater Command. Cells were cultivated in a medium composed of 10% FBS and 90% RPMI-1640 medium under the conditions of 5% CO2, 37 °C and 95% humidity to reach about 80% confluence prior to the subsequent assays.
Cell Transfections
CircRIMS and miR-613 were overexpressed in KYSE450 cells by transfecting KYSE450 cells (106) with either 1μg CircRIMS expression vector or 50 nM miR-613 mimic. Mimic of miR-613 and negative control (NC) miRNA were provided by Genecopoeia (Guangzhou, China). The expression vector of CircRIMS was constructed with pcDNA3.1 expression vector provided by Invitrogen (Shanghai, China). NC experiments were performed by transfecting empty vector or NC miRNA into the same number of cells. In all transfections untransfected cells were control (C) cells. The transfected cells were kept for 48h in fresh medium to perform the following experiments.
RNA Preparations
Total RNA isolation from tissues and cells was carried out using Ribozol (VWR), followed by digestion with DNase I (Invitrogen) until OD 260/280 ratio reached about 2.0, which indicated pure RNA. RNA integrity analysis was performed through 5% urea-PAGE gel electrophoresis.
RT-qPCR Assay
StaRT Reverse Transcription kit (AnyGenes) was used to prepare cDNA samples through reverse transcriptions (RTs) using RNA samples as template. PowerTrack SYBR Green Master Mix (Rhenium) was used to perform qPCRs to analyze the expression of CircRIMS. The endogenous control of CircRIMS was 18S RNA. Expression of mature miR-613 was analyzed using All-in-One™ miRNA qRT-PCR Detection Kit (Genecopoeia) by adding poly (A), followed by RTs and qPCRs with poly (T) as reverse primer. U6 was used as the internal standard for normalizing gene expression of miR-613. Ct values of target genes were normalized to endogenous controls using 2−ΔΔCT method.
The primer sequences were listed as follows: CircRIMS forward: 5ʹ- −3ʹ TACAGTTGGAGCAGGTGGAAAG; reverse: 5ʹ- −3ʹ CGATGCCCCCAAACTACATTCAT. 18S RNA forward: 5ʹ-CTTTCGATGGTAGGTAGTGGCCT-3ʹ; reverse: 5ʹ-CAATGATCCTTCCGCAGGTTCACCTAC-3ʹ. miR-613 forward: 5ʹ-AGGAATGTTCCTTCT-3ʹ; reverse: 5ʹ-GTGCAGGGTCCGAGGT-3ʹ. U6 forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; reverse: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Luciferase Activity Assay
The wild-type or mutant seed sequence at the predicted region of CircRIMS was synthesized and cloned into the pGL3 Luciferase Reporter vectors (Promega). Cells were co-transfected with miR-613 mimics, or mimic control, together with pGL3 vectors, which contained the wild type (WT) or mutation (Mut) binding region of CircRIMS. Dual-Luciferase Assay was used to harvest the cells for detecting the activity of luciferase (Promega) at 48 hours following the transfection.
Fluorescence in situ Hybridization (FISH)
Alexa Fluor 488-labeled miR-613 probes and Alexa Fluor 555-labeled CircRIMS probes were designed and synthesized by RiboBio (Guangzhou, China). The FISH experiment was carried out by a fluorescent in situ Hybridization Kit (RiboBio). A total of 1×105 cells were seeded onto autoclaved glass slides and cultured for a period of 24 hours. Cells was fixed by 4% paraformaldehyde, then permeabilized by 0.5% Triton X-100, and cultured overnight at a temperature of 37 C. Finally, the slides were incubated with DAPI and observed under a fluorescence microscope (Leica)
Methylation-Specific PCR (MSP)
Quick Genomic DNA Extraction Kit (Clinisciences) was used to isolate genomic DNA from KYSE450 cells with transfections. EZ DNA Methylation-Gold™ Kit (ZYMO RESEARCH) was used to convert genomic DNA samples. The methylation of miR-613 was analyzed through routine PCRs and MSPs, which were all performed using 2x Taq mix (Invitrogen).
BrdU Assay
Cells with transfections were cultivated in a 96-well plate with 4000 cells in a 0.1 mL medium per well. Cells were cultivated under the aforementioned conditions for 48h, followed by incubation with BrdU (10 mM) for 48h. After that, cells were fixed and incubated with peroxidase-coupled anti-BrdU-antibody (Sigma–Aldrich) for 48h. After washing, peroxidase substrate was used to incubate the cells for 2h. Cell proliferation was analyzed by measuring OD values at 450 nm.
Establishing Xenografts in Mice
Twenty BALB/c nude mice (6 weeks old, Beijing Vital River Laboratory Animal Technology Co. Ltd) were randomly divided into four groups (n = 5/group): C: subcutaneously injected with KYSE450 cells in the dorsal region; CircRIMS: subcutaneously injected with circRIMS overexpression KYSE450 cells in the dorsal region; miR-613: subcutaneously injected with miR-613 overexpression KYSE450 cells in the dorsal region; CircRIMS + miR-613: subcutaneously injected with circRIMS and miR-613 overexpression KYSE450 cells in the dorsal region. The volume of xenografts was recorded. Mice were euthanized 4 weeks after cell injection, and the tumor was collected and further analyzed. All experiments involving animals were approved by the Ethics Committee of the General Hospital of Eastern Theater Command and followed by Guidelines for the ethical review of laboratory animal welfare People s Republic of China National Standard GB/T 35892 2018.
Western Blotting
Protease inhibitor cocktail was added to radioimmunoprecipitation assay (RIPA) lysate (Beyotime); cells and tumor tissues are lysed with this mixed solution. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to the polyvinylidene fluoride (PVDF) membrane, sealed with Blocking Buffer (Beyotime), and detected with primary anti-DNMT1 (Abcam) and ki67 (Abcam). GAPDH (Abcam) was used as an internal control of the same membrane. The blots were incubated in horseradish peroxidase conjugated secondary antibody (Cell Signaling Technology) for a duration of 2 hours, and the immune response signal was visualized by the ECL detection system (Beyotime).
Statistical Analysis
Heml 1.0 software was used to plot heatmaps to present the differential expression of CircRIMS and miR-613 in paired tissue samples. ANOVA Tukey’s test was used to compare independent cell transfection groups. Correlations were analyzed by Pearson’s correlation coefficient. The expression levels of CircRIMS in ESCC tissues were detected by RT-qPCR assay. With the median expression level of CircRIMS in ESCC tissues as a cutoff value, the 64 ESCC patients were grouped into high and low (n=32) level groups. Survival curves of both groups were plotted and compared by Log rank test. P<0.05 was statistical significant.
Results
Poor Prognosis of ESCC Patients Was Correlated with the High Expression Level of CircRIMS in ESCC Tissues
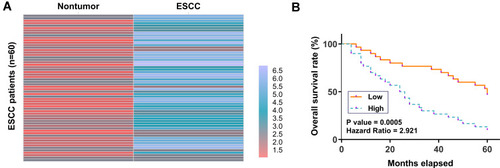
ESCC and paired non-tumor tissue samples were subjected to RNA isolation and RT-qPCR to determine the differential expression of CircRIMS in ESCC, which is expressed as a heatmap plotted using Heml 1.0 software. Compared to non-tumor tissues, ESCC tissues exhibited higher expression levels of CircRIMS (). Survival analysis showed that patients in high CircRIMS level groups experience worse overall survival compared to patients in low CircRIMS level group ().
Figure 1 Poor prognosis of ESCC patients was correlated with the high expression level of CircRIMS in ESCC tissues. ESCC and paired non-tumor tissue samples were subjected to RNA isolation and RT-qPCR to determine the differential expression of CircRIMS in ESCC, which is expressed as a heatmap plotted using Heml 1.0 software (A). With the median expression level of CircRIMS in ESCC tissues as a cutoff value, the 64 ESCC patients were grouped into high and low (n=32) level groups (B).
MiR-613 Was Under-Expressed in ESCC and Inversely Correlated with CircRIMS
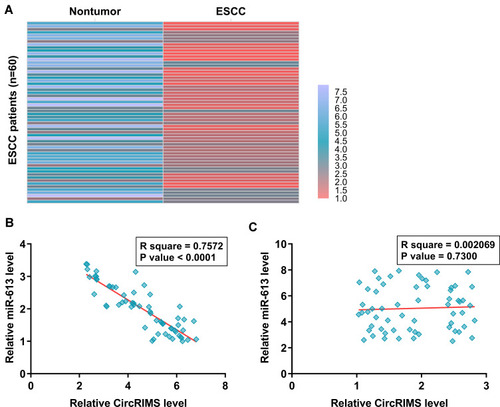
The expression of miR-613 in paired ESCC and non-tumor tissues from the 64 ESCC patients was also analyzed by RNA isolation and RT-qPCR. Heatmap analysis showed that expression levels of miR-613 were lower in ESCC tissues compared to non-tumor tissues (). Pearson’s correlation coefficient analysis showed that CircRIMS and miR-613 were inversely and significantly correlated across ESCC tissue samples (, p<0.05), but not non-tumor tissues ().
Figure 2 MiR-613 was under-expressed in ESCC and inversely correlated with CircRIMS. ESCC and paired non-tumor tissue samples were subjected to RNA isolation and RT-qPCR to determine the differential expression of miR-613 in ESCC, which is expressed as a heatmap plotted using Heml 1.0 software (A). Pearson’s correlation coefficient analysis was performed to analyze the correlations between CircRIMS and miR-613 ESCC tissue samples (B) and non-tumor tissues (C).
CircRIMS Overexpression Decreased the Expression of miR-613 in KYSE450 Cells Through Methylation
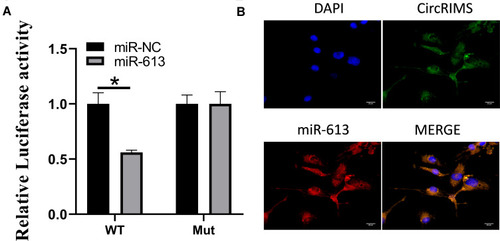
Luciferase activity assay in KYSE450 cells was carried out to demonstrate the reaction between CircRIMS and miR-613. The findings indicated that miR-613 evidently downregulated the luciferase activity co-transfected with pGL3- 3 UTR of CircRIMS; however, it does not include the pGL3-3 UTR-mut (). Moreover, the FISH experiment was used to detect the location of CircRIMS and miR-613. As illustrated in , co-location of circNELL2 and miR-127-5p in cytoplasm was observed.
Figure 3 CircRIMS directly targets miR-613. Luciferase activities were used to investigate the targeting of circNELL2 and miR-127-5p (A). FISH experiment using the probe of miR-613 and CircRIMS were carried out to evaluate the co-location of them (B). *p<0.05.
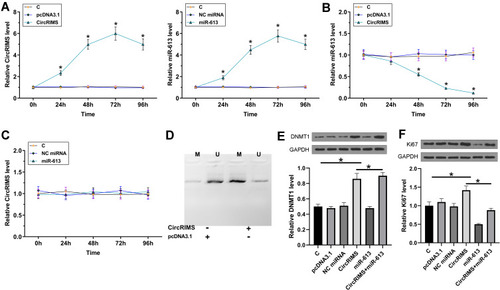
To study the crosstalk between CircRIMS and miR-613, KYSE450 cells were transfected with either CircRIMS expression vector or miR-613 mimic, followed by the confirmation of the overexpression of CircRIMS and miR-613 by RT-qPCR every 24h until 96h. It was observed that CircRIMS and miR-613 were significantly overexpressed between 24h and 96h post-transfection (, p<0.05). In addition, CircRIMS overexpression significantly decreased the expression of miR-613 (), while overexpression of miR-613 failed to significantly affect the expression of CircRIMS (). MSP was performed to analyze the role of CircRIMS in regulating the methylation of miR-613 gene. Compared to cells transfected with empty pcDNA3.1 vector, cells transfected with CircRIMS expression vector exhibited increased methylation of miR-613 ().
Figure 4 CircRIMS overexpression decreased the expression of miR-613 in KYSE450 cells through methylation. To study the crosstalk between CircRIMS and miR-613, KYSE450 cells were transfected with either CircRIMS expression vector or miR-613 mimic, followed by the confirmation of the overexpression of CircRIMS and miR-613 by RT-qPCR every 24h until 96h (A). The effects of CircRIMS overexpression on the expression of miR-613 (B), and the effects of miR-613 overexpression on the expression of CircRIMS (C) were analyze by RT-qPCR. MSP was performed to analyze the role of CircRIMS in regulating the methylation of miR-613 gene (D). The expression of DNMT1 (E) and Ki67 (F) was detected by Western blot assay. M, methylated PCR products; U, un-methylated PCR products; *p<0.05.
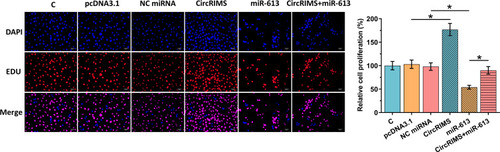
CircRIMS Overexpression Reduced the Inhibitory Effects of miR-613 Overexpression on Cell Proliferation
The role of CircRIMS and miR-613 in regulating the proliferation of KYSE450 cells was analyzed using the BrdU assay. CircRIMS overexpression significantly increased cell proliferation, while miR-613 overexpression significantly decreased cell proliferation. Moreover, CircRIMS overexpression reduced the inhibitory effects of miR-613 overexpression on cell proliferation (, p<0.05).
Figure 5 CircRIMS overexpression reduced the inhibitory effects of miR-613 overexpression on cell proliferation. The role of CircRIMS and miR-613 in regulating the proliferation of KYSE450 cells was analyzed by BrdU assay. Experiments were repeated 3 times and mean±values of three biological replicates were present an compared. *p<0.05.
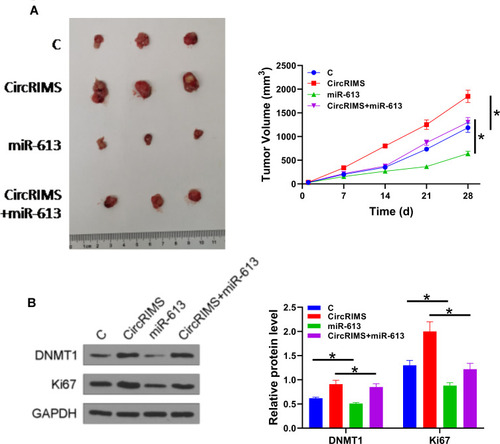
CircRIMS Overexpression Reduced the Tumor Inhibitory Effects of miR-613 on Cell Proliferation in vivo
To further investigate the role of CircRIMS in ESCC, we used a xenograft animal model. After 28 d of monitoring, we found that the growth rate of tumor volume was remarkably promoted in the CircRIMS group while miR-613 group showed restrained tumor growth (). To determine the role and relationship of circLPAR3 and miR-613, we performed Western blot assay to test DNMT1 and Ki67 expression. The results showed that CircRIMS expression obviously promoted DNMT1 and Ki67 levels in tumors () while miR-613 reversed. Importantly, we found that the expression of DNMT1 also improved in CircRIMS + miR-613group compared with C group, indicating that circLPAR3 might downregulate miR-613 through methylation thereby increasing the tumor growth of ESCC.
Figure 6 CircLPAR3 increased the tumor growth of ESCC by downregulating miR-613 through methylation. A xenograft tumour model was established by subcutaneously injecting transfected KYSE450 cells. Tumour image was shown in each group (A). The expression of DNMT1 and Ki67 was detected by Western blot assay (B). Experiments were repeated 3 times and mean±values of three biological replicates were present an compared. *P < 0.05.
Discussion
The present study explored the role of CircRIMS in ESCC and analyzed its crosstalk with miR-613, which is a critical player in ESCC.Citation15 We found that CircRIMS was upregulated in ESCC and it could downregulate miR-613 through methylation to increase cell proliferation both in vitro and in vivo.
Clinical treatment of ESCC is challenged by the poor prognosis, which is mainly caused by the low early diagnostic rate.Citation16 Due to the lack of sensitive biomarkers, the early diagnosis of ESCC is unlikely to be significantly improved in the near future. In this study, we showed that the expression levels of CircRIMS were closely correlated with the poor survival of ESCC patients, suggesting that CircRIMS may serve as a potential prognostic biomarker for ESCC. Therefore, monitoring the expression of CircRIMS may guide the determination of treatment approaches for ESCC, thereby improving the survival of patients. However, clinical trials are needed to further test our hypothesis.
A previous study reported that CircRIMS played an oncogenic role in gastric cancer.Citation14 CircRIMS is overexpressed in gastric cancer and overexpression of CircRIMS promotes the in vitro metastasis of gastric tumors by sponging hsa-miR-148a-5p and hsa-miR-218-5p.Citation14 It has also been reported that miR-613 is under-expressed in ESCC and overexpression of miR-613 targets G6PD to suppress the invasion and migration of ESCC cells.Citation15 Based on our understanding, the role of CircRIMS in other types of cancers is unknown, as well as the possible relationship between CircRIMS and miR-613.
In this study, we reported the overexpression of CircRIMS in ESCC. Moreover, CircRIMS overexpression significantly increased the proliferation of ESCC cells both in vivo and in vitro. Therefore, CircRIMS may play oncogenic roles by increasing cell proliferation. Moreover, our study showed that overexpression of miR-613 decreased tumor growth by inhibiting cell proliferation. We also identified the reaction between CircRIMS and miR-613 by luciferase activity assay and FISH experiment and further found that miR-613 could reverse the effect of CircRIMS in promoting cell proliferation and tumor growth.
As a member of the DNA methyltransferases (DNMTs), DNMT1 plays an important role in mediating gene expression and chromatin structure, by preserving existing DNA methylation during DNA replication. MSP was performed to illustrate that CircRIMS overexpression exhibited increased methylation of miR-613. We then found that CircRIMS significantly improved the expression of DNMT1 both in vivo and in vitro. We therefore speculated that CircRIMS may downregulate miR-613 through methylation in ESCC, thereby inhibiting cell proliferation.
Conclusion
In conclusion, CircRIMS is overexpressed in ESCC and it may downregulate miR-613 through methylation to promote cancer cell proliferation.
Disclosure
The authors declare that they have no conflicts of interest in this work.
Acknowledgments
The authors would like to thank the participants in this study.
References
- Higuchi K, Koizumi W, Tanabe S, et al. Current management of esophageal squamous-cell carcinoma in Japan and other countries. Gastrointest Cancer Res. 2009;3(4):153–161.19742141
- Short MW, Burgers KG, Fry VT. Esophageal cancer. Am Fam Physician. 2017;95(1):22–28.28075104
- Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. 2018;41(3):210–215. doi:10.1016/j.asjsur.2016.10.00527986415
- Sohda M, Kuwano H. Current status and future prospects for esophageal cancer treatment. Ann Thorac Cardiovasc Surg. 2017;23(1):1–11. doi:10.5761/atcs.ra.16-0016228003586
- Wu SG, Zhang WW, Sun JY, Li FY, Lin Q, He ZY. Patterns of distant metastasis between histological types in esophageal cancer. Front Oncol. 2018;8:302. doi:10.3389/fonc.2018.0030230135855
- Ai D, Zhu H, Ren W, et al. Patterns of distant organ metastases in esophageal cancer: a Population-Based Study. J Thorac Dis. 2017;9(9):3023–3030. doi:10.21037/jtd.2017.08.7229221275
- Huang M, Shen A, Ding J, Geng M. Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol Sci. 2014;35(1):41–50. doi:10.1016/j.tips.2013.11.00424361003
- Bozic I, Allen B, Nowak MA. Dynamics of targeted cancer therapy. Trends Mol Med. 2012;18(6):311–316. doi:10.1016/j.molmed.2012.04.00622595628
- Joo WD, Visintin I, Mor G. Targeted cancer therapy–are the days of systemic chemotherapy numbered? Maturitas. 2013;76(4):308–314. doi:10.1016/j.maturitas.2013.09.00824128673
- Kato H, Arao T, Matsumoto K, et al. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. Int J Oncol. 2013;42(4):1151–1158. doi:10.3892/ijo.2013.183023426935
- Ma S, Paiboonrungruan C, Yan T, Williams KP, Major MB, Chen XL. Targeted therapy of esophageal squamous cell carcinoma: the NRF2 signaling pathway as target. Ann N Y Acad Sci. 2018;1434(1):164–172. doi:10.1111/nyas.1368129752726
- Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi:10.1016/j.gde.2017.11.00729245064
- Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi:10.7150/ijms.2804730745810
- Lin J, Zhang Y, Zeng X, Xue C, Lin X. CircRNA CircRIMS acts as a MicroRNA sponge to promote gastric cancer metastasis. ACS Omega. 2020;5(36):23237–23246. doi:10.1021/acsomega.0c0299132954174
- Su X, Gao C, Feng X, Jiang M. miR-613 suppresses migration and invasion in esophageal squamous cell carcinoma via the targeting of G6PD. Exp Ther Med. 2020;19(4):3081–3089. doi:10.3892/etm.2020.854032256796
- Barret M, Prat F. Diagnosis and treatment of superficial esophageal cancer. Ann Gastroenterol. 2018;31(3):256–265. doi:10.20524/aog.2018.025229720850
