 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Epithelial–mesenchymal transition (EMT) is involved in normal developmental cellular processes, but it may also be co-opted by a subset of cancer cells, to enable them to invade and form metastases at distant sites. Several gene transcription factors regulate EMT, including Snail1, Snail2, Zeb1, Zeb2, and Twist; ongoing studies continue to identify and elucidate other drivers. Specific micro ribonucleic acids (RNAs) have also been found to regulate EMT, including the microRNA-200 (miR-200) family, which targets Zeb1/Zeb2. Cancer “stem cells” – with the ability to self-renew and to regenerate all the cell types within the tumor – have been found to express EMT markers, further implicating both cancer stem cells and EMT with metastasis. Microenvironmental cues, including transforming growth factor-β, can direct EMT tumor metastasis, such as by regulating miR-200 expression. In human tumors, EMT markers and regulators may be expressed in a subset of tumor cells, such as in cells at the invasive front or tumor–microenvironment interface, though certain subtypes of cancer can show widespread mesenchymal-like features. In terms of therapeutic targeting of EMT in patients, potential areas of exploration could include targeting the cancer stem cell subpopulation, as well as microRNA-based therapeutics that reintroduce miR-200. This review will examine evidence for a role of EMT in invasion and metastasis, with the focus being on studies in lung and breast cancers. We also carry out analyses of publicly-available gene expression profiling datasets in order to show how EMT-associated genes appear coordinately expressed across human tumor specimens.
Introduction
Metastasis is the primary cause of death in cancer patients.Citation1 Tumor invasion and metastasis, whereby cancer cells escape from the primary tumor mass and colonize at distant sites, involve multiple steps, including localized invasion, intravasation, transport to other organs, extravasation, formation of micrometastasis, and colonization.Citation2 Epithelial–mesenchymal transition (EMT), a process by which epithelial cells acquire characteristics of mesenchymal cells, is largely thought to play an important role in invasion and metastasis.Citation2 EMT is a natural process involved, for example, with development and wound healing, and it may be co-opted by at least a subset of tumor cells that acquire the ability to invade and metastasize.
This review will examine evidence for a role of EMT in invasion and metastasis, with the focus being on studies in lung and breast cancers. We will consider this topic, from both the perspective of experimental studies and the perspective of analyses of human tumor specimens. In addition, we will make use of molecular profiling datasets of breast and lung tumors, which are available in the public domain, in order to assess how EMT-associated genes may appear coordinately expressed across human tumors.
Epithelial-mesenchymal transition (EMT)
During EMT, epithelial cells lose their cell polarity and molecular expression, enabling cell-cell adhesion, and they gain migratory and invasive properties; numerous other cellular changes may also be associated with cells undergoing EMT.Citation3 Many processes involving EMT occur during embryogenesis; for example, neural crest cells undergo EMT in order to migrate away from the neural tube and to differentiate into bone, smooth muscle, peripheral neurons and glia, as well as melanocytes.Citation4 During wound healing, epithelial cells differentiate into myofibroblasts that rebuild the extracellular matrix and facilitate wound contraction.Citation5 When wound healing processes go awry in certain contexts, excess fibrous connective tissue can lead to organ fibrosis.Citation6
The molecular players involved in EMT, include Specific markers that distinguish an epithelial cell from a mesenchymal cell, as well as regulators that can drive a cell towards a mesenchymal or epithelial state. Cells undergoing EMT typically show both an increase in protein abundances of vimentin, N-cadherin, fibronectin, integrin αvβ6, and a decrease in E-cadherin, desmoplakin, cytokeratins, and occludin.Citation5 Several transcriptional suppressor families regulate EMT, including the zinc-finger proteins Snail 1 and Snail2, the two-handed zinc-fnger 8EF1 family factors (δEF1/Zeb1 and SIP1/Zeb2), and the basic helix-loop-helix factors, Twist and E12/E47.Citation7–Citation11 Evidence also suggests that signals derived from the cellular microenvironment can regulate EMT,Citation12,Citation13 such as through cell-cell contacts mediated by families of transmembrane receptors and ligands expressed on adjacent cells.Citation14
Over the last few years, posttranscriptional regulation of EMT has become an emerging paradigm.Citation15 Recently, Specific micro ribonucleic acids (RNAs) (miRNAs or miRs) - small, noncoding RNAs that posttranscriptionally regulate gene expression - have been found to regulate EMT, the most notable example being the regulation of Zeb1 and Zeb2 by the microRNA-200 (miR-200) family, where loss of miR-200 leads to EMT.Citation13,Citation16–Citation18 Other miRNAs that regulate Zeb1/2 include miR-205 and miR-192/215,Citation16,Citation19 and miRNAs that regulate Snail 1 or Snail2 include miR-1, miR-29b, miR-30c, miR-34, and miR-203.Citation20 Other EMT-associated genes involved in protein translation include: Y-box binding protein-1 (YB-1), which directly activates cap-independent translation of messenger RNAs (mRNAs) encoding Snail1 and other transcription factors related to EMT;Citation21 and heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1), which binds a transforming growth factor-β (TGFβ)-activated translation element in the transcripts of EMT genes DAB2 (disabled-2) and ILEI (interleukin-like EMT inducer).Citation22 Moreover, EMT initiates widespread changes in alternative splicing of gene transcripts, in large part through down-regulation of epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2).Citation23
Role of EMT in tumor invasion and metastasis
Since many normal cellular processes may be co-opted by cancer cells to their own advantage, there is much evidence that EMT aids tumor invasion and metastasis. Conceptually, this would seem plausible, as EMT would enable tumor epithelial cells to lose their cell polarity and cell–cell adhesive interactions and junctions, allowing the cells to escape from the primary tumor. Mesenchymal-like cancer cells could more effectively invade surrounding tissues and migrate to distant sites in ways that can reflect cell migration during development. Recently, EMT has been associated with the subset of tumor cells believed to be highly tumorigenic, also referred to as “cancer stem cells,”Citation24,Citation25 which would fit with the notion of a small percentage of tumor cells having the potential for invasion and metastases.Citation1
Much of the evidence for an EMT role in invasion and metastasis can be found in experimental studies, where EMT can be readily induced, for example, in vitro in a variety of cancer or immortalized cell lines.Citation7–Citation11 In our own studies, we have made use of a mouse model of human lung adenocarcinoma, driven by mutant K-ras and Tp53, where the tumors metastasize to sites commonly involved in lung cancer patients, and where metastasis is driven entirely by repression of the miR-200 family.Citation13 In our system as well as others, microenvironmental cues, including TGFβ, can direct tumor metastasis by regulating miR-200 expression. Studies in breast cancer and other cancers also show a similar role for miR-200.Citation17 Using cell lines derived from our mouse model, we have been able to identify additional genes with roles in the miR-200 pathway or EMT, including MIR34A, Jagged 2 (JAG2), and VEGFR1.Citation10,Citation26,Citation27
There is increasing support for the hypothesis that most tumors contain a subpopulation of cells, often referred to as tumor-initiating cells or “cancer stem cells,” with the ability to self-renew and to regenerate all the cell types within the tumor.Citation28–Citation32 These cancer stem cells, which can be isolated from the bulk tumor cells using Specific cell surface and other markers, have also been found to express EMT markers.Citation24,Citation25 Moreover, inducing EMT in immortalized human mammary epithelial cells (for example, by forced expression of Twist or Snail transcription factors), results in the cells also acquiring traits associated with stem cells, such as the expression of stem cell markers and an increased ability to form mammospheres.Citation25 In addition, over-expression of miR-200c causes both normal mammary stem cells and cancer-associated stem cells to lose their defining characteristics.Citation33 The above would indicate that the cancer EMT and cancer stem cell theories may coincide, where the subpopulation of tumor cells with the ability to form metastases do so, in part, through the use of processes associated with EMT.
EMT as observed in human tumor specimens
While experimental models can help establish cause-and-effect relationships between genes and pathways, studies of human tumor specimens are also needed in order to help ground experimental observations as being relevant in the setting of human patients. However, the manifestation of EMT in this setting may be contrary to the expectations of some, which has hindered widespread acceptance of the idea of cancer-associated EMT among clinicians in particular.Citation12 For one thing, tumor metastases established at distant sites appear to be more epithelial than mesenchymal, suggesting to some that EMT has not occurred, though an alternative explanation is that cancer-associated EMT is a transient state, and that mesenchymal-like cells can revert to an epithelial state upon tumor formation.Citation12 Another complication in human tumor studies of EMT is that EMT can appear manifested in only a subset of tumor cells,Citation11 for example, at the tumor–host interface. Analyses that average molecular signals across all cells within the tumor may, therefore, miss patterns distinctive to only a subpopulation of cells.
In Specific contexts, EMT-associated features can be observed in human tumors, such as in cells at the invasive front of cancers.Citation34 In addition, Specific subsets of human breast cancer show widespread mesenchymal features; these subsets include the metaplastic subtype,Citation35 as well as the expression-based claudin-low subtype.Citation24 Both metaplastic and claudin-low breast cancers also express markers of breast cancer stem cells.
EMT has also been related to therapy resistance in cancer, with both preclinical and clinical evidence. After neoadjuvant chemotherapy in breast cancer, the remaining tumor cells have been found to be enriched in stem cell and EMT markers,Citation24,Citation36 indicating that different therapies might be needed to target this population. Similarly, chemotherapy-treated lung tumors show enrichment for lung cancer stem cells (with CD133+ marker).Citation37 Functional studies also provide evidence for EMT’s involvement in chemoresistance; for example, a recent study that inhibited ZEB1 in docetaxel-resistant human lung adenocarcinoma cells, thereby significantly enhancing their chemosensitivity.Citation38 In another recent study, a panel of lung cancer cell lines was probed using a gene signature of EMT, and the cell lines that appeared more mesenchymal also showed greater resistance to epidermal growth factor receptor and PI3K/Akt pathway inhibitors.Citation39
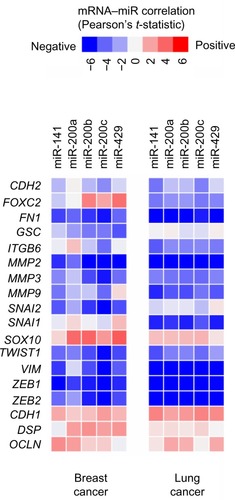
Global molecular analyses of human tumors, including gene expression profiling, have added a tremendous amount of data into the public domain. In this review, we have surveyed two sizeable datasets on breast and lung cancer to see whether EMT-associated genes appear coordinately expressed across tumor specimens. For breast cancer, we had previously assembled a compendium of nine separate gene expression array datasets (n = 1,340),Citation40 and recently we have carried out the same for lung adenocarcinoma (n = 1,492, representing eleven datasets),Citation41–Citation51 thereby giving us robust sample numbers for correlation analyses. With these data, we do find that genes canonically involved in EMT are, in fact, coordinately expressed with respect to each other, with a subset of tumors in each case looking relatively more mesenchymal-like as compared to the rest of the tumors (). Data from The Cancer Genome Atlas include profiling of both microRNAs and mRNAs,Citation52,Citation53 and in these datasets (for both breast and lung cancer), we see that lower expression of miR-200 family members is coordinate with EMT overall (), with miR-200 showing a strong anticorrelation with its target genes ZEB1 and ZEB2 in particular.
Figure 1 EMT-associated genes appear coordinately expressed across human tumor specimens of both breast and lung cancer.
Notes: Two gene expression profiling datasets are represented, (A) one being a “compendium” of published data on human breast cancers,Citation40 and (B) the other being a compendium of data on human lung cancers.Citation41–Citation51 Using a panel of canonical EMT markers as shown (from the review article by Lee et alCitation4), we have “scored” each of the tumor profiles for “EMT-ness” (ie, similarity to mesenchymal cells). Yellow denotes relatively high mRNA expression; blue indicates lower mRNA expression. For each dataset, a subset of tumors appears to be relatively more mesenchymal-like as compared to the rest of the tumors. Genes represented in the breast cancer dataset are limited to those featured on the U133A array platform.
Abbreviations: n, number; EMT, epithelial–mesenchymal transition; mRNA, messenger ribonucleic acid.
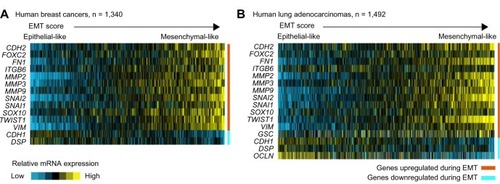
Figure 2 In both human breast cancer and lung cancer, lower expression of miR-200 family members is correlated with EMT marker expression.
Notes: Matrix of expression correlations between individual miR-200 family members and canonical genes encoding EMT markers (the list from Lee et al,Citation4 plus ZEB1 and ZEB2). Red indicates a positive correlation between microRNA and mRNA; blue indicates a negative correlation. Data are from The Cancer Genome Atlas (n = 503 human breast cancers and n = 159 human lung squamous cancers).Citation52,Citation53Abbreviations: mRNA, messenger ribonucleic acid; EMT, epithelial–mesenchymal transition.
Another potential use of public expression data is to uncover previously unknown or unappreciated correlations involving EMT. To this end, we have “scored” each of the tumor profiles for “EMT-ness” (ie, similarity to mesenchymal cells) by applying the following equation to the normalized values,
using a list of markers provided in another review.Citation4 Some of the strongest correlates of EMT, as found in both breast and lung cancer, are provided in ; however, many more genes could be implicated by using less stringent statistical cutoffs than what might be used for fitting within the printed page; out of 12,000 genes represented in both breast and lung datasets, over 4,000 have at least a nominally significant correlation with EMT (P < 0.01, Pearson’s correlation) in both datasets. The genes most up or down with EMT () include many that are related to the extracellular matrix or to cell adhesion. The genes most down with EMT (ie, high in cells that appear more epithelial-like) include grainyhead-like 2 (GRHL2), which was recently found to play a major role in the suppression of oncogenic EMT in breast cancer cells.Citation54
Table 1 Top gene correlates of EMT phenotype in human breast and lung tumors
One might suggest that if EMT were involved in metastases, then tumors showing EMT-like characteristics ought to show worse outcomes; however, such a survival correlation may be difficult to observe, given the reasons noted above, such as the presence of tumor cell subpopulations. For most gene array datasets in particular, the stroma content of the tumor samples may also contribute to mesenchymal-associated patterns. Notably, we do not see robust survival correlations for EMT markers in our tumor compendium datasets. At the same time, however, low miR-200 levels have been found elsewhere to be part of a larger microRNA expression profile that predicts poor outcome in early-stage lung cancer patients.Citation55 Regarding survival correlations involving mesenchymal-like or stem cell-like breast cancer cells, results have been somewhat mixed;Citation56 however, the claudin-low breast cancer subtype has been associated with worse outcome compared to other subtypes.Citation57 Another potential issue with analysis of array data in particular, would be the fact that expression values in this case are not absolute but relative; in contrast, ERCC1 mRNA can be used to normalize QT-PCR (quantitative polymerase chain reaction) values, and RNA sequencing can provide measures of both absolute and relative abundances.
Potential for targeting EMT.in cancer therapy and management
Molecular biology studies of EMT have shed light onto the processes of invasion and metastases, and the hope is that these basic biology findings can be eventually translated into new therapeutic approaches. Based on our discussion, one could see at least two potential areas of focus for targeting EMT: targeting the cancer stem cell subpopulation of the tumor, and miRNA-based therapeutics to reintroduce miR-200 (a master regulator of EMT) into tumor cells. Molecular pathways associated with EMT and stem cells in breast cancer, which have been suggested for targeting, include Notch, Wnt, and TGFβ.56 In addition, one study carried out a chemical screen for compounds showing selective toxicity for breast cancer stem cells, with top hits including salinomycin.Citation58 The appeal for the use of miRNAs as therapeutics is that they are small and might be more easily delivered into the cell; current challenges, on the other hand, include getting sufficient quantities of the therapeutic agents into the tumor, while minimizing toxicity and off-target effects, though work in this area is ongoing.Citation59
While we are not aware of ongoing clinical trials that directly target or evaluate EMT status in patients, there are trials currently evaluating stem cell markers pre- and/or posttherapy, as related to treatment or outcome. In breast cancer, current Phase II trials evaluating the CD44 stem cell marker include NCT01688609Citation60 (involving treatment of HER2+ cancers with lapatinib and trastuzumab) and NCT01372579Citation61 (involving treatment of triple-negative cancers in the neoadjuvant setting with carboplatin and eribulin mesylate). There are also a number of trials evaluating therapies that target cancer stem cell-associated pathways, including Notch; these include NCT01193881,Citation62 a trial in advanced nonsmall cell lung cancer involving gamma-secretase inhibitor RO4929097.
Current research and future directions
It would be most desirable for us to be able to translate, in the near term, our understanding of EMT biology into improved treatment approaches for cancer. However, at the same time, exploring the biology even further in order to catalogue and elucidate the key players and drivers of EMT would represent an investment that could have long-term rewards in improved targeting of metastasis in patients. The publicly-available molecular datasets, including those from The Cancer Genome Atlas, can be a tool in screening for novel EMT-associated genes – though only a fraction of EMT correlates may turn out to be key drivers, and these would need to be established using functional studies.
One avenue of research that can be further expanded upon is the role of the tumor microenvironment in initiating EMT and metastasis. It should be stated that we favor a model where diverse microenvironmental cues, rather than acquired genetic alterations alone, contribute to a subset of tumor cells that eventually evade and metastasize.Citation13 To this end, better experimental systems – over conventional two-dimensional cell cultures on plastic, or even over three-dimensional cultures in Matrigel™ (BD Biosciences, San Jose, CA, USA) – are needed in order to better mimic elements of the tumor microenvironment. Recent examples of such model systems include synthetic polymer-based scaffolds,Citation63 as well as ex vivo three-dimensional models using a natural matrix, in order to form a barrier between the endothelial and epithelial spaces, allowing lung cancer cell lines to be able to form lung nodules with intact vasculature.Citation64
Conclusion
Over time, we have learned a great deal about the elements involved in EMT, as well as those involved in invasion and metastases; individual findings, as made from both experimental and clinical studies, are coming together to give us a more complete picture. A number of diverse theories and observations surrounding cancer cell behavior can potentially fall under the umbrella of cancer-associated EMT, which may include aspects of cancer stem cells and tumor microenvironmental influences. More questions remain, including those pertaining to what the key drivers of EMT are (both within and outside of the cancer cell) over the natural course of the disease. Better experimental models are needed in order to study the role of the tumor microenvironment. Analysis of molecular data on human tumors, combined with results from experimental studies, can identify new or underappreciated players. There is also a need to better map out the extent of tumor heterogeneity and to characterize the distinct cancer cell subpopulations. We believe that by increasing our knowledge in the above areas, we can increase the potential for making an impact in the clinical setting.
Acknowledgments
Grant support: this study was supported in part by P30 CA125123 (CJC) and K08 CA151651 (DLG) from the National Institute of Health.
Disclosure
The authors report no conflicts of interest in this work.
References
- HanahanDWeinbergRAThe hallmarks of cancerCell20001001577010647931
- WeinbergRAThe Biology of Cancer1st edNew York, NYGarland Science2006
- BoyerBVallésAEdmeNInduction and regulation of epithelial-mesenchymal transitionsBiochem Pharmacol20006081091109911007946
- LeeJMDedharSKalluriRThompsonE WThe epithelial-mesenchymal transition: new insights in signaling, development, and diseaseJ Cell Biol2006172797398116567498
- WeberCELiN YWaiPYKuoPCEpithelial-mesenchymal transition, TGF-β, and osteopontin in wound healing and tissue remodeling after injuryJ Burn Care Res201233331131822561306
- López-NovoaJMNietoMAInflammation and EMT: an alliance towards organ fibrosis and cancer progressionEMBO Mol Med200916–730331420049734
- WellnerUSchubertJBurkUCThe EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAsNat Cell Biol200911121487149519935649
- EgerAAignerKSondereggerSDeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cellsOncogene200524142375238515674322
- PeinadoHOlmedaDCanoASnail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype?Nat Rev Cancer20077641542817508028
- YangYAhnYHGibbonsDLThe Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in miceJ Clin Invest201112141373138521403400
- AignerKDampierBDescovichLThe transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarityOncogene200726496979698817486063
- GaoDVahdatLTWongSChangJCMittalVMicroenvironmental regulation of epithelial-mesenchymal transitions in cancerCancer Res201272194883488923002209
- GibbonsDLLinWCreightonCJContextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expressionGenes Dev200923182140215119759262
- BrabletzTJungASpadernaSHlubekFKirchnerTOpinion: migrating cancer stem cells – an integrated concept of malignant tumour progressionNat Rev Cancer20055974474916148886
- WuCYTsaiYPWuMZTengSCWuKJEpigenetic reprogramming and post-transcriptional regulation during the epithelial-mesenchymal transitionTrends Genet201228945446322717049
- GregoryPABertAGPatersonELThe miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1Nat Cell Biol200810559360118376396
- BurkUSchubertJWellnerUA reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cellsEMBO Rep20089658258918483486
- ParkSMGaurABLengyelEPeterMEThe miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2Genes Dev200822789490718381893
- WangBHerman-EdelsteinMKohPE-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-betaDiabetes20105971794180220393144
- LamouilleSSubramanyamDBlellochRDerynckRRegulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAsCurr Opin Cell Biol201325220020723434068
- EvdokimovaVTognonCNgTTranslational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transitionCancer Cell200915540241519411069
- ChaudhuryAHusseyGSRayPSJinGFoxPLHowePHTGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEINat Cell Biol201012328629320154680
- WarzechaCCJiangPAmirikianKAn ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transitionEMBO J201029193286330020711167
- CreightonCJLiXLandisMResidual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating featuresProc Natl Acad Sci U S A200910633138201382519666588
- ManiSAGuoWLiaoMJThe epithelial-mesenchymal transition generates cells with properties of stem cellsCell2008133470471518485877
- AhnYHGibbonsDLChakravartiDZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expressionJ Clin Invest201212293170318322850877
- RoybalJDZangYAhnYHmiR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1Mol Cancer Res201191253521115742
- SchepersAGSnippertHJStangeDELineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomasScience2012337609573073522855427
- ChenJLiYYuTSA restricted cell population propagates glioblastoma growth after chemotherapyNature2012488741252252622854781
- BertoliniGRozLPeregoPHighly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatmentProc Natl Acad Sci U S A200910638162811628619805294
- DriessensGBeckBCaauweASimonsBDBlanpainCDefining the mode of tumour growth by clonal analysisNature2012488741252753022854777
- Al-HajjMWichaMSBenito-HernandezAMorrisonSJClarkeMFProspective identification of tumorigenic breast cancer cellsProc Natl Acad Sci U S A200310073983398812629218
- ShimonoYZabalaMChoRWDownregulation of miRNA-200c links breast cancer stem cells with normal stem cellsCell2009138359260319665978
- PatersonELKazenwadelJBertAGKhew-GoodallYRuszkiewiczAGoodallGJDown-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progressionNeoplasia201315218019123441132
- HennessyBTGonzalez-AnguloAMStemke-HaleKCharacterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristicsCancer Res200969104116412419435916
- LiXLewisMTHuangJIntrinsic resistance of tumorigenic breast cancer cells to chemotherapyJ Natl Cancer Inst2008100967267918445819
- LiuY PYangCJHuangMSCisplatin selects for multidrug-resistant CD133+ cells in lung adenocarcinoma by activating Notch signalingCancer Res201373140641623135908
- RenJChenYSongHChenLWangRInhibition of ZEB1 reverses EMT and chemoresistance in docetaxel-resistant human lung adenocarcinoma cell lineJ Cell Biochem201311461395140323255418
- ByersLADiaoLWangJAn epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistanceClin Cancer Res201319127929023091115
- KesslerJDKahleKTSunTA SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesisScience2012335606634835322157079
- BeerDGKardiaSLHuangCCGene-expression profiles predict survival of patients with lung adenocarcinomaNat Med20028881682412118244
- BhattacharjeeARichardsWGStauntonJClassification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclassesProc Natl Acad Sci U S A20019824137901379511707567
- TomidaSTakeuchiTShimadaYRelapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosisJ Clin Oncol200927172793279919414676
- ChitaleDGongYTaylorBSAn integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumorsOncogene200928312773278319525976
- SheddenKTaylorJMEnkemannSADirector’s Challenge Consortium for the Molecular Classification of Lung AdenocarcinomaGene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation studyNat Med200814882282718641660
- ZhuCQDingKStrumpfDPrognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancerJ Clin Oncol201028294417442420823422
- BildAHYa oGChangJTOncogenic pathway signatures in human cancers as a guide to targeted therapiesNature2006439707435335716273092
- TangHXiaoGBehrensCA 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patientsClin Cancer Res20131961577158623357979
- OkayamaHKohnoTIshiiYIdentification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomasCancer Res201272110011122080568
- BotlingJEdlundKLohrMBiomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validationClin Cancer Res201319119420423032747
- HouJAertsJden HamerBGene expression-based Classification of non-small cell lung carcinomas and survival predictionPLoS One201054e1031220421987
- Cancer Genome Atlas Research NetworkComprehensive genomic characterization of squamous cell lung cancersNature2012489741751952522960745
- Cancer Genome Atlas NetworkComprehensive molecular portraits of human breast tumoursNature20124907418617023000897
- CieplyBRileyPPiferPMSuppression of the epithelial-mesenchymal transition by Grainyhead-like-2Cancer Res20127292440245322379025
- PatnaikSKKannistoEKnudsenSYendamuriSEvaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resectionCancer Res2010701364520028859
- CreightonCJChangJCRosenJMEpithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancerJ Mammary Gland Biol Neoplasia201015225326020354771
- PratAParkerJSKarginovaOPhenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancerBreast Cancer Res2010125R6820813035
- GuptaPBOnderTTJiangGIdentification of selective inhibitors of cancer stem cells by high-throughput screeningCell2009138464565919682730
- Nana-SinkamS PCroceCMClinical applications for microRNAs in cancerClin Pharmacol Ther20139319810423212103
- National Cancer Institute (NCI)Lapatinib Ditosylate, Trastuzumab, Paclitaxel, and Surgery in Treating Patients With Breast Cancer Available from http://clinicaltrials.gov/show/NCT01688609. NLM identification NCT01688609Accessed July 12, 2013
- Northwestern UniversityCarboplatin and Eribulin Mesylate in Triple Negative Breast Cancer Patients Available from http://clinicaltrials.gov/ct2/show/NCT01372579?term=NCT01372579&rank=1. NLM identifier NCT01372579Accessed July 12, 2013
- National Cancer Institute (NCI)RO4929097 and Erlotinib Hydrochloride in Treating Patients With Stage IV or Recurrent Non-Small Cell Lung Cancer Available from http://clinicaltrials.gov/ct2/show/NCT01193881?term=NCT01193881&rank=1. NLM identifier NCT01193884Accessed July 12, 2013
- GillBJGibbonsDLRoudsariLCA synthetic matrix with independently tunable biochemistry and mechanical properties to study epithelial morphogenesis and EMT in a lung adenocarcinoma modelCancer Res201272226013602322952217
- MishraDKSakamotoJHThrallMJHuman lung cancer cells grown in an ex vivo 3D lung model produce matrix metalloproteinases not produced in 2D culturePLoS One201279e4530823028922