Abstract
Background
Exosomes contain abundant circular RNAs (circRNAs), playing an important role in intercellular communication. However, the function and underlying molecular mechanism of exosomal circRNAs in foot metastatic melanoma remain unclear.
Methods
Twelve differentially expressed exosomal circRNAs between patients with metastatic and primary foot melanoma were screened through high-throughput sequencing, and their expression levels were detected by the real-time reverse transcriptase-polymerase chain reaction (RT-qPCR). CircRNA102927 silencing and overexpression A2058 cell line was constructed, and the effects of circRNA102927 on cell proliferation, apoptosis, migration, invasion, and epithelial-mesenchymal transition (EMT) were assessed using cell counting kit-8 (CCK-8), flow cytometry, wound healing, Transwell, and Western blot assays, respectively.
Results
Twelve differentially expressed exosomal circRNAs were screened and ROC curve showed that six circRNAs could be used as the diagnostic biomarkers for metastatic melanoma. Melanoma-secreted exosomes induced the differentiation of CD4+ T cells into Treg cells. CircRNA102927 was highly expressed in metastatic melanomas. Functionally, circRNA102927 silencing inhibited proliferation, EMT, migration, and invasion in metastatic melanoma cells, while promoting apoptosis. Meanwhile, overexpression of circRNA102927 had the opposite effects.
Conclusion
Our investigation suggests that silencing exosomal circRNA102927 may suppress foot melanoma metastasis by inhibiting invasiveness, EMT and promoting apoptosis.
Introduction
Melanoma is one of the most invasive and dangerous forms of skin cancer, accounting for approximately 90% of skin cancer-related deaths despite constituting only 1% of all skin cancers.Citation1 Melanoma tends to spread easily and progress rapidly. Its progression encompasses several stages, including precursor nevi, primary melanomas, and metastatic melanomas.Citation2 Although immune checkpoint inhibitors and targeted therapies greatly enhance melanoma treatment outcomes, metastatic spread and recurrence remain major challenges.Citation3,Citation4 Acral melanoma, a subtype that occurs in hairless skin tissue, such as the palms of the hands, soles of the feet, or under the fingernails and toes, typically has a poorer prognosis than other melanoma subtypes. This poorer prognosis is often due to delayed diagnosis, which makes acral melanoma more susceptible to metastasis.Citation5 Therefore, it is crucial to delve deeper into the diagnostic markers and molecular mechanisms of acral melanoma, particularly focusing on metastasis.
Exosomes, characterized by a lipid bilayer and a diameter ranging from about 30 to 100 nm, are pervasive in various body fluids, including blood, sweat, urine, tears, and ascites.Citation6,Citation7 These small vesicles play a crucial role in mediating transduction signals among cancer cells. They act as carriers, facilitating the transfer of diverse bioregulators, such as proteins, lipids, nucleic acids, miRNAs, lncRNAs, and circRNAs, thereby promoting tumor proliferation, metastasis, immune evasion, and influencing the tumor microenvironment.Citation8,Citation9 Moreover, exosomes have a significant advantage as diagnostic biomarkers because they are present in various body fluids, can be collected non-invasively, and can be used in clinical trials.Citation10 Recent studies have shown that exosomes in saliva and the isolation of small extracellular vesicles using a bead-assisted platform have great potential and flexibility for tumor diagnosis.Citation11,Citation12 circRNA (Circular RNA) lacks the 5′ cap and 3′ polytope tail, forming a closed ring structure.Citation13 Tumor-derived exosomes contain an abundance of circRNAs, which is crucial for tumor metastasis.Citation14 Exosomes from cancer-associated fibroblasts enhance tumorigenesis, metastasis, and chemoresistance in colorectal cancer through the upregulation of circ0067557, which targets Lin28.Citation15 Circ0001715 sponges miR-205-5p to enhance TREM2 expression, which in turn promotes M2 macrophage polarization and lung adenocarcinoma cell proliferation and metastasis.Citation16 Additionally, hypoxia-induced exosomal circPLEKHM1 serves as a prognostic biomarker for patients with non-small cell lung cancer and facilitates macrophage polarization and tumor metastasis.Citation17 In recent years, an increasing number of studies have shown that circRNAs play an important role in the metastatic mechanisms of melanoma. CircROR1 is highly expressed in metastatic melanoma cells and tissues, which facilitates metastasis in vitro and tumor growth in vivo.Citation18 Through its interaction with p70S6K2, Circ-GLI1 promotes metastasis in melanoma by activating the Hedgehog/GLI1 and Wnt/β-catenin pathways and increasing Cyr61 expression.Citation19 In addition, circ0001591, circRNA0082835, and circ0079593, etc, have been shown to be involved in the metastatic process of melanoma.Citation20–23 However, there is limited research on exosomal circRNAs concerning the metastatic mechanisms of foot melanoma. Therefore, it is essential to investigate the regulatory mechanisms of exosomal circRNAs in acral melanoma metastasis.
High-throughput RNA sequencing (RNA-seq) is widely employed to investigate lncRNA function, search for candidate drug targets, and identify biomarkers for cancer classification and diagnosis.Citation2 In this study, we screened 12 differentially expressed exosomal circRNAs between metastatic and primary foot melanoma using RNA-seq sequencing. CircRNA102927 expression was elevated in metastatic melanoma and it promoted the progression of metastatic melanoma. CircRNA102927 silencing impeded tumor development. This investigation provides new insights and potential therapeutic strategies for metastatic melanoma.
Materials and Methods
Clinical Specimens
Three patients with foot metastatic melanoma (zhuan yi, ZY) and two patients with foot primary melanoma (yuan fa, YF) were enrolled in this study (Supplementary Table 1). Before this study, every patient enrolled in this study had given written informed consent.
circRNA Sequencing Analysis
The extraction of total RNA from foot of three ZY and two YF patients was performed using TRIzol reagent (Thermo Fisher Scientific, Massachusetts, USA). Subsequently, NanoDrop ND-1000 (Thermo Fisher Scientific) was employed for the quantification of total RNA in each sample. To selectively retain circular RNAs post-elimination of non-circular counterparts, RNase R (Illumina, San Diego, California, USA) was utilized. For RNA expression profiling, the Arraystar Human Circular RNA Array (Arraystar, Rockville, Maryland, USA) was employed. CircRNAs were amplified and transcribed into fluorescent cDNAs using the random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The arrays were subsequently subjected to scanning by the Agilent Scanner G2505C, and the obtained array images were processed through the Agilent Feature Extraction software (version 11.0.1.1). Normalization of chips was carried out with the limma package in R software (version 4.1.3). Differentially expressed circRNAs were identified through the edgeR R package, applying criteria of |logFC| > 1 and P < 0.05. For visualization, volcano plot and heatmap of the differentially expressed circRNAs were generated using ggplot2 and pheatmap packages, respectively. Furthermore, the microarray IDs of the differentially expressed circRNAs were converted to obtain circBank IDs. Subsequently, exons of the genes included in the differentially expressed circRNAs were analyzed in the CircInteractome database (https://circinteractome.irp.nia.nih.gov/).
Gene Ontology (GO) Enrichment Analysis
DAVID (Database for Annotation, Visualization, and Integrated Discovery; https://david.ncifcrf.gov/home.jsp; version 6.8) is an online analysis tool for annotation, visualization and integrated discovery, providing typical batch annotation and gene-GO term enrichment analysis to highlight the most relevant GO terms associated with related genes. The GO annotation analysis of the up-regulated genes and down-regulated differentially expressed circRNAs was conducted in the DAVID database, which included molecular function (MF), biological process (BP), and cellular component (CC). A P < 0.05 was set as the cutoff criteria. The GOplot package in R was used to generate bubble plots, visualizing the most important GO terms and their related genes.
Exosome Isolation and Identification
Exosomes were isolated from the serum of patients by using VEX Exosome Isolation Reagent (Vazyme, Nanjing, China). Briefly, following coagulation, the blood was initially centrifuged at 3500 rpm for 10 min at 4°C to obtain serum. Serum was then subjected to an additional centrifugation at 4600 rpm for 30 min at room temperature to eliminate residual cells and debris. Subsequently, 1/5 of the sample volume of VEX Exosome Isolation Reagent was added to the serum sample. The mixture was inverted and allowed to stand at 4°C for 30 min. After the incubation, the mixture was centrifuged at 9600 rpm for 5 min, resulting in the presence of serum exosomes in the sediment at the bottom of the tube. Subsequently, exosome marker proteins, including CD9 and Tsg101, were detected by Western blot.
Western Blot Assay
Total protein extraction was conducted using RIPA buffer containing protease inhibitor (Solarbio, Beijing, China). Following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Roche, Basel, Switzerland). To prevent nonspecific binding, PVDF membranes were blocked by incubation with 5% skim milk. The membranes were then exposed to primary antibodies (dilution of 1:2000) and incubated overnight at 4°C. Primary antibodies included anti-CD9 (Abcam, Cambridge, UK), anti-Tsg101 (Abcam), anti-FOXP3 (Abcam), anti-TGF-β (Abcam), anti-GATA-3 (Abcam), anti-E-cadherin (Abcam), anti-N-cadherin (Abcam), anti-Vimentin (Abcam), and GAPDH (Abcam). Following three washes with TBST, PVDF membranes were treated with HRP-conjugated secondary antibodies for 1h. Finally, ECL chemiluminescence reagent (Amersham, Little Chalfont, UK) was applied to visualize the protein blots.
Real-Time Reverse Transcriptase-Polymerase Chain Reaction (RT-qPCR)
Total RNA extraction was performed by the TRIZOL reagent (Thermo Fisher Scientific). To specifically preserve circular RNAs, RNase R was employed. Then,1 μg of RNA was subjected to reverse transcription using Prime-Script RT Reagent Kit (Thermo Fisher Scientific). The ABI 7500 real-time PCR equipment (Thermo Fisher Scientific) was used for quantitative expression measurement, employing SYBR Green Mix Taq (Takara, Dalian, China). GAPDH was employed as the internal reference, and the Ct values obtained were analyzed using the 2−ΔΔCt method, calculated as ΔΔCt = [Ct (target gene) – Ct (GAPDH)]. The primer sequences employed in this study are detailed in Supplementary Table 2.
Analysis of the Potential of Six circRNAs as a Biomarker to Distinguish Between ZY and YF
With the pROC package in R software, the receiver operating characteristic (ROC) curve was created to evaluate the predictive value of six circRNAs in distinguishing between foot metastatic melanoma and foot primary melanoma. The area under the curve (AUC) value of the ROC curve exceeded the threshold of 0.8, indicating statistical significance.
CD4+ T Cells Co-Cultured with Exosomes
A total of 5 mL of peripheral blood was acquired from health controls, and patients with metastatic melanoma and primary melanoma. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque density gradient centrifugation. Following that, CD4+ and Treg cells were separated from PBMCs using human CD4 and Treg cell MACS kits (Miltenyi Biotec, Bergisch Gladbach, Germany). The data were obtained and analyzed using FACS. Naïve CD4+ T cells were incubated in T cell media supplemented with 10% FBS and 100 IU/mL IL-2. To activate the cells, plate-bound OKT3 was introduced at a 1 μg/mL concentration. Then, the cells were cultivated for 7 days with or without the supplementation of 10 μg/mL exosomes.
Enzyme-Linked Immunosorbent Assay (ELISA)
Interleukin-10 (IL-10), IL-4, and IL-17 levels were measured using ELISA kits (Esebio Biotechnology Co., Ltd., Shanghai, China) as per the manufacturer’s instructions. 50 µL of serum from different groups was transferred to a sample plate, followed by 50 µL of IL-10, IL-4, and IL-17 HRP-labeled reagents in each well, which were then incubated for 30 min. The chromogenic reagent was then added, and the reaction was halted with a stop solution, followed by a 10-min incubation in the dark. Absorbance at 450 nm was read using a microplate reader (DALB, Shanghai, China). Cytokine release was indicated in unit of pg/mL.
Cell Culture
Normal human primary melanocytes, human malignant melanoma cells A375 (from primary lesion), and A2058 (from metastatic lesion) were purchased from Icellbioscience Biotechnology Co., Ltd. (Shanghai, China). Cells were grown in DMEM medium (Thermo Fisher Scientific) containing 10% fetal bovine serum (Thermo Fisher Scientific) in a humidified environment at 37°C with 5% CO2. Human primary melanocytes were verified by immunofluorescence, and A375 and A2058 cells were verified by short tandem repeat (STR) analysis. All cells were regularly examined for mycoplasma contamination.
Cell Transfection
A2058 cells were used for transfection. The short hairpin RNA (shRNA) sequences, including sh-circRNA102927-1, sh-circRNA102927-2, sh-circRNA102927-3, and a blank control short hairpin RNA (sh- circRNA) was designed and synthesized by Shanghai Integrated Biotech Solutions Co., Ltd (Shanghai, China). These sequences were then incorporated into the pSuper-retro-puro vector using the Lipofectamine 2000 reagent (Thermo Fisher Scientific) in accordance with the manufacturer’s guidelines. In addition, to generate a stable circ102927 overexpression vector, full-length circRNA102927 cDNA was synthesized and cloned into the pCDNA3.1-circRNA-EF1-ZsGreen overexpression vector, containing front and back circular frames to facilitate RNA circularization. An empty vector without the circRNA102927 sequence was employed as a negative control. The silencing and overexpression efficiency was evaluated using RT-qPCR after 48 h transfection.
Cell Counting Kit-8 (CCK8)
In the 96-well plate, each well was planted with 1×104 transfected A2058 cells. They were treated with 10 µL of CCK-8 reagent (Solarbio) at 0, 24, 48, and 72 h, and then cells were incubated for 2 h at 37 °C. Subsequently, the optical density (OD) at 450 nm was assessed utilizing a microplate reader (DALB) to measure cell viability.
Wound Healing Assay
A total of 1×104/well transfected cells were seeded in the six-well plate and cultured until reaching 90% confluence. A 200-μL pipette tip was then employed to generate a scratch wound. Subsequently, A2058 cells were incubated in the serum-free medium for 24 h. Photographs were taken with an Olympus BX51 microscope (Olympus, Tokyo, Japan) at 0 and 24 h, and the wound healing was analyzed by the ImageJ software.
Transwell Assay
Cell invasion was assessed with transwell chambers (BD Biosciences, New Jersey, USA), which were previously coated with matrigel. A total of 1×105 cells transfected cells were planted in each superior chamber added with 100 μL serum-free medium, while the lower chambers contained 600 µL of medium with 20% FBS. Following 48 h of incubation, cells in the lower chamber were fixed with 4% paraformaldehyde for 30 min, followed by treatment with a 0.2% Triton X-100 (Sigma, Missouri, USA) solution for 15 min. Subsequently, the cells were stained with 0.1% crystal violet for 30 min and observed under a microscope (Olympus).
Cell Apoptosis Analysis
Using the Annexin V-FITC apoptosis kit (Sigma), cell apoptosis was measured. After washing with cold PBS, cells were incubated for 15 min at room temperature in the dark with 100 μL of 1× binding buffer that contained 5 μL of Annexin V-FITC and 10 μL of propidium iodide. Using a flow cytometer (BD), apoptosis was examined.
Statistical Analyses
GraphPad Prism 7.0 (GraphPad Software, San Diego, California, USA) was used for the statistical analysis, and the findings were shown as means ± standard deviation (SD). One-way analysis of variance (ANOVA) and Tukey’s test were utilized for assessing multiple groups, while the Student’s t-test was used to compare two groups. P < 0.05 was used as the statistical significance threshold.
Results
Differentially Expressed circRNAs Between ZY and YF
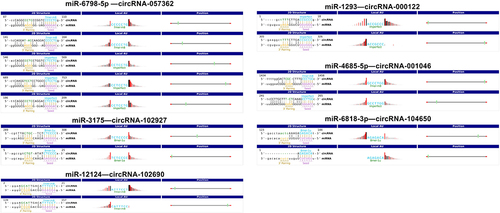
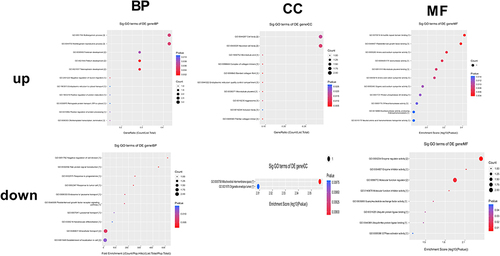
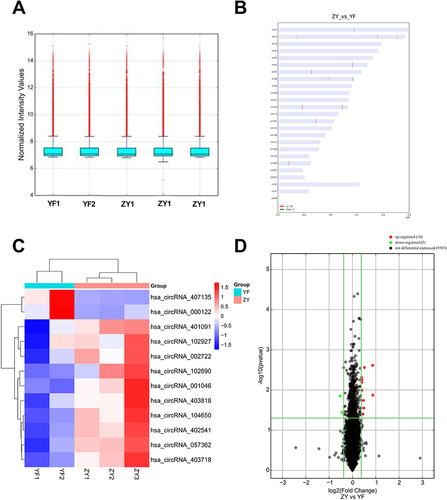
Exosomes were isolated from the serum of patients with ZY and YF, and then they were subjected to circRNAs sequencing. Firstly, we normalized the read counts for each sample, which showed that the median values were highly consistent across all samples (). In addition, the distribution of circRNA host gene chromosomes were analyzed (). There were 12 differentially expressed circRNAs were screened and 10 circRNA expressions were increased and 2 circRNAs expressions were decreased (Supplementary Table 3). Heatmap () and volcano map () were used for the visalization of 12 differentially expressed circRNAs. The exons contained in the 12 circRNAs were analyzed in the CircInteractome database, and a total of six circRNA exon sequences were obtained. CircRNAs are known to act as competing endogenous RNAs, binding to regulatory miRNAs and modulating their expression at the post-transcriptional level. Further analysis focused on identifying the downstream binding miRNAs of the six differentially expressed circRNAs (). Bioinformatics analysis revealed that several miRNAs are targets of specific circRNAs, each with complementary binding sites: miR-6798-5p was the target of circRNA057362, miR-3175 was the target of circRNA102927, miR-12124 was the target of circRNA102690, miR-1293 was the target of circRNA000122, miR-4685-5p was the target of circRNA001046, and miR-6818-3p was the target of circRNA104650. Additionally, GO enrichment analysis uncovered the functional roles of these circRNAs. In the BP terms, 10 up-regulated circRNAs were mainly involved in pallium development (GO: 0021543), telencephalon development (GO: 0021537), and multi-organism reproductive process (GO: 0044703). In the CC terms, 10 up-regulated circRNAs were primarily associated with banded collagen fibril (GO: 0098643), fibrillar collagen trimer (GO: 0005583), and neuronal cell body (GO: 0043025). In the MF terms, 10 up-regulated circRNAs were mostly implicated in armadillo repeat domain binding (GO: 0070016), platelet-derived growth factor binding (GO: 0048407), and amino acid: sodium symporter activity (GO: 0005283). The 2 down-regulated circRNAs, mainly related to intracellular transport (GO: 0046907), enzyme regulator activity (GO: 0030234), and molecular function regulator (GO: 0098772; ).
Figure 1 Identification of differentially expressed exosomal circRNAs between foot metastatic melanoma (ZY) and primary foot melanoma (YF). (A) Boxplot of chip background correction. (B) Distribution of circRNA host gene chromosomes. (C) Heatmap for cluster analysis of 12 differentially expressed circRNAs. (D) Volcano map for 12 differentially expressed circRNAs.
Six circRNAs Served as the Biomarkers for Distinguishing Between ZY and YF
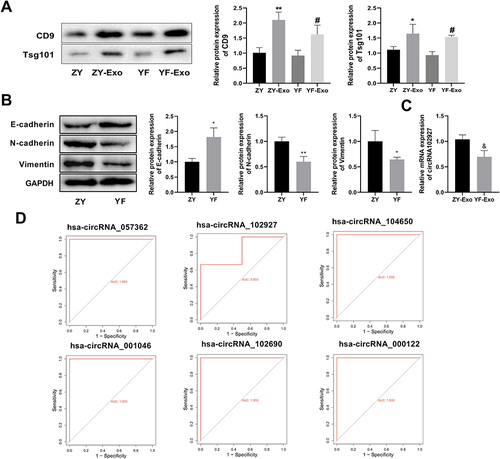
Protein expression of CD9 and Tsg101 was assessed to confirm the presence of exosomes post-extraction (). The findings unveiled a substantial increase in the expression of CD9 and Tsg101 in the ZY-Exo group compared to the ZY group, and a similar considerable elevation was noted in the YF-Exo group compared to YF, which indicated that the exosome was successfully extracted. In addition, the expression levels of epithelial-mesenchymal transition (EMT)-related proteins in the ZY and YF groups were evaluated by Western blot. The data showed that compared with the ZY group, the expression of E-cadherin was elevated, while protein levels of N-cadherin and Vimentin were reduced in the YF group (). RT-qPCR showed that compared to the YF-Exo group, the circ102927 expression in the ZY-Exo group was elevated (). Then, the diagnostic efficacy of six circRNAs (circRNA057362, circRNA102927, circRNA104650, circRNA001046, circRNA102690, and circRNA000122) between ZY and YF was explored by ROC curve. The outcome indicated that the AUC values of six circRNAs were greater than 0.8, suggesting that they could be used as the biomarkers to distinguish ZY and YF ().
Figure 4 Six circRNAs could be used as a biomarker to distinguish foot metastatic melanoma (ZY) and primary foot melanoma (YF). (A) Western blot was used to detect the expression of exosomal marker proteins CD9 and Tsg101 in the ZY, ZY-Exo, YF, and YF-Exo groups. (B) Protein levels of key regulators of EMT, including E-cadherin, N-cadherin, and Vimentin, which were measured by Western blot in the ZY and YF groups. (C) The expression of circRNA102927 in ZY and YF exosomes was determined by RT-qPCR. (D) The ROC curve profile illustrated the diagnostic efficacy of six circRNAs in ZY and YF. *P < 0.05, **P < 0.01 vs ZY group; #P < 0.05 vs YF group; &P < 0.05 vs ZY-Exo group.
Melanoma Exosomes Induced CD4+ T Cells Differentiation into Treg Cells
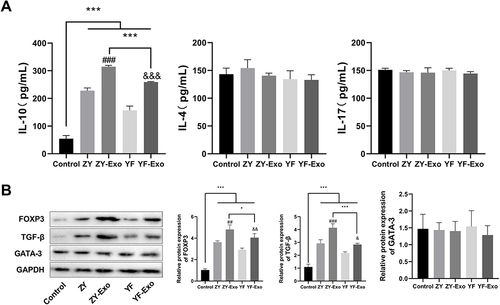
During CD4+ T cells differentiation into Treg cells, the clearance of tumor cells by the killer immune cell population was inhibited, ultimately promoting tumor progression.Citation24 We explored the effects of melanoma exosomes on CD4+ T cells and Treg cells. CD4+ T cells were cultured in vitro and then were subjected to melanoma exosome induction. The levels of IL-10 (Treg cell marker), IL-4 (T-helper 1 cell marker), and IL-17 (T-helper 17 cell marker) were assessed by ELISA. The results indicated that the levels of IL-4 and IL-17 in each group did not exhibit significant differences. However, in contrast to the Control group, IL-10 expression was notably higher in the ZY, ZY-Exo, YF, and YF-Exo groups. Additionally, IL-10 expression was substantially elevated in the ZY-Exo group comparison to the ZY group, and it was also considerably higher in the YF-Exo group relative to the YF group. Moreover, IL-10 level was obviously elevated in the ZY-Exo group as opposed to the YF-Exo group (). Subsequently, Western blot assay was conducted to evaluate the Treg regulatory protein expressions, including FOXP3 and TGF-β, as well as the Th1 regulatory protein GATA-3. Results revealed that in the ZY, ZY-Exo, YF, and YF-Exo groups, FOXP3 and TGF-β expression levels were markedly elevated compared to those in the Control group. However, no substantial disparity in the expression of GATA-3 was observed. Furthermore, FOXP3 and TGF-β protein expressions in the ZY-Exo group were markedly elevated compared to those in the ZY group. In comparison with the YF group, the YF-Exo group demonstrated significantly elevated expression of FOXP3 and TGF-β in relation to the ZY group. In addition, in the ZY-Exo group, the protein expression of FOXP3 and TGF-β were substantially elevated in comparison to the YF-Exo group. The above results indicate that both the ZY and YF exosomes could induce CD4+ T cell differentiation to Treg, with ZY exosomes exhibiting a more pronounced effectiveness in this process ().
Figure 5 Exosomes derived from melanoma promoted the differentiation of CD4+ T cells into T Treg cells. (A) The concentrations of Treg cell marker IL-10, Th1 cell marker IL-4, and Th17 marker IL-17 were assessed using ELISA. (B) Western blotting was carried out to assess the expression levels of Treg regulatory proteins, including FOXP3 and TGF-β, along with the Th1 regulatory protein GATA-3. *P < 0.05, ***P < 0.001 vs Control group; ##P < 0. 01, ###P < 0.001 vs ZY or YF group; &P < 0.05, &&P < 0. 01, &&&P < 0. 001 vs YF-Exo group.
The Expression Levels of circRNAs Were Detected by RT-qPCR
RT-qPCR was used to detect the expression levels of circRNAs ( and ). Since no corresponding gene name or CDS sequence was found for circRNA403718, we assessed the expression levels of the other 11 circRNAs. The results showed that compared with human primary melanocytes, the expression levels of circRNA403818, circRNA057362, circRNA402541, circRNA002722, circRNA104650, circRNA001046, circRNA401091, circRNA102690, and circRNA102927 were significantly increased, while circRNA000122 and circRNA407135 expression levels were significantly decreased in A375 cells (primary) and A2058 (metastatic) cells. Furthermore, the expression levels of circRNA403818, circRNA057362, circRNA402541, circRNA104650, circRNA001046, circRNA102690, and circRNA102927 were higher in A2058 cells than that in A375 cells. In contrast, circRNA000122 and circRNA407135 expressions were lower in A2058 cells than that in A375 cells.
Figure 6 RT-qPCR was used to assess the expression levels of circRNAs in human primary melanocytes, human malignant melanoma cells A375 (primary) and A2058 (metastatic). *P < 0.05, **P < 0.01, ***P < 0.001 vs Control group; #P < 0.05, ##P < 0. 01 vs A375 group.
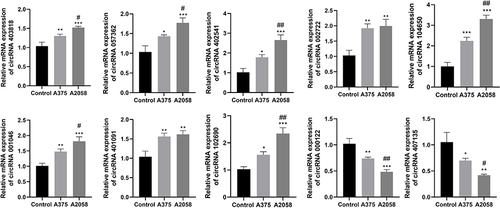
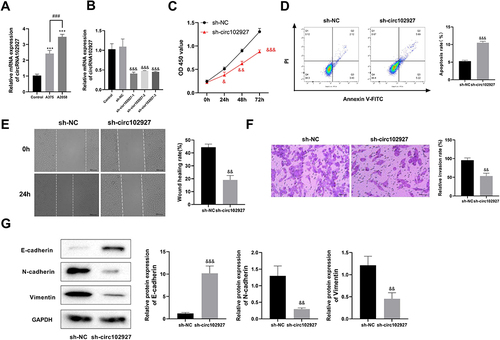
Figure 7 Silencing circ102927 inhibited the proliferation, migration, invasion, and EMT of foot metastatic melanoma (ZY) cells and promotes apoptosis. (A) The levels of circRNA102927 expression in A375 and A2058 cells were assessed using RT-qPCR. (B) RT-qPCR was used to determine the circRNA102927 silencing efficacy in A2058 cells. (C) A2058 cell proliferation was assessed by CCK8 after sh-NC and sh-circ102927 treatment. (D) The apoptosis of A2058 cells was assessed using Annexin V-FITC in the sh-NC and sh-circ102927 groups. (E) A2058 cell migration was detected by wound healing assay after 24 h transfection with sh-NC and sh-circ102927. (F) Transwell was used to determine cell invasion. (G) EMT-related protein expressions (E-cadherin, N-cadherin, and Vimentin) were measured by Western blot. ***P < 0.001 vs Control group; ###P < 0.001 vs A375 group; &P < 0.05, &&P < 0.01, &&&P < 0.001 vs sh-NC group.
Silencing circRNA102927 inhibited ZY cell proliferation, migration, invasion, and EMT, while promoting apoptosis
Since circ102927 might act as a sponge for miR-3175, which plays an important role in tumor cell proliferation, invasion, apoptosis, EMT, and metastasis,Citation17,Citation25,Citation26 it was selected for further exploration. Functional exploration was first performed by knockdown of circRNA102927 in A2058 cells. RT-qPCR results showed that circRNA102927 was successfully knocked down (). Considering that sh-circRNA102927-1 demonstrated the most effective silencing efficiency, it was chosen for further exploration. Cell proliferation was assessed using the CCK-8 assay. The findings demonstrated that circRNA102927 silencing inhibited A2058 cell proliferation (). Flow cytometry showed that circRNA102927 silencing boosted apoptosis in ZY cells. (). Furthermore, circ102927 silencing suppressed cell migration and invasion ( and ) according to the wound healing and Transwell assays. Additionally, protein level of E-cadherin was significantly elevated, and the protein expressions of N-cadherin and Vimentin were decreased after circ102927 knockdown (). The above findings indicate that the suppression of circ102927 hinders the processes of proliferation, migration, invasion, and EMT in ZY cells, while advancing apoptosis.
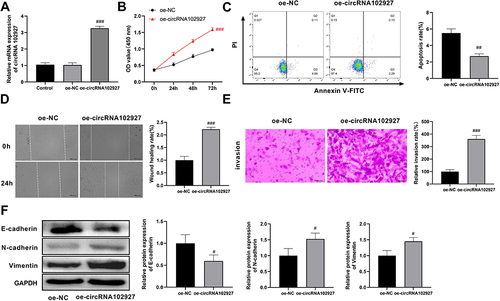
circRNA102927 overexpression promoted ZY cell proliferation, migration, invasion, and EMT, while suppressing apoptosis
Subsequently, we assessed the effects of circRNA102927 overexpression on the A2058 cell behaviors. RT-qPCR showed that the expression level of circRNA102927 was significantly elevated in the oe-circRNA102927 group compared with the oe-NC group, suggesting that circRNA102927 was effectively overexpressed (). The results from CCK-8 showed that circRNA102927 overexpression promoted the proliferation of ZY cells (). Conversely, circRNA102927 overexpression inhibited cell apoptosis (). Simultaneously, migration and invasion of ZY cells were enhanced after circRNA102927 overexpression ( and ). Furthermore, circRNA102927 overexpression decreased the protein expression of E-cadherin, and promoted the protein levels of N-cadherin and Vimentin (). Together, circRNA102927 overexpression facilitates ZY cell proliferation, migration, invasion, and EMT, while inhibiting apoptosis.
Figure 8 Upregulation of circRNA102927 led to enhanced proliferation, migration, invasion, and EMT in foot metastatic melanoma (ZY) cells, along with reduced apoptosis. (A) After A2058 cells were transfected with oe-circRNA102927, overexpression efficiency was assessed by RT-qPCR. (B) The proliferation of A2058 cells was measured by CCK8 after treatment with oe-NC or oe-circRNA102927. (C) Flow cytometry was used to assess the effect of circRNA102927 overexpression on apoptosis in metastatic melanoma cells. (D) A2058 cell migration was detected after circRNA102927 overexpression by wound healing assay. (E) The Transwell assay was employed to determine cell invasion upon circRNA102927 upregulation. (F) Western blot was performed to detect the effect of circRNA102927 overexpression on the expression levels of EMT-related proteins (E-cadherin, N-cadherin, and Vimentin) in A2058B cells. #P < 0.05, ##P < 0.01,###P < 0.001 vs oe-NC group.
Discussion
Metastasis is a fundamental characteristic of malignant tumors, involving the escape of malignant cells from the primary tumor site, migration through the lymphatic and/or blood circulation, and eventual spread to distant sites.Citation27 Surgical resection and adjuvant therapy are effective in curing primary tumors, but metastatic tumors, accounting for over 90% of cancer-related mortality, remain largely incurable due to their resistance to existing therapeutic agents.Citation28,Citation29 Patients frequently overlook melanoma of the foot, and lesions in this area are prone to misdiagnosis and metastasis.Citation30 This is attributed to the extended duration of the disease, resulting in an overall poorer prognosis compared to melanomas in other areas.Citation31 In the present investigation, we identified six differentially expressed exosomal circRNAs, which could distinguish between metastatic melanoma and primary melanoma. Furthermore, we found that downregulating circRNA102927 suppressed EMT, proliferation, migration, and invasion, while promoting apoptosis in metastatic melanoma. The opposite effects were observed with the overexpression of circRNA102927.
Exosomes are small vesicles of endocytic origin carrying various active constituents, which are involved in tumor progression.Citation32 Exosomes exert a crucial function in tumor metastasis by transferring specific information between cells. They are implicated in the regulation of the EMT and remodeling of the extracellular matrix.Citation33–35 Additionally, exosomes can promote tumor angiogenesis, immune escape, contribute to the formation of pre-metastatic niches, and influence organotropic metastasis.Citation36,Citation37 circRNA, due to its resistance to degradation by nucleic acid exonucleases and its abundance and stability in exosomes, holds promise as a valuable biomarker for multiple tumors, such as liver, colorectal, and gastric cancers.Citation38–41 circRNAs also serve as miRNA sponges during gene regulation and influence protein function, which participates in various tumor-related molecular processes, such as tumor cell proliferation, invasion, and metastasis.Citation42 In this investigation, we collected three patients with foot malignant melanomas with left inguinal lymph node or lung metastasis and two patients with foot primary melanomas for exosomal circRNAs RNA-seq. In total, 12 differentially expressed circRNAs were screened that might play an important role in metastasis of foot melanoma. Furthermore, circRNA057362, circRNA102927, circRNA104650, circRNA001046, circRNA102690, circRNA000122, and circRNA102927 could be used as the biomarkers to distinguish primary melanoma from metastatic melanoma.
In tumor tissues, exosomes act as messengers between tumor cells and immune cells, evading the host immune response through various strategies, which include suppressing tumor antigens, inhibiting regulatory immune cells, and secreting immunosuppressive molecules.Citation43 Tregs are a subset of T cells, playing a pivotal role in tumor immune escape. It can suppress CD4+ helper T cells and CD8+ cytotoxic T cells activation and differentiation, thereby inducing responsiveness to self-antigens and antigens expressed by tumors.Citation44,Citation45 Moreover, Tregs promote cancer cell survival by impeding the infiltration of effector T cells in tumor-associated immunity therapy.Citation46 The research has shown that exosomal circGSE1 promotes the amplification of Tregs by targeting the miR-324-5p/TGFBR1/Smad3 axis, thereby promoting the progression of HCC.Citation47 Exosomal circCCAR1 induces anti-PD1 resistance and CD8+ T-cell dysfunction in HCC.Citation48 In this study, melanoma exosomes induced the differentiation of CD4+ T cells to Treg cells and enhanced the expression of IL-10, FOXP3, and TGF-β, which promoted melanoma metastasis. The forkhead/winged helix transcription factor (Foxp3) is specifically expressed in Tregs, and its sustained expression is essential for the maintenance of intact Tregs inhibition.Citation49 Tregs release suppressive cytokines, such as IL-10, TGF-β, and IL-35, which impede immune activity through IL-10 and related pathways.Citation50 Additionally, Tregs hinder the function of CD8+ T cells and dendritic cells (DCs) by utilizing membrane-bound TGF-β, effectively modulating the body’s antitumor immune response.Citation51 Altogether, we found that exosomes secreted by foot melanoma may promote Tregs cell expansion by promoting the expression levels of IL-10, FOXP3, and TGF-β, which in turn promote tumor progression, especially in terms of metastasis.
EMT in cancer is a critical event associated with metastasis. Through the induction of EMT and the promotion of blood vessel permeability, the interaction between tumor cells and TME plays a crucial role in augmenting metastasis and intravasation.Citation52 During the process of EMT, tumor cells undergo a transition from epithelial to mesenchymal properties, which involves the downregulation of E-Cadherin and loss of cell polarity, accompanied by the upregulation of N-cadherin, twist, snail, and Vimentin.Citation53,Citation54 Exosomal circRNAs affect metastasis of multiple tumors by regulating the EMT process. hsa_circ_0009143 (circRNA_PVT1) is up-regulated in cervical cancer and can target miR-1286 via an exosomal pathway to induce EMT in cervical cancer cells, promoting tumor metastasis.Citation55 By modulating the microRNA-653-5p/paired box 6 axis, exosomal circular RNA hsa_circ_007293 facilitates papillary thyroid carcinoma cell proliferation, migration, invasion, and EMT transition.Citation56 In this investigation, circRNA102927 knockdown inhibited EMT in foot metastatic melanoma cells by promoting E-cadherin protein expression, whereas decreasing N-cadherin and Vimentin expression. CircRNA102927 overexpression promoted the process of EMT. Furthermore, reduced sensitivity to apoptotic stimuli plays an important role in successful metastasis of tumor cells.Citation57 Studies have suggested that tumor exosomes influence tumor metastasis by regulating cell proliferation, migration, and invasion.Citation58,Citation59 We found that overexpression of circRNA102927 facilitated the development of metastatic melanoma and silencing circRNA102927 also inhibited proliferation, migration, and invasion of A2058 cells, and promoted apoptosis, all of which contributed to the inhibition of foot melanoma metastasis.
However, there are some limitations to this study. First, the sample size was small, with data collected from only 2 YF patients and 3 ZY patients. Second, bioinformatics analysis revealed that miR-3175 was a target of circRNA102927, which required further experimental verification. Then, although six circRNAs with diagnostic values were identified, only the function of circRNA102927 was investigated. The mechanisms underlying circRNA102927, as well as the functions and mechanisms of the other circRNAs, will be further explored in future studies.
Conclusion
Our investigation showed that circRNA102927 was highly expressed in foot metastatic melanoma. Knockdown of circRNA102927 inhibited proliferation, migration, invasion and EMT, and promoted apoptosis in metastatic melanoma cells, which inhibits tumor metastasis. Overexpressing circRNA102927 resulted in the opposite effects. Our investigation provides new insights and potential therapeutic targets for metastatic foot melanoma.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital. And our study complies with the Declaration of Helsinki. Before this study, every patient enrolled in this study had given the written informed consent.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
Data Sharing Statement
The data and materials supporting the findings of this study are available from the corresponding authors upon request.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
- Wang C, Tan S, Liu WR, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18(1):134. doi:10.1186/s12943-019-1061-8
- Boutros A, Croce E, Ferrari M, et al. The treatment of advanced melanoma: current approaches and new challenges. Crit Rev Oncol Hematol. 2024;196:104276. doi:10.1016/j.critrevonc.2024.104276
- Shirley CA, Chhabra G, Amiri D, Chang H, Ahmad N. Immune escape and metastasis mechanisms in melanoma: breaking down the dichotomy. Front Immunol. 2024;15:1336023. doi:10.3389/fimmu.2024.1336023
- Nunes LF, Quintella Mendes GL, Koifman RJ. Acral melanoma: a retrospective cohort from the Brazilian National Cancer Institute (INCA). Melanoma Res. 2018;28(5):458–464. doi:10.1097/CMR.0000000000000476
- Kok V, Yu CJIJON. Cancer-derived exosomes: their role in cancer biology and biomarker development. International Journal of Nanomedicine. 2020;15:8019–8036. doi:10.2147/IJN.S272378
- Liu J, Ren L, Li S, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. 2021;11(9):2783–2797. doi:10.1016/j.apsb.2021.01.001
- Dai J, Su Y, Zhong S, et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5(1):145. doi:10.1038/s41392-020-00261-0
- Li C, Hou X, Zhang P, et al. Exosome-based tumor therapy: opportunities and challenges. Curr Drug Metab. 2020a;21(5):339–351. doi:10.2174/1389200221666200515103354
- Jafari A, Karimabadi K, Rahimi A, et al. The emerging role of exosomal miRNAs as biomarkers for early cancer detection: a comprehensive literature review. Technology in Cancer Research & Treatment. 2023;22:15330338231205999. doi:10.1177/15330338231205999
- Kozhevnikova D, Chernyshev V, Yashchenok A. Progress in isolation and molecular profiling of small extracellular vesicles via bead-assisted platforms. Biosensors. 2023;1:1.
- Rajendran P, Sekar R, Zahra HA, et al. Salivaomics to decode non-coding RNAs in oral cancer. A narrative review. Noncoding RNA Res. 2023;8(3):376–384. doi:10.1016/j.ncrna.2023.05.001
- He Y, Huang Q, Ge Y, et al. The role of circular RNA in tumor microenvironment and immunotherapy. Int J Biol Macromol. 2023;242:124929. doi:10.1016/j.ijbiomac.2023.124929
- Long F, Lin Z, Li L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20(1):26. doi:10.1186/s12943-021-01318-6
- Yang C, Zhang Y, Yan M, et al. Exosomes derived from cancer-associated fibroblasts promote tumorigenesis, metastasis and chemoresistance of colorectal cancer by upregulating circ_0067557 to target Lin28. BMC Cancer. 2024;24(1):64. doi:10.1186/s12885-023-11791-5
- Chen M, Cao C, Ma JJTC. Tumor-related exosomal circ_0001715 promotes lung adenocarcinoma cell proliferation and metastasis via enhancing M2 macrophage polarization by regulating triggering receptor expressed on myeloid cells-2. Thoracic Cancer. 2024;15(3):227–238. doi:10.1111/1759-7714.15182
- Kim S, Bae WJ, Ahn JM, et al. MicroRNA signatures associated with lymph node metastasis in intramucosal gastric cancer. Mod Pathol. 2021;34(3):672–683. doi:10.1038/s41379-020-00681-x
- Sun L, Bin S, Huang C, Wang Q. CircROR1 upregulates CCNE1 expression to promote melanoma invasion and metastasis by recruiting KAT2A. Exp Dermatol. 2024;33(4):e15071. doi:10.1111/exd.15071
- Chen J, Zhou X, Yang J, et al. Circ-GLI1 promotes metastasis in melanoma through interacting with p70S6K2 to activate Hedgehog/GLI1 and Wnt/β-catenin pathways and upregulate Cyr61. Cell Death & Disease. 2020;11(7):596. doi:10.1038/s41419-020-02799-x
- Durante G, Comito F, Lambertini M, Broseghini E, Dika E, Ferracin M. Non-coding RNA dysregulation in skin cancers. Essays Biochem. 2021;65(4):641–655. doi:10.1042/EBC20200048
- Lu J, Li Y. Circ_0079593 facilitates proliferation, metastasis, glucose metabolism and inhibits apoptosis in melanoma by regulating the miR-516b/GRM3 axis. Mol Cell Biochem. 2020;475(1–2):227–237. doi:10.1007/s11010-020-03875-8
- Sun Y, Hou Z, Luo B, et al. Circular RNA circRNA_0082835 promotes progression and lymphatic metastasis of primary melanoma by sponging microRNA miRNA-429. Bioengineered. 2021;12(1):4159–4173. doi:10.1080/21655979.2021.1953822
- Yin D, Wei G, Yang F, Sun XJH, toxicology E. Circular RNA has circ 0001591 promoted cell proliferation and metastasis of human melanoma via ROCK1/PI3K/AKT by targeting miR-431-5p. Human & Experimental Toxicology. 2021;40(2):310–324. doi:10.1177/0960327120950014
- Lee GR. The Balance of Th17 versus Treg Cells in Autoimmunity. Int J Mol Sci 19. 2018;1:1.
- Qi A, Han J, Jia F, Liu C. miR-3175 and miR-134 affect proliferation, invasion and apoptosis of glioma cells through PI3K/AKT signaling pathway. J BUON. 2019;24(6):2465–2474.
- Zhong X, Tang J, Li H, et al. MiR-3175 promotes epithelial-mesenchymal transition by targeting Smad7 in human conjunctiva and pterygium. FEBS Lett. 2020;594(7):1207–1217. doi:10.1002/1873-3468.13698
- Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5(1):28. doi:10.1038/s41392-020-0134-x
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi:10.1016/j.cell.2011.09.024
- Venetis K, Piciotti R, Sajjadi E, et al. Breast cancer with bone metastasis: molecular insights and clinical management. Cells. 2021;10(6):1377. doi:10.3390/cells10061377
- Nam KW, Bae YC, Nam SB, Kim JH, Kim HS, Choi YJ. Characteristics and treatment of cutaneous melanoma of the foot. Arch Plast Surg. 2016;43(01):59–65. doi:10.5999/aps.2016.43.1.59
- Adams B, Peng P, Williams MJTJOF, Foot ASOPOTACO, Surgeons A. Melanoma of the foot is associated with advanced disease and poorer survival. The Journal of Foot and Ankle Surgery: Official Publication of the American College of Foot and Ankle Surgeons. 2018;57(1):52–55. doi:10.1053/j.jfas.2017.07.018
- Li K, Chen Y, Li A, Tan C, Liu X. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2019;144(7):1486–1495. doi:10.1002/ijc.31774
- Luo Y, Zhu Q, Xiang S, et al. Downregulated circPOKE promotes breast cancer metastasis through activation of the USP10-Snail axis. Oncogene. 2023;42(44):3236–3251. doi:10.1038/s41388-023-02823-2
- Nag S, Bhattacharya B, Dutta S, et al. Clinical theranostics trademark of exosome in glioblastoma metastasis. ACS Biomater Sci Eng. 2023;9(9):5205–5221. doi:10.1021/acsbiomaterials.3c00212
- Yoon J, Byun H, Kim S, Jung D, Lee SJN-CRR. Exosomal LINC00853 promotes progression of gastric cancer via the MAP17/PDZK1/AKT signaling pathway. Non-Coding RNA Research. 2024;9(3):876–886. doi:10.1016/j.ncrna.2024.03.011
- Gautam SK, Batra SK, Jain M. Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma. Mol Cancer. 2023;22(1):118. doi:10.1186/s12943-023-01813-y
- Guo Y, Ji X, Liu J, et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. 2019;18(1):39. doi:10.1186/s12943-019-0995-1
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Research. 2015;25(8):981–984. doi:10.1038/cr.2015.82
- Meng H, Niu R, Huang C, Li JJC. Circular RNA as a novel biomarker and therapeutic target for HCC. Cells. 2022;1:11.
- Wang D, Wang S, Jin M, et al. Hypoxic exosomal circPLEKHM1-mediated crosstalk between tumor cells and macrophages drives lung cancer metastasis. Adv Sci (Weinh). 2024;11(22):e2309857. doi:10.1002/advs.202309857
- Zheng R, Zhang K, Tan S, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Molecular Cancer. 2022;21(1):49. doi:10.1186/s12943-021-01471-y
- Lin J, Wang X, Zhai S, et al. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J Hematol Oncol. 2022;15(1):128. doi:10.1186/s13045-022-01348-7
- Tuo B, Chen Z, Dang Q, et al. Roles of exosomal circRNAs in tumour immunity and cancer progression. Cell Death Dis. 2022;13(6):539. doi:10.1038/s41419-022-04949-9
- Li C, Jiang P, Wei S, Xu X, Wang J. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol Cancer. 2020b;19(1):116. doi:10.1186/s12943-020-01234-1
- Li MO, Rudensky AY. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol. 2016;16(4):220–233. doi:10.1038/nri.2016.26
- Brown CC, Gottschalk RA. Volume control: turning the dial on regulatory T cells. Cell. 2021;184(15):3847–3849. doi:10.1016/j.cell.2021.06.015
- Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022;113(6):1968–1983. doi:10.1111/cas.15365
- Hu Z, Chen G, Zhao Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22(1):55. doi:10.1186/s12943-023-01759-1
- Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5(1):15179. doi:10.1038/srep15179
- Langhans B, Nischalke HD, Kramer B, et al. Role of regulatory T cells and checkpoint inhibition in hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68(12):2055–2066. doi:10.1007/s00262-019-02427-4
- Wang C, Huang X, Wu Y, Wang J, Li F, Guo GJIJOBS. Tumor cell-associated exosomes robustly elicit anti-tumor immune responses through modulating dendritic cell vaccines in lung tumor. International Journal of Biological Sciences. 2020a;16(4):633–643. doi:10.7150/ijbs.38414
- Dou R, Liu K, Yang C, et al. EMT-cancer cells-derived exosomal miR-27b-3p promotes circulating tumour cells-mediated metastasis by modulating vascular permeability in colorectal cancer. Clinical and Translational Medicine. 2021;11(12):e595. doi:10.1002/ctm2.595
- Paolillo M, Schinelli S. Extracellular matrix alterations in metastatic processes. Int J Mol Sci. 2019;20(19):4947. doi:10.3390/ijms20194947
- Whiteside TL. The role of tumor-derived exosomes in epithelial mesenchymal transition (EMT). Transl Cancer Res. 2017;6(S1):S90–S92. doi:10.21037/tcr.2017.02.13
- Wang H, Wei M, Kang Y, Xing J, Zhao Y. Circular RNA circ_PVT1 induces epithelial-mesenchymal transition to promote metastasis of cervical cancer. Aging. 2020b;12(20):20139–20151. doi:10.18632/aging.103679
- Lin Q, Qi Q, Hou S, et al. Exosomal circular RNA hsa_circ_007293 promotes proliferation, migration, invasion, and epithelial-mesenchymal transition of papillary thyroid carcinoma cells through regulation of the microRNA-653-5p/paired box 6 axis. Bioengineered. 2021;12(2):10136–10149. doi:10.1080/21655979.2021.2000745
- Townson JL, Naumov GN, Chambers AF. The role of apoptosis in tumor progression and metastasis. Curr Mol Med. 2003;3(7):631–642. doi:10.2174/1566524033479483
- Jiang C, Zhang N, Hu X, Wang HJMC. Tumor-associated exosomes promote lung cancer metastasis through multiple mechanisms. Mol Cancer. 2021;20(1):117. doi:10.1186/s12943-021-01411-w
- Liu K, Gao X, Kang B, Liu Y, Wang D, Wang Y. The role of tumor stem cell exosomes in cancer invasion and metastasis. Front Oncol. 2022;12:836548. doi:10.3389/fonc.2022.836548