Abstract
The identification of all epigenetic modifications implicated in gene expression is the next step for a better understanding of human biology in both normal and pathological states. This field is referred to as epigenomics, and it is defined as epigenetic changes (ie, DNA methylation, histone modifications and regulation by noncoding RNAs such as microRNAs) on a genomic scale rather than a single gene. Epigenetics modulate the structure of the chromatin, thereby affecting the transcription of genes in the genome. Different studies have already identified changes in epigenetic modifications in a few genes in specific pathways in cancers. Based on these epigenetic changes, drugs against different types of tumors were developed, which mainly target epimutations in the genome. Examples include DNA methylation inhibitors, histone modification inhibitors, and small molecules that target chromatin-remodeling proteins. However, these drugs are not specific, and side effects are a major problem; therefore, new DNA sequencing technologies combined with epigenomic tools have the potential to identify novel biomarkers and better molecular targets to treat cancers. The purpose of this review is to discuss current and emerging epigenomic tools and to address how these new technologies may impact the future of cancer management.
Introduction
Sequencing the human genome marked not the end of the field of genomics, but its beginning. Scientists now stand in a unique position in the history of medicine to define human disease, armed with the technological advancements and the data that has resulted from the Human Genome Project.Citation1,Citation2 Using this information, some progress has already been made toward launching individualized and personalized medicine, especially for certain types of cancer. For example, the launch of different types of genetic tests to predict disease (preventive medicine) and the development of the drug imatinib (Gleevec®) for blood tumorsCitation3 were major breakthroughs. In addition, biomarkers that are able to subgroup tumors based on aggressivity, thus aiding in clinical decisions, were also identified.Citation4 A precise molecular characterization of human cancers will allow a better understanding of the basis for disease susceptibility and environmental influence, better diagnosis and prognosis, and the refinement of individualized treatment for optimal therapeutic efficacy.
Genomes from various individuals have already been sequenced,Citation5,Citation6 allowing genomic comparisons. Projects such as the HapMapCitation7 that identified variations in the human genome, and ENCODECitation8 that is analyzing the functional elements in the genome, are helping in the understanding of complex disease phenotypes. Such projects have the potential to help elucidate the information encoded by human genomes and aid in the treatment of diseases such as cancer.
One of the main issues in genomic science is understanding how gene expression is regulated. To understand the mechanisms that are implicated in gene regulation, the genes that are expressed in each cell type of the body and how changes in their expression will impact in the development of diseases represent major challenges. In addition, environmental factors such as the exposure to chemical compounds during life, smoking, and nutrition can clearly affect and change the expression of genes.Citation9 Thus, in the post-genomic era, studies of how human genes are regulated and the mechanisms that are implicated in this process are of major importance for our understanding of normal processes and diseased states.
The information beyond the genome sequence was recently coined as the epigenome.Citation10 The epigenome is defined as the group of modifications that can occur at a genomic level that will not change the sequence of the bases of the DNA but can change the DNA conformation and, as a consequence, change the expression of genes. Epigenetics is the study of these modifications in the DNA.Citation11 The following are the main epigenetic modifications that occur in the DNA molecule: 1) binding of different proteins to the DNA such as histones and methyl-binding proteins, 2) addition of chemical groups in the bases of the DNA such as methyl (CH3), and 3) microRNAs and other noncoding RNAs that can regulate the expression of genes through various mechanisms.
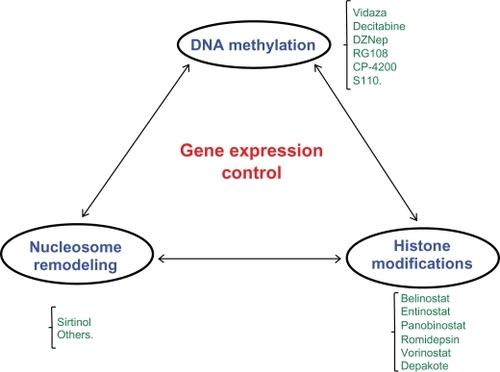
While epigenetics has garnered more attention, it is not a new field, and studies dating from the 1980s have shown the promise of using drugs that affect these mechanisms to treat diseases, especially cancers.Citation12,Citation13 In the last decade, we have been facing an overwhelming increase in drugs affecting epigenetic mechanisms that have been developed to treat different types of cancer (see and ). Examples are a growing number of DNA methylation inhibitors, histone modification inhibitors, and small molecules that target chromatin-remodeling proteins. 5-Azacytidine was the first inhibitor of an enzyme implicated in epigenetic modifications described.Citation14 This drug inhibits the DNA methyltransferase (DNMT) enzyme that is responsible for adding methyl groups to cytosines located in both DNA and RNA molecules. Another example of a DNMT inhibitor is 5-aza-2′-deoxycytidine, which is incorporated just in the DNA molecule. The DNA methyltransferase covalently binds to these nucleotide analogs, and this sequestration affects its normal function. These compounds can also affect the way proteins implicated in cell regulation are able to bind to the DNA/RNA substrates. 5-Azacytidine was first tested in myelodysplastic syndrome and leukemia, and it showed promising results in patients with both diseases.Citation15,Citation16 Since 5-azacytydine and other epigenetic drugs are not very specific, side effects are a major problem. A challenge faced by researchers in this field is two-fold: design more specific drugs and drugs that have fewer side effects since they have a global effect in the epigenome of the cells. To overcome this issue, new DNA sequencing technologies (second and third generation) combined with epigenomic tools have emerged. It is becoming clear that these technologies may facilitate the identification of better molecular targets for drug development and biomarker identification for cancer management. In that regard, the main purpose of this review is to discuss the emergence of epigenomic tools derived from new DNA sequencing technologies and how they may affect the management of cancer in the future.
Table 1 Drugs developed using epigenetics and epigenomics tools
Figure 1 Epigenomics and cancer therapy. Schematic representation of the three main epigenomic/epigenetic components that are implicated in gene expression control in human cells (blue). Some drugs that are in current use, under development, and in clinical trials for each epigenetic mechanism of gene regulation are shown in green. The drugs presented here are just a representative list for each mechanism, and some drugs under development and in clinical trials at the moment are not shown.
Cancer genomics – now and then
Genomics is defined as the study of entire genomes of organisms, including extrachromosomal DNA such as the mitochondrial genetic material. This field includes intensive efforts to determine the entire DNA sequence of organisms, using fine-scale genetic mapping and DNA sequencing with current and emerging technologies. In contrast, investigating the roles of single genes is a primary focus of genetics. Single gene research does not fall into the definition of genomics unless the aim is to verify the effect that a gene may have on the entire genome’s networks and pathways. Genomics has been the main focus in molecular biology, especially after the completion of the sequencing of genomes from several organisms. Genomics tools have already helped in the understanding of several aspects of the genome of cancer cells when compared to normal controls. One important example is the identification of the gene HER2/neu (ErbB-2), which is an oncogene mapped to human chromosome 17 that is over-expressed, or amplified, in ∼30% of breast cancer tumors.Citation17 Identification of this molecular characteristic culminated in the development of the drug trastuzumab (Herceptin®).Citation18 Breast cancer patients that are HER2/neu (ErbB-2) positive have increased survival rates when treated with this drug.Citation18
One question that has recently emerged after the sequencing of entire human genomes and comparisons between them is ‘What have we learned from sequencing human genomes?’ It is clear that important advances were generated after the sequencing of the first human genome a decade ago. These advancements were mainly applied to basic science such as improvements in DNA sequencing technologies, a decrease in the cost for sequencing the DNA molecule, and also the development of new methods to analyze molecular changes in diseases; for example, there was an increase in the number of predictive and prognostic genetic tests for cancers and other diseases. In contrast, for applied medicine and patients, there have been a disappointingly small number of drugs and therapies that were discovered and developed using genomics tools. Hence, we have learned that the sequence of human genomes is just a starting point to generate and accumulate the basic information needed for more complex and deeper analyses.
Recently, cancer genomes were sequenced and compared with normal cells for leukemia, breast, lung, and other tumor types, using second-generation DNA sequencing technologies.Citation19–Citation25 The purpose was to identify mutations that could give rise to new biomarkers and new therapies for these types of cancers. In addition, the 1000 Genome Project was recently launchedCitation26 with the objective of sequencing the genome of thousands of individuals in a small period of time. In parallel, companies are starting to provide whole genome sequencing services, with the aim of understanding the individual’s susceptibility for diseases, including different cancers.Citation27 As a result, a large number of individuals will have their genomes sequenced in the years to come. But is this enough to understand how the human cell machinery works and to determine how cancer or any other disease arises? It is likely that the technology will be beneficial to genomic science in the future by paving new ways of approaching diseases, especially cancers. Even though the genomic information will be of importance to identify mutations and other chromosomal abnormalities (ie, insertions or deletions in the genomic DNA of diseased cells), more studies will be necessary to completely understand human genomes. In this regard, the big challenge will be to identify and catalog all the genes that are present in normal cells and their defects (mutations, deletions, insertions, amplifications, fusion proteins, etc) in tumors. There is growing evidence that epigenomics will contribute to this understanding, and in the following sections, I will discuss drugs based on epigenetic mechanisms that are currently in use, in clinical trials, and under development. Furthermore, I will also discuss how this field could aid in the development of better therapies for cancer and in the identification of new biomarkers.
Epigenomics and new therapies
Epigenomics is becoming more important as the technologies for genome-wide epigenetic modification analyses improve. The best-known epigenetic marker is DNA methylation, and some genes are already described with this epigenetic change in different tumor types.Citation28 DNA methylation occurs in normal cells mainly in regions that are intergenic, and loss of methylation (hypomethylation) in these areas was first reported in the 1980s.Citation29 Loss of DNA methylation or global hypomethylation is an early event in cancer, and when it occurs in repetitive regions of the genome, the consequences can be chromosomal instability, as previously described.Citation30 On the other hand, gain of DNA methylation (hypermethylation) in the promoter region of different genes can lead to a decrease in the expression of this gene (also known as downregulation) and can be one of the causes for cancer development.Citation31 It is hypothesized that DNA methylation may affect gene expression by blocking transcription after methyl-binding and other proteins form a complex that blocks the access for transcription factors to the gene promoter.Citation32 However, there are other models suggesting that DNA methylation is the consequence instead of the cause of gene inactivation.Citation33 In this case, defects in the transcription factors such as mutations will leave promoter regions ‘opened’ and more susceptible to the action of the DNMTs.Citation33 Consequently, these regions of the DNA will be methylated.Citation33
Different genes are already described as hypermethylated in the initiation and progression of several types of tumors.Citation34 The enzymes responsible for the control of DNA methylation in eukaryotic cells are the DNMTs.Citation35 There are three enzymes already described: DNMTs 1, 2, and 3. However, DNMT1 may be the most important, especially in diseases such as cancer.Citation36 Different types of drugs that target DNMTs have already been developed. The idea behind using a drug that blocks the enzymes that control DNA methylation is that in cancer, there is an increase in their activity.Citation37 Drugs that block DNMTs include 5-azacytidine, 5-aza-2′-deoxycytidine, small molecule inhibitors, and others (see for more details). 5-Azacytidine (Vidaza®) was approved by the Food and Drug Administration (FDA) since it increased the survival of patients with myelodysplastic syndrome, and many patients on Vidaza became transfusion independent, showing the potential for this type of therapy.Citation38
Histone marks were recently described as an important epigenetic modification to control gene expression in normal cells.Citation39 Histone modifications, such as methylation, acetylation, ADP-ribosylation, ubiquitination, phosphorylation, and others, to histone tails alter chromatin structure. However, a complete understanding of the precise molecular mechanisms by which these alterations to histone tails influence DNA-histone interactions remains elusive. There are two main hypothesis on how histone modifications can affect chromosome function: 1) they may alter the electrostatic charge of the histone, resulting in a structural change in histones or their binding to DNA; or 2) these modifications are binding sites for protein recognition motifs, such as the bromodomains or chromodomains, that recognize acetylated lysines or methylated lysines, respectively.Citation40 The existence of these modifications and recognition motifs led to the ‘histone code’ hypothesis proposed by Strahl and Allis.Citation41 Overall, post-translational modifications of histones create an epigenetic mechanism for the regulation of a variety of normal and disease-related processes, including cancers. Drugs affecting histone modifications have been already developed and showed promising results in the treatment for different tumor types (see for more details).
Based on this knowledge, different types of histone deacetylase inhibitors, DNMT inhibitors, and small molecules that block enzymes that are implicated in these epigenetic mechanisms have been developed. In addition, combining conventional therapies to drugs that affect epigenetic mechanisms is becoming common. For example, clinical trials using a combination of DNMT inhibitors with conventional chemotherapy were well tolerated in cancer patients and showed encouraging results when compared with chemotherapy alone.Citation42
Drugs targeting chromatin and nucleosome remodeling proteins are also a promising therapeutic strategy to treat human cancers (). Some of these proteins are deregulated in cancer, such as sirtuin 1 (SIRT1). It was recently shown that targeting this protein with drugs such as Salermide or Sirtinol can lead to the reactivation of pro-apoptotic genes that are epigenetically repressed exclusively in cancer cells.Citation43 These drugs are also promising as an anticancer agent, providing molecular evidences that SIRT1 might be involved in human tumorigenesis.Citation43 Chromatin-remodeling proteins are important for proper gene expression, and new drugs targeting these proteins will be developed generating more effective therapies against cancer. All the epigenetic/epigenomic mechanisms and some of the drugs that have been tested for cancer therapies are represented in .
The impact of new technologies on cancer research
Second-generation DNA sequencing technologies have been used to identify and detect genetic and genomic changes in tumors when compared with normal cells. In the epigenomics field, these technologies have been helpful in identifying regions of the DNA that are differentially methylated and have different histone marks.Citation44 The identification of proteins that are responsible for wrapping the nucleosome of tumor cells was also possibleCitation44 (see ). The development of new technologies to study cancer epigenomes will be crucial for the identification of defects in tumor cells.
Table 2 Different types of technologies to uncover epigenomic changes in cancer
Thirty years ago, sodium bisulfite was first described as a reagent that could be used to detect DNA methylation in specific regions of the DNA.Citation45 This discovery has revolutionized the way we have been analyzing DNA methylation changes in cancer cells from different tumor types.Citation45 This technology allowed the analyses of specific regions of the DNA, the so-called gene-by-gene analyses (), to evaluate the percentages of DNA methylation and correlate it to gene expression. In addition to sodium bisulfite treatment, digestion with methylation-sensitive restriction enzymes and several different methods using restriction enzymes combined with polymerase chain reactionCitation46 have been used for years. Some limitations of these methods include the low number of dinucleotide CGs or CpGs that can be analyzed at a time (). Analyses of individual genes and/or regions of the DNA may also be applied to evaluate histone marks and nucleosome packaging, with the use of antibodies against specific marks in the histone proteins. These procedures, however, are very laborious and restricted to the region(s) of interest. Recent methods were developed for epigenomic analyses, which can evaluate epigenetic changes on a global level in the genome of tumor cells (). Examples include chromatin immunoprecipitation combined with DNA sequencing (ChiP–Seq) and high-throughput DNA methylation analyses after sodium bisulfite treatment using new DNA sequencing technologies (). Second-generation DNA sequencing methods are mainly based on pyrosequencing and emulsion polymerase chain reaction combined with beads that are embedded in slides with small pores (for more information on second-generation DNA sequencing, see ).
A wave of new sequencing technologies, named third-generation DNA sequencing, have been developed with the promise of sequencing genomes, transcriptomes, and epigenomes faster and with lower costs. Some of these technologies are based on the so-called nanopores (). These pores are small holes that could be biological or solid, in which the DNA can pass and be detected in a controlled manner.Citation47 These technologies rely on the detection of single molecules, and labeling of the sequencing substrate is sometimes required. It is possible that these new sequencers developed using nanotechnology could read long stretches of DNA in a greater or comparable way to the technologies that are currently available. A recent report has demonstrated that it is already possible to detect DNA methylation changes without the use of the reagent sodium bisulfite (which degrades the DNA and usually requires high amounts of starting material).Citation48 This is possible in a single molecule real time sequencing reaction with nanodetectors.Citation48 It is becoming clear that the new technologies under development will be of importance for research in epigenomics. This will have a positive impact on cancer research, facilitating the identification of new biomarkers and drug targets.
Implications for cancer management
The stratification of patients based on their tumor profile and/or specific biomarkers is becoming the best way to subgroup individuals with the same tumor characteristics. This field is also known as personalized or individualized medicine, and its objective is to associate the best treatment for each specific patient or group of patients. Personalized medicine involves the systematic use of molecular information about each individual patient to select or optimize preventative and therapeutic care. These new approaches are changing the way in which pharmaceutical companies try to identify and test new cancer drugs. The idea of a blockbuster drug that could treat a broad spectrum of tumor types has become unlikely; cancer is a complex disease, and even a specific cancer type, such as breast cancer, has a variety of subclasses with completely different pathological and molecular features.
Epigenomics is a unique approach to cancer research since it can help in the identification of groups of patients with the same epigenetic changes and characteristics in their tumors. The epigenetic drugs used today are unspecific and have side effects such as the ones that occur in conventional chemotherapy. This can vary from patient to patient, depending on the dosage that is prescribed. Importantly, lower dose treatments can reduce the side effects. The identification of DNA methylation and histone modifications associated with cancer may have important clinical utility in the future. The development of new technologies to uncover these changes in a high-throughput fashion will have a major impact as discussed above and shown in and .
Table 3 Some examples of epigenetic and epigenomic changes in single genes or group of genes and their potential impact in cancer management
Some advancement in the field of epigenetics and epigenomics has already led to the identification of specific biomarkers to manage the disease (see for more details). A variety of genes have been described as hypermethylated and/or hypomethylated in cancers, and this feature has shown some clinical significance in specific tumor types.Citation49,Citation50 Genes such as GSTP1, which is hypermethylated in a high percentage of prostate cancers, has been used as a biomarker for this disease in body fluids and biopsy specimens.Citation51 In addition, groups of genes from the same pathway and/or network have shown the same epigenetic changes in tumors. Examples include genes implicated in cell adhesion, DNA repair, and apoptosis that can be downregulated by DNA methylation. Downregulation of genes associated with cell adhesion and migration can increase the risk of the tumor cells to metastasize to a secondary site in the body.Citation52,Citation53 For example, our group has already shown that the adhesion molecule ADAM23 is highly methylated in breast tumors, and this feature is correlated to metastases and a poor prognosis in breast tumors.Citation54,Citation55 A classic example of a DNA repair gene downregulated by DNA methylation is the MGMT gene, which is silenced by epigenetic mechanisms in brain tumors.Citation56 Tumors that do not express the gene MGMT are more sensitive to radiotherapy and chemotherapy with temozolomide, and this molecular feature has been used in clinical decisions and disease management for glioblastomas.Citation56,Citation57 Changes in the profile of histone modifications have also been used to evaluate and manage the risk of prostate cancer recurrence in patients.Citation58
Another group of genes in which epigenetic changes can be monitored to manage cancer risk and progression is the microRNA (miR) gene family.Citation59 Some studies have already shown that miRs can be regulated by epigenetic mechanisms (). Changes in DNA methylation have been reported in specific cancer types for different miRs.Citation60 Additionally, miRs are associated with important embryonic gene pathways in cancer, and this connection between embryonic development and cancer should be carefully examined for drug development in the future.Citation61 Since miRs regulate hundreds to thousands of protein-coding genes by incomplete base-pairing,Citation62–Citation64 allowing them to affect networks and pathways of genes, it will be of importance to monitor miR expression changes mediated by epigenetics during cancer initiation and progression.
In the case of drug development and new therapies, it is likely that the future of epigenetic therapy will include the use of multiple drugs that individually have little effect in epigenetic silencing but that might be expected to have synergistic and/or additional effects when combined. For example, a recent study using histone deacetylase inhibitors and high-dose chemotherapy both in vitro and in vivo indicated that this combination might overcome chemoresistance, achieve durable remission, and improve survival of patients with Burkitt lymphoma.Citation65 A major problem with the use of current epigenetic drugs is that they are nonspecific and can reactivate genes randomly. A concern is that they can cause a whole-genome hypomethylation, increase the number of chromosomal abnormalities, and affect the tumorigenic phenotype of cancer cells as previously described.Citation66,Citation67 However, evidence that DNA methylation inhibitors act only in dividing cells, leaving nondividing cells unaffected, has already been reported.Citation68 In addition, it seems that these drugs activate genes that have become abnormally silenced in cancer.Citation69,Citation70
The question we face today in the epigenomics field applied to cancer research is ‘How to manage cancer with the new technologies and tools that are becoming available?’ The new technologies under development will facilitate the identification of better epigenetic markers and may aid in the development of more specific therapies. These drugs will be focused in a group of genes and not the entire epigenome (see for epigenetic drugs). The other question is ‘What will be the impact of epigenomics in the clinics?’ In other words, how could we translate the discoveries from basic science to the patient’s bedside? In this regard, the FDA has already approved a few epigenetic drugs for different tumor types, and some of these are very promising ( and ). Molecular biomarkers, such as genes or groups of genes with changes in epigenetic modifications in tumors, have also been used to guide clinical decisions. Examples are epigenetic changes in GSTP1 in prostate cancer, p16ink4A in different types of cancer, and MGMT in brain tumors (see ). There are companies (ie, Oncomethylome Sciences, Epigenomics AG, Sequenom, and others) already offering a test with a panel of epigenetic markers covering different genomic regions for cancers (see for more details). More recently, the company Exact Sciences released a combined test for four methylation markers for early detection of colon cancer with a 100% sensitivity. The advantages of using epigenetic markers for early detection of cancer and disease monitoring is that the test can be done in a small tumor sample or even in body fluids such as stool, blood, spinal fluid, and urine. Depending on the combination of markers obtained after the tests, clinicians are able to predict the appearance of the disease and also group cancer patients based in tumor aggressiveness and other clinical features, facilitating clinical decisions. Clearly, the impact of epigenomics in cancer management is expected to increase with the advent and development of new technologies.
Table 4 Examples of companies offering predictive epigenetic-based tests for cancer diagnosis and monitoring
Conclusions and future directions
In conclusion, the burgeoning fields of genomics and epigenomics comprise essential facets of modern cancer research. The FDA has already approved some epigenetic drugs, and others are in clinical trials and under development, demonstrating that this field already affects the way we manage cancer. In addition, single genes and groups of genes from the same pathway have been identified as differentially methylated in cancers, and some have been used as molecular biomarkers in order to identify patients with a better or a worse prognosis. Histone modification changes have also been used as markers to monitor cancer patients. Clearly, epigenetic changes in tumors will affect the decisions that are made in the clinics for the patients, especially treatment regimens and disease progression monitoring. Future directions include the discovery of new biomarkers and the development of more efficient drugs against different tumor types with the evolving technologies and the emergence of a new generation of DNA sequencers. Based on the information discussed here, growing evidence indicates that new epigenomic tools will increasingly affect the way we monitor and manage cancer in the future.
Financial disclosure
The author is the founder of the web-based company Genomic Enterprise and works as a consultant (www.genomicenterprise.com). Genomic Enterprise has the purpose of ensuring the integration of genetic, epigenetic, and genomic information to interested customers. Genomic Enterprise’s objectives include the development of business-to-business (B2B), business-to-consumer (B2C), and consumer-to-business (C2B) services in the scientific field.
Acknowledgements
The author acknowledges Dr Christopher A Hamm, Carl Radosevich, and Kelly Arndt for critically reading this manuscript. The author also thanks the Children’s Memorial Research Center, The Falk Brain Tumor Foundation, The Maeve McNicholas Memorial Foundation, and the Avon Foundation (Grant # 01-2009-037) for their financial support.
Disclosure
The author reports no conflicts of interest in this work.
References
- VenterJCAdamsMDMyersEWThe sequence of the human genomeScience200129155071304135111181995
- LanderESLintonLMBirrenBInitial sequencing and analyses of the human genomeNature2001409682286092111237011
- DrukerBJTalpazMRestaDJEfficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemiaN Engl J Med2001344141031103711287972
- LossosISCzerwinskiDKAlizadehAAPrediction of survival in diffuse large-B-cell lymphoma based on the expression of six genesN Engl J Med2004350181828183715115829
- LevySSuttonGNgPCThe diploid genome sequence of an individual humanPLoS Biol2007510e25417803354
- WangJWangWLiRThe diploid genome sequence of an Asian individualNature20084567218606518987735
- International HapMap ConsortiumA haplotype map of the human genomeNature200543770631299132016255080
- ENCODE Project ConsortiumIdentification and analysis of functional elements in 1% of the human genome by the ENCODE pilot projectNature2007447714679981617571346
- López-MauryLMargueratSBählerJTuning gene expression to changing environments: from rapid responses to evolutionary adaptationNat Rev Genet2008858359318591982
- CallinanPAFeinbergAPThe emerging science of epigenomicsHum Mol Genet200615Spec No 1R95R10116651376
- EstellerMEpigenetics in cancerN Engl J Med2008358111148115918337604
- WilsonVLJonesPAMomparlerRLInhibition of DNA methylation in L1210 leukemic cells by 5-aza-2′-deoxycytidine as a possible mechanism of chemotherapeutic actionCancer Res1983438349334966190553
- JonesPAAltering gene expression with 5-azacytidineCell19854034854862578884
- JonesPATaylorSMCellular differentiation, cytidine analogs and DNA methylationCell198020185936156004
- PintoAZagonelV5-Aza-2′-deoxycytidine (Decitabine) and 5-azacytidine in the treatment of acute myeloid leukemias and myelodysplastic syndromes: past, present and future trendsLeukemia19937Suppl 151607683358
- KaronMSiegerLLeimbrockS5-Azacytidine: a new active agent for the treatment of acute leukemiaBlood19734233593654125239
- SlamonDJGodolphinWJonesLAStudies of the HER-2/neu proto-oncogene in human breast and ovarian cancerScience198924449057077122470152
- PaikSKimCWolmarkNHER2 status and benefit from adjuvant trastuzumab in breast cancerN Engl J Med2008358131409141118367751
- LeyTJMardisERDingLDNA sequencing of a cytogenetically normal acute myeloid leukaemia genomeNature20084567218667218987736
- StephensPJMcBrideDJLinMLComplex landscapes of somatic rearrangement in human breast cancer genomesNature200946272761005101020033038
- LeeWJiangZLiuJThe mutation spectrum revealed by paired genome sequences from a lung cancer patientNature2010465729747347720505728
- PleasanceEDStephensPJO’MearaSA small-cell lung cancer genome with complex signatures of tobacco exposureNature2010463727818419020016488
- PleasanceEDCheethamRKStephensPJA comprehensive catalogue of somatic mutations from a human cancer genomeNature2010463727819119620016485
- BeroukhimRMermelCHPorterDThe landscape of somatic copy-number alteration across human cancersNature2010463728389990520164920
- DalglieshGLFurgeKGreenmanCSystematic sequencing of renal carcinoma reveals inactivation of histone modifying genesNature2010463727936036320054297
- ButlerDHuman genome at ten: science after the sequenceNature201046573011000100120577184
- KayeJThe regulation of direct-to-consumer genetic testsHum Mol Genet200817R2R180R18318852208
- EstellerMEpigenetic gene silencing in cancer: the DNA hypermethylomeHum Mol Genet200716Spec No 1R50R5917613547
- FeinbergAPVogelsteinBHypomethylation distinguishes genes of some human cancers from their normal counterpartsNature1983301589589926185846
- CostaFFPaixãoVACavalherFPSATR-1 hypomethylation is a common and early event in breast cancerCancer Genet Cytogenet2006165213514316527607
- JonesPABaylinSBThe fundamental role of epigenetic events in cancerNat Rev Genet20023641542812042769
- EstellerMCancer epigenomics: DNA methylomes and histone-modification mapsNat Rev Genet20078428629817339880
- TurkerMSGene silencing in mammalian cells and the spread of DNA methylationOncogene200221355388539312154401
- JonesPABaylinSBThe epigenomics of cancerCell2007128468369217320506
- BestorTHThe DNA methyltransferases of mammalsHum Mol Genet20009162395240211005794
- RobertsonKDDNA methylation, methyltransferases, and cancerOncogene200120243139315511420731
- EadsCADanenbergKDKawakamiKCpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpressionCancer Res199959102302230610344733
- FenauxPMuftiGJHellstrom-LindbergEEfficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III studyLancet Oncol200910322323219230772
- BernsteinBEMikkelsenTSXieXA bivalent chromatin structure marks key developmental genes in embryonic stem cellsCell2006125231532616630819
- KustatscherGLadurnerAGModular paths to ‘decoding’ and ‘wiping’ histone lysine methylationCurr Opin Chem Biol200711662863517988933
- StrahlBDAllisCDThe language of covalent histone modificationsNature20004036765414510638745
- SekeresMAListAFCuthbertsonDPhase I combination trial of lenalidomide and azacitidine in patients with higher-risk myelodysplastic syndromesJ Clin Oncol201028132253225820354132
- LaraEMaiACalvaneseVSalermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effectOncogene200928678179119060927
- BrownCGThe DNA sequencing renaissance and its implications for epigenomicsEpigenomics20091158
- WangRYGehrkeCWEhrlichMComparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residuesNucleic Acids Res1980820477747907443525
- KanedaATakaiDKaminishiMMethylation-sensitive representational difference analysis and its application to cancer researchAnn N Y Acad Sci200398313114112724218
- RheeMBurnsMANanopore sequencing technology: research trends and applicationsTrends Biotechnol2006241258058617055093
- FlusbergBAWebsterDRLeeJHDirect detection of DNA methylation during single-molecule, real-time sequencingNat Methods20107646146520453866
- EhrlichMCancer-linked DNA hypomethylation and its relationship to hypermethylationCurr Top Microbiol Immunol200631025127416909914
- EstellerMRelevance of DNA methylation in the management of cancerLancet Oncol20034635135812788407
- WoodsonKO’ReillyKJHansonJCThe usefulness of the detection of GSTP1 methylation in urine as a biomarker in the diagnosis of prostate cancerJ Urol2008179250851118076912
- YuJChengYYTaoQCheungKFMethylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancerGastroenterology2009136264065119084528
- KimDSKimMJLeeJYAberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic featuresCancer2007110122785279217960794
- CostaFFVerbisckNVSalimACEpigenetic silencing of the adhesion molecule ADAM23 is highly frequent in breast tumorsOncogene20042371481148814661055
- VerbisckNVCostaETCostaFFADAM23 negatively modulates αvβ3 integrin activation during metastasisCancer Res2009695546555219549921
- EstellerMGarcia-FoncillasJAndionEInactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agentsN Engl J Med2000343191350135411070098
- HegiMEDiserensACGorliaTMGMT gene silencing and benefit from temozolomide in glioblastomaN Engl J Med200535210997100315758010
- SeligsonDBHorvathSShiTGlobal histone modification patterns predict risk of prostate cancer recurrenceNature200543570461262126615988529
- WurdingerTCostaFFMolecular therapy in the microRNA eraPharmacogenomics J20077529730417189960
- LujambioACalinGAVillanuevaAA microRNA DNA methylation signature for human cancer metastasisProc Natl Acad Sci U S A200810536135561356118768788
- CostaFFSeftorEABischofJMEpigenetically reprogramming metastatic tumor cells with an embryonic microenvironmentEpigenomics20091238739820495621
- BartelDMicroRNAs: target recognition and regulatory functionsCell2009136221523319167326
- AmbrosVThe functions of animal microRNAsNature2004431700635035515372042
- PassettiFFerreiraCGCostaFFThe impact of microRNAs and alternative splicing in pharmacogenomicsPharmacogenomics J20099111319156160
- Richter-LarreaJARoblesEFFresquetVReversion of epigenetically-mediated BIM silencing overcomes chemoresistance in Burkitt lymphomaBlood2010 [Epub ahead of print].
- IssaJPDNA methylation as a therapeutic target in cancerClin Cancer Res20071361634163717363514
- HammCAXieHCostaFFGlobal demethylation of rat chondrosarcoma cells after treatment with 5-aza-2-deoxycytidine results in increased tumorigenicityPLoS One2009412e834020019818
- LiangGGonzalesFAJonesPAAnalysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidineCancer Res200262496196611861364
- de SmetCde BackerOFaraoniIThe activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylationProc Natl Acad Sci U S A19969314714971538692960
- SaitoYJonesPAEpigenetic activation of tumor suppressor microRNAs in human cancer cellsCell Cycle20065192220222217012846
- GimsingPHansenMKnudsenLMA phase I clinical trial of the histone deacetylase inhibitor belinostat in patients with advanced hematological neoplasiaEur J Haematol200881317017618510700
- TanJCangSMaYNovel histone deacetylase inhibitors in clinical trials as anti-cancer agentsJ Hematol Oncol2010435
- PlimackERKantarjianHMIssaJPDecitabine and its role in the treatment of hematopoietic malignanciesLeuk Lymphoma20074881472148117701577
- AppletonKMackayHJJudsonIPhase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumorsJ Clin Oncol200725294603460917925555
- FiskusWWangYSreekumarACombined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cellsBlood2009114132733274319638619
- KakihanaMOhiraTChanDInduction of E-cadherin in lung cancer and interaction with growth suppression by histone deacetylase inhibitionJ Thorac Oncol20094121455146520009910
- GuptaMAnsellSMNovakAJInhibition of histone deacetylase overcomes rapamycin-mediated resistance in diffuse large B-cell lymphoma by inhibiting Akt signaling through mTORC2Blood2009114142926293519641186
- ChenSYeJKijimaIEvansDThe HDAC inhibitor LBH589 (panobinostat) is an inhibitory modulator of aromatase gene expressionProc Natl Acad Sci U S A201010724110321103720534486
- KauhJFanSXiaMc-FLIP degradation mediates sensitization of pancreatic cancer cells to TRAIL-induced apoptosis by the histone deacetylase inhibitor LBH589PLoS One201054e1037620442774
- BruecknerBGarcia BoyRSiedleckiPEpigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferasesCancer Res200565146305631116024632
- BruecknerBRiusMMarkelovaMRDelivery of 5-azacytidine to human cancer cells by elaidic acid esterification increases therapeutic drug efficacyMol Cancer Ther2010951256126420442313
- ChuangJCWarnerSLVollmerDS110, a 5-aza-2′-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growthMol Cancer Ther2010951443145020442312
- GrantCRahmanFPiekarzRRomidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumorsExpert Rev Anticancer Ther2010107997100820645688
- Duenas-GonzalezACandelariaMPerez-PlascenciaCValproic acid as epigenetic cancer drug: preclinical, clinical and transcriptional effects on solid tumorsCancer Treat Rev200834320622218226465
- SiegelDHusseinMBelaniCVorinostat in solid and hematologic malignanciesJ Hematol Oncol200923119635146
- HrzenjakAKremserMLStrohmeierBSAHA induces caspase-independent, autophagic cell death of endometrial stromal sarcoma cells by influencing the mTOR pathwayJ Pathol2008216449550418850582
- AlcaínFJVillalbaJMSirtuin inhibitorsExpert Opin Ther Pat200919328329419441904
- HermanJGGraffJRMyöhänenSMethylation-specific PCR: a novel PCR assay for methylation status of CpG islandsProc Natl Acad Sci U S A19969318982198268790415
- FragaMFEstellerMDNA methylation: a profile of methods and applicationsBiotechniques200233363264912238773
- WuJSmithLTPlassCHuangTHChIP-chip comes of age for genome-wide functional analysisCancer Res200666146899690216849531
- ParkPJChIP-seq: advantages and challenges of a maturing technologyNat Rev Genet2009101066968019736561
- WangYHayakawaJLongF‘Promoter array’ studies identify cohorts of genes directly regulated by methylation, copy number change, or transcription factor binding in human cancer cellsAnn N Y Acad Sci2005105816218516394135
- van SteenselBHenikoffSEpigenomic profiling using microarraysBiotechniques200335234635035235435635712951776
- MardisERThe impact of next generation sequencing technology on geneticsTrends Genet2008313314118262675
- KircherMKelsoJHigh-throughput DNA sequencing – concepts and limitationsBioessays201032652453620486139
- BrantonDDeamerDWMarzialiAThe potential and challenges of nanopore sequencingNat Biotechnol200826101146115318846088
- EidJFehrAGrayJReal-time DNA sequencing from single polymerase moleculesScience2009323591013313819023044
- DrmanacRSparksABCallowMJHuman genome sequencing using unchained base reads on self-assembling DNA nanoarraysScience20103275961788119892942
- RyanDRahimiMLundJToward nanoscale genome sequencingTrends Biotechnol200725938538917658190
- WettergrenYOdinENilssonSp16INK4a gene promoter hypermethylation in mucosa as a prognostic factor for patients with colorectal cancerMol Med2008147–841242118418463
- LeungRCLiuSSChanKYPromoter methylation of death-associated protein kinase and its role in irradiation response in cervical cancerOncol Rep20081951339134518425396
- LiuMTaketaniTLiRLoss of p73 gene expression in lymphoid leukemia cell lines is associated with hypermethylationLeuk Res200125644144711337015
- GazinCWajapeyeeNGobeilSAn elaborate pathway required for Ras-mediated epigenetic silencingNature200744971651073107717960246
- SeniskiGGCamargoAAIerardiDFADAM33 gene silencing by promoter hypermethylation as a molecular marker in breast invasive lobular carcinomaBMC Cancer200998019267929
- HermanJGUmarAPolyakKIncidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinomaProc Natl Acad Sci U S A19989512687068759618505
- ZhangHLiYLaiMThe microRNA network and tumor metastasisOncogene201029793794819935707