Abstract
Purpose
The protease inhibitor S (PiS) and Z (PiZ) variants have been stated as the only genetic cause of chronic obstructive pulmonary disease (COPD) in Caucasians. However, its frequency in admixed populations is low. We aimed to identify genetic susceptibility between PiS (rs17580) and PiZ (rs28929474) polymorphisms with COPD related to tobacco smoking and biomass-burning smoke as well as to determine its frequencies in Mestizo and Amerindian populations from Mexico.
Patients and Methods
One thousand and eight hundred seventy-eight subjects were included in two comparisons of cases and controls, (1) smokers with and without COPD (COPD-S, n=399; SWOC, n=1106); (2) Biomass-burning smoke-exposed subjects with and without COPD (COPD-BS, n=98; BBES, n=275). In addition, 2354 Mexican subjects identified as Mestizos (n=1952) and Amerindian (n=402) were included. The population structure was evaluated using 59 informative ancestry markers.
Results
The AT genotype of rs17580 is associated with COPD in both comparisons (COPD-S vs SWOC p<0.001, OR=2.16; COPD-BS vs BBES p<0.0001, OR=11.50). The population of the Mexico-North has a greater Caucasian contribution (54.7%) compared to the center (46.9%) and southeast (42.7%).
Conclusion
The rs17580, AT genotype, is associated with COPD in Mexican-Mestizo smokers and exposed to biomass-burning smoke. The rs17580 AT is more frequent in the Mexican-Mestizo population of the North of the country, which has a high Caucasian component.
Introduction
Alpha-1 antitrypsin (A1AT) is a protein produced mainly by hepatocytes and secreted to the systemic circulation, acts as a neutrophil elastase inhibitor.Citation1 At the pulmonary level, it is one of the main enzymes responsible for maintaining the protease-antiprotease balance.Citation2 Variants in the protein are assigned according to their electrophoresis migration velocity in a magnetic field with varying pH gradients. Leading researchers in this area designated these proteins as M (medium) for those with medium velocity, F (fast) for those with fast migration, and S (slow) for those with slow migration. Once new variants were discovered, the anodic ones were assigned the first letters of the alphabet, while the cathodic ones were assigned the last letters. The normal genotype, present in >90% of healthy individuals (94–96% in Caucasians), is called PiMM and is characterized by serum levels of approximately 150–350 mg/100 Ml.Citation3 The A1AT protein is encoded by the SERPINA1 gene, located on the long arm of chromosome 14,Citation4 it harbors hundreds of genetic polymorphisms.Citation5 The most widely studied, since they are associated with liver and lung diseases, are PiS (Glu264Val, rs17580), and PiZ (Glu342Lys, rs28929474),Citation5 both are single nucleotide polymorphisms (SNPs), the first is found in exon 3 of the gene, where adenine (A) is replaced by thymine (T), at the protein level, instead of glutamic acid (Glu = E) Valine is found (Val = V); The PiZ polymorphism is located in exon 5, it is a change of guanine (G) for adenine (A), in the protein, glutamic acid (Glu = E) is replaced by lysine (Lys = K).Citation6 This latter polymorphism causes the accumulation of A1AT polymers in the endoplasmic reticulum of hepatocytes.Citation7,Citation8 In European populations, the deficiency of A1AT (PiZ allele) increases the risk of chronic obstructive pulmonary disease (COPD).Citation9
Table 1 Description of Demographic Variables of the COPD Patients and Expositional Comparison Groups
Table 2 Genotype and Allele Frequency Comparisons in COPD Patient Groups and Smokers/Biomass-Burning Smoke-Exposed Subjects
Table 3 Ancestral Contribution of the Mexican-Mestizo Regions Under the Study
Table 4 Genotype and Allele Frequencies (%) in Mestizo and Amerindian Populations of Mexico
Epidemiological studies have estimated the frequency of this polymorphism, indicating that A1AT deficiency alleles are not exclusive to European Caucasians, as it exists in some populations in North Africa (Egypt = 28.6%), Latin America (Venezuela = 7.8%), populations in Australia and New Zealand (12.2% and 26.0%, respectively), North America (Canada = 12.9%) and Asia (Tajikistan = 15.3%). On the other hand, the PiS allele is found in higher frequency and more distributed in Caucasian, European, African, Asian, and American populations compared to PiZ.Citation10 Regarding genetic association studies in lung diseases, compared with PiZ, the PiS allele is not strongly associated as a risk factor for COPD.Citation11 We aimed to establish whether there is a genetic association between the PiS and PiZ polymorphisms of the SERPINA1 gene in COPD related to tobacco smoking and related to exposure to biomass-burning smoke, as well as the frequencies of both variants in Mestizo and the indigenous populations of Mexico.
Patients and Methods
Participants
Case and Controls Groups
One thousand and eight hundred seventy-eight Mexican-Mestizo individuals with at least two previous generations of those born in Mexico, over 40 years old, with a diagnosis of COPD related to tobacco smoking (COPD-S, n = 399; tobacco index ≥5 packages/year, as we have previously shownCitation12,Citation13) or related to exposure to biomass-burning smoke (COPD-BS, n = 98; exposure years to biomass-burning smoke ≥10 years) were included. The diagnosis was based on clinical history, physical examination, spirometry data, as well as the criteria established by the American Thoracic Society (ATS).
In the smokers without COPD group (SWOC, n = 1106) individuals without evidence of lung disease and with normal spirometry values were included.
Participants included in the COPD-S, SWOC, and COPD-BS groups were recruited from the Department of Smoking Research and COPD of the Instituto Nacional de Enfermedades Respiratorias Ismael Cosio Villegas (INER) at Mexico City.
Finally, a group of biomass-burning smoke-exposed subjects (BBES, n = 275), clinically healthy, with no history of active or passive smoking, was included. The participants of this group are part of the national program for equality between women and men with the COPD Early Diagnosis campaign in women residents of rural populations, mainly in the northern highlands of the state of Oaxaca, and suburban areas of the Tlalpan mayoralty of Mexico City.Citation14
Functional maneuvers were taken using identical portable, battery-operated, ultrasound transit-time-based EasyOne™ spirometers (Medical Technologies, Chelmsford, MA, USA). The calibration was checked daily using a 3 L syringe at 588 mmHg of pressure (corresponding to the altitude of Mexico City). The reference values established by Pérez-Padilla for the Mexican population were used.Citation15 Post-bronchodilator spirometry was performed by administering 400 mg of nebulized salbutamol with the use of a spacer. Those individuals diagnosed with bronchial asthma, bronchiectasis, active tuberculosis, lung cancer, cystic fibrosis, allergic alveolitis, or idiopathic pulmonary fibrosis were excluded. Subjects with different ancestry to the Mexican were not included.
Populational Study
The individuals included in this analysis were 2354, clinically healthy, not biologically related to each other, or with the participants in the case–control comparisons, older than 40 years. Individuals of Mexican-Mestizo origin were characterized and classified according to their ancestry (with at least two previous generations born in the same state of the country) in three regions of the Mexican Republic: (1) North, includes the states of Baja California Norte, Baja California, Coahuila, Sonora, Nuevo León, Tamaulipas, Durango, San Luis Potosí, Sinaloa, Jalisco, Nayarit, Aguascalientes, and Zacatecas), (2) Central, states of Colima, Guanajuato, Michoacán, Mexico City, State of Mexico, Hidalgo, Morelos, Puebla, Querétaro, Tlaxcala) and (3) Southeast, states of Guerrero, Campeche, Chiapas, Oaxaca, Quintana Roo, Veracruz, Tabasco, and Yucatán. A depicting map of Mexican states and their location regarding grouping is included in Supplementary Figure 1.
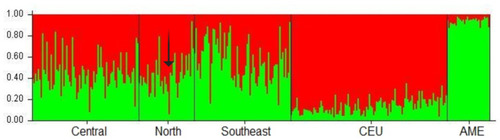
Figure 1 Population structures of the Mexican Mestizo groups and the reference populations (CEU and AME) The arrow indicates an individual with both risk genotypes (AT and GA for PiS and PiZ, respectively) whose Caucasian contribution reaches 0.935 and only 0.065 for Native American ancestry.
Individuals having Amerindian ancestral component were recruited through campaigns carried out by the INER in rural locations, with the prior authorization of the corresponding authorities. Historical and linguistic studies were employed to characterize participants and they had to have at least two generations with the same Amerindian ancestry, the groups included in the study were: the Otomi (from the Mezquital Valley, State of Hidalgo),Citation16 the Mazahua (northwest of the State of Mexico) and the Maya group (State of Yucatán).
Ethics Approval and Informed Consent
This study was approved by the Institutional Committees for Research, Ethics in Research, and Biosecurity of the Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas (INER). This study was conducted in accordance with the Declaration of Helsinki.
Each individual conducted a questionnaire on anthropometric data and hereditary pathology background; They voluntarily agreed to participate in the study and signed the written informed consent letter, which was previously approved by the INER Institutional Committees (approbation protocol codes: B20-08, B35-09, B09-10, B10-12 and C13-13).
Genomic DNA Extraction and Concentration Adjustment
A sample of 6 mL of peripheral blood in EDTA tubes was drawn from each participant. From the blood sample, leukocytes were separated, then genomic DNA was obtained using the commercial BD tract DNA isolation kit (Maxim Biotech, San Francisco CA, USA). The DNA was quantified by ultraviolet light absorption microspectrophotometry at 260 nm using a NanoDrop 2000 spectrophotometer device (Thermo Scientific, Wilmington, DE, USA), protein contamination was determined at 280 nm, and a contaminant-free sample was considered when the ratio 260/280 was between 1.7 and 2.0. Each sample was adjusted to 25 ng/µL for subsequent genotyping.
Genotyping of PiS and PiZ Alleles
Samples were genotyped by allelic discrimination using pre-designed Taqman probes, in a real-time PCR thermal cycler (7300 Real-Time PCR Systems, Applied Biosystems, CA, USA), the probes used were C____594695_20 for rs17580 (PiS), and C__34508510_10 for the rs28929474 (PiZ) (Applied Biosystems, CA, USA).
The PCR assay conditions were: Pre-reading 50°C, 1 minute; absolute quantification: 50°C, 2 minutes, 1 cycle; 95°C, 10 minutes, 1 cycle; 95°C, 15 seconds, 60°C 1 minute, 40 cycles; post-reading 50°C, 1 minute. Genotypes assignment was carried out, taking into account allelic discrimination and confirmed by absolute quantification; Also, three controls without template (contamination controls) were included for each genotyping plate, and 1% of the samples included in the study were genotyped in duplicate as an allele allocation control. Data interpretation was performed through Sequence Detection Software (SDS v 1.4, Applied Biosystems, CA, USA). The fluorophores employed for both SNPs were VIC for allele A and FAM for allele B.
All experiments were performed following the relevant guidelines and regulations. The STREGA (STrengthening the REporting of Genetic Association) guidelines were considered in the design of this genetic association study.
Population Structure Analysis
To evaluate the population structure of the participants included, 711 subjects (~30%) Mexican-Mestizos (central = 604, North 39, and Southeast = 68) were analyzed with 59 ancestry-informative markers (AIMs) that previously in our workgroup were selected and evaluated for different studies,Citation17 allele frequencies of these SNPs among Caucasian population vs Mexican-Mestizo, as well as Amerindian vs Mexican-Mestizos, presented a difference >40%. Caucasians (CEU, n = 120) of the International HapMap Project version 3, release R2 were taken as reference population,Citation18 and the Amerindian population of the Mexican-Mestizo genomic diversity project made up of people from the state of Oaxaca in Mexico (AME, n=30).Citation19 The population structure was generated using STRUCTURE v2.3.4Citation20 under unsupervised conditions.
Statistical Analysis
The statistical program SPSS v 15.0 (IBM, NY, USA) was used to describe the study population, showing the median and minimum and maximum values of each quantitative variable; for sex and regional distribution percentages were presented. Observance to Hardy–Weinberg equilibrium (HWE) was confirmed with Finetti v.3.0.8 software.Citation21 Genotype analysis was carried out with Pearson’s chi-squared and Fisher’s exact tests using Epi Info v. 7.1 software,Citation22 2x2 contingency tables were made to estimate the genetic association to COPD for PiS and PiZ. While the codominant model for the association analysis of genotypes in case comparisons using Epidat 3.1Citation23 was performed. The Bonferroni correction was employed to adjust the p-value using Plink v. 1.07.Citation24 To evaluate the effect of SNPs on pulmonary function variables, binary logistic regression was performed considering several comparison groups. First, the lung function values (%) were compared in a dominant model; then, they were stratified in cases and controls. On the other hand, the levels of the variables of lung function and age at diagnosis were stratified by genotypes through the Kruskal–Wallis test. The graphics were prepared using R studio.Citation25 The comparisons were made considering exposure to two different Ambiental risk factors. Then, we compare COPD-S vs SWOC and COPD-BS vs BBES.
The frequencies’ report of the PiS and PiZ variants in the Mexican-Mestizo and Amerindian populations was carried out after the evaluation of population ancestry, using Structure v2.3.4 under unsupervised conditions, with 10,000 Burn, 50,000 repetitions and the 2 reference populations previously described.Citation26 Subsequently, allele and genotype frequencies, expressed as a percentage, were calculated for PiS and PiZ in each of the 6 populations included in the study.
Results
The demographic characteristics of cases and control groups are described in ; there is a statistically significant difference when comparing the age of both groups as for sex, there are no significant differences. For cigarette consumption, the COPD-S group has more years of consuming cigarettes, as well as a higher number of cigarettes per day regarding the SWOC group; since the pulmonary function measures were used in the study groups classification, we found a statistically significant difference in their values, when comparing between both groups. Regarding biomass-burning exposed subjects, patients having COPD are older than exposed without the disease; in both groups, female sex is the most frequent, all subjects in the BBES group are women in comparison to ~92% in the COPD-BS group. Besides, we found differences in the biomass-burning smoke exposure variables.
Hardy–Weinberg Equilibrium (HWE)
We found an HWE deviation (p<0.01) in the rs28929474 in the SWOC group. For this reason, we only considered relevant the results observed in rs17580 in both case–control comparisons. Supplementary Table 1 includes HWE in case and control groups and Supplementary Table 2 shows HWE in population included groups, while Supplementary Figure 2 shows the De Finetti plot of the alleles in case and control groups of comparisons.
Genetic Association with COPD and Lung Function
For both polymorphisms, the homozygous genotype for the common allele is the most frequent (>87%); at the level of alleles, variant G of rs28929474 and A of rs17580 are the most frequent (>93%).
The analysis of the differences in genotype frequencies for both comparisons (smokers with and without COPD and subjects exposed to biomass-burning smoke with and without COPD) was performed using the dominant model.
For the rs28929474 variant, there is no statistically significant association in any of the comparisons; however, for rs17580 the frequency of AT + TT is higher in the COPD-S group (12.28%) compared to the SWOC group (6.24%), this difference was statistically significant (p <0.001, OR = 2.10, CI 95% = 1.43–3.09). Regarding the allele frequencies analysis, the minor allele (T) is found in a smaller proportion in smokers without COPD (3.16%), concerning the frequency found in the COPD-S group (6.14%), this difference is statistically significant (p <0.001, OR = 2.0, 95% CI = 1.37–2.91).
In the comparison of subjects exposed to biomass-burning smoke, for rs17580 the frequency of AT + TT is higher in the COPD-BS group (11.22%) in comparison to the BBES group (1.09%), this difference was statistically significant (p <0.0001, OR = 11.46, 95% CI = 3.12–42.03). Concerning the analysis of allele frequencies, the minor allele (T) is found in a smaller proportion in subjects exposed to biomass-burning smoke without COPD (0.55%), with respect to the frequency found in the COPD-BS group (5.61%), this difference is statistically significant (p <0.0001, OR = 10.84, 95% CI = 2.99–39.28). The allele and dominant model frequencies of rs28929474 (PiZ) and rs17580 (PiS) for the case and control groups are shown in . Supplementary Table 3 shows full-length frequencies of data for both comparisons.
There was no relationship between the presence of heterozygous genotypes with the lung function variables; no significant differences were found when comparing the percentages of lung function variables stratified by genotypes. In the same way, there is no significant difference between genotypes and the age at diagnosis.
Frequencies of the rs28929474 and rs17580 polymorphisms in Mestizo and Amerindian populations in Mexico
A 59 AIMs panel was used to evaluate the population structure analysis in ~ 30% (n = 711) of the Mexican mestizo individuals included in the study. The results of the Caucasian and Amerindian ancestral contributions are described in ; Interestingly, subjects in the North region have the highest Caucasian contribution among the three regions studied (~ 55%), while individuals from the Southeast carry the most considerable Amerindian contribution (> 57%).
On the other hand, our results show that the subjects of the Central region present 46.9% Caucasian and 53.1% Amerindian contribution (). The graph shows the central (n = 604), North (n = 39), and the southeast region (n = 68), all of them were selected randomly. On average, the Mexican-mestizo contribution presents CEU 0.480 and 0.519 AME contribution.
Allele and genotype frequencies of PiS and PiZ in Mexican Mestizo and Amerindian population are presented in . In the North region, the T allele of rs17580 (PiS) is found in higher frequency (5.7%) concerning the central and Southeast (2.2% and 1.6% respectively) regions, with statistically significant difference (North vs. central, p = 0.0003 and North vs. Southeast, p = 0.0016). The same behavior is observed in the Otomi, Maya, and Mazahua populations (2.4%, 2.6, and 0.5% respectively), we only found a statistically significant difference when comparing North vs. Mazahua (p = 2.0E-06). The homozygous genotype with the risk variant (TT) is almost non-existent in the 6 populations analyzed, except for the central area where it is located in 0.07% of the population.
The heterozygous genotype (AT) is below 5.5% in 5 of 6 populations, except in the North where it is found in 11.4%; this correlates with the ancestral population analysis, that revealed that this population has a more significant Caucasian contribution (54.7%) compared to the rest of the Mexican mestizos. Supplementary Table 4 shown statistically significant differences in genotype frequencies for the heterozygous genotype were found in the comparisons: North vs. central, North vs. Southeast and North vs. Mazahua (p = 0.0002, p = 0.0013 and p = 3.0E-06, respectively) as well as in the comparison central vs. Mazahua (p = 0.0125).
Regard to the rs28929474 (PiZ) variant, the risk allele (A) is found in 2.7% of the Northern population and 2.6% of the Maya population, while in the central and southeast of the country it is below 2%, whereas in the Otomi and Mazahua groups the A allele does not exist.
For this allele, a statistically significant difference was found in the comparison between North vs. central and vs. Southeast (p = 0.0238 and p = 0.0381, respectively) (Supplementary Table 4). The low frequency of the A allele is reflected in genotypes where the homozygous with the risk allele (AA) is absent in the six populations while the heterozygous (GA) is 5.4% in the North population and 5.3 in Maya, for the rest of the populations it is below 2.5%, while in the Otomi and the Mazahua it was not detected. Like with the PiS variant, the mestizo population of the North (with the highest Caucasian contribution) has the higher frequencies. However, statistical differences were found in comparisons of the populations of the North vs. central and Southeast (p = 0.0229 and p = 0.0367, respectively).
Subsequently, the population structure of eight Mexican mestizo individuals, with at least one risk allele for PiS or PiZ, randomly selected from the North and central regions were analyzed. An average of 0.590 of Caucasian contribution and 0.410 of Amerindian contribution was obtained that, when compared with the average for the Mexican mestizo of the three regions of the country, these subjects have a higher Caucasian contribution regarding the Mexican mestizo (CEU = 0.480 and AME = 0.519). Of these 8 individuals, one of them presented both risk genotypes, (AT in PiS and GA in PiZ), finding that this subject has 0.935 Caucasian and 0.065 Amerindian contributions (indicated with an arrow in ).
Taking these data into account, we can propose that the frequency of risk alleles for Mexican mestizos has a latitudinal distribution. But for the indigenous populations analyzed, this does not apply, especially for the Otomi and Mazahua groups, where there is no risk allele (A) for PiZ, obtaining 100% homozygous individuals for the common variant (GG).
Discussion
In the current study, we have assessed the genetic susceptibility associated with PiS and PiZ variants evaluated by molecular techniques that let us know the state of genotypes for each protein variant. Previously, among smokers, we found the rs17580 AT genotype associated with a decreased FEV1/FVC ratio compared to subjects with the homozygous AA genotype,Citation27 and recently, two SNPs (rs709932 and rs1303) in the SERPINA1 gene too, forming a haplotype shaped by both common alleles (CT haplotype) associated with reduced risk for COPD.Citation28
However, investigations have focused on the PiS and PiZ variants; individuals SS, SZ, and ZZ express serum concentrations of 85%, 25%, and 15% of the protein, respectively, compared to the normal levels (MM phenotype).Citation29 This characterization based on the protein (phenotype) assumes that both variants are in the homozygous state at gene-level.
Since each variant corresponds to different locations into the gene, it should be taken into consideration the possibility of heterozygous state for each locus, eg an individual which harbor two-copies (one of each parent) of the A allele of the rs17580, at protein level expresses the “normal” protein. In contrast, a person with two copies of the T allele produces an altered protein (deficient, PiS). A third possibility is to carry one allele T and other A (TA, heterozygous), in this case, one copy/gene/chromosome encodes for a normal protein while the other for a deficient; the same happens with alleles and genotypes for rs28929474. In consequence, genotypes more than phenotypes should be conducted in the COPD pathogenesis mechanisms elucidation, where PiS and PiZ variants are supposed to be involved.
Also, multiple studies have evaluated the participation of genetics factors in the COPD pathogenesis,Citation30 including PiS and PiZ variants, mostly applying this phenotype-based concept and leaving out the variability for each polymorphism and the potential contribution to COPD and/or its clinical features.
Some reports about COPD secondary to wood smoke (primary source employed to heat meals, which we are grouping as biomass); describe that biomass smoke exposure is associated with an airway-predominant COPD phenotype; while COPD tobacco-smoking related is associated with an emphysema-predominant phenotype.Citation31,Citation32 Also, lung morphology in necropsies from women with COPD exposed to biomass smoke shown more lung fibrosis and pigment deposition and thicker pulmonary arterial intima than smokers.Citation33
In the present study, we observe that the minor allele (T) of rs17580 (PiS) is associated with COPD in both comparisons, tobacco smoking, and exposure to biomass-burning smoke. This polymorphism was reported by our workgroup associated with lower values in lung function among smokers,Citation27 but not with the genetic susceptibility to the disease, probably due to small sample size; current analysis in addition to two-comparisons with two different risk exposure factors includes a better case–control ratio, almost 1:3 in each comparison. These results lead us to the question about the frequency of risk alleles among Mexican people, in which ancestral contribution has been marked by an admixture between Caucasian and indigenous (Amerindian) populations.Citation34
Some populations have national registries for subjects carrying PiZ (homozygous) who, without being smokers develop COPD, such is the case of the Swedish registry where 28% of its non-smoking population undergoes from the disease.Citation35 Those who carry the PiZ allele commonly have a more significant decrease in lung function, and if they are smokers, a high probability of mortality compared to those who are not PiZ carriers.Citation36,Citation37
In our population-level study, the PiS and PiZ variants are distributed latitudinally, being the northern region of the country (with the highest Caucasian contribution) where both variants are most frequents compared to the central and Southeast regions. On the other hand, the 2011 National (Mexican) Addiction Survey reports that northern Mexico has a prevalence of active tobacco smoking of around 26%, higher when compared to the south (nearly 18% prevalence).Citation38 Our findings are significant since, by the first time, we identified the rs17580 GA genotype associated with COPD in smokers and biomass-burning smoke-exposed subjects. Though T allele and TA genotype frequencies are low in the Mexican population, those from the North region (which high Caucasian component and have higher tobacco consumption) have an increased risk of developing COPD. Unfortunately, in the southern region even though Amerindian contribution predominates, this exposed group was women in charge of household chores, among which is cooking, an activity in which they use wood (primarily) to start and maintain the combustion of biomass to heat food, as we have previously reported.Citation39 For that reason, the biomass burning-smoke group had a predominance of females.
The current study is not exempt from limitations; maybe the most important are the differences found in the packs-years history and exposure index to biomass-burning smoke in controls’ groups; a matched comparison should resolve this potential confounding, but, the sample size would be reduced considerably.
The distribution of the risk alleles in Amerindian groups cannot be generalized since the complexity around these populations, such as lifestyle and ways of relating.
In general, are composed by individuals who mostly retain customs and ideals of the culture; sometimes it is difficult for a person not born within the community to join them; as a result, they can only have offspring among subjects of the same ethnic origin, possibly for this reason there is a decrease in genetic variability in these population groups. This is best noted with the PiZ allele (rs28929474 A allele), which in the Otomi and Mazahua population is absent. However, in the Maya group, it is very similar to the frequency found in Mexican Mestizos in the north of the country. For indigenous populations, it is recommended to continue studying other groups present in Mexico since data from the Mexican Institute of Statistics, Geography, and Informatics of 2010 reports that there are about 15.7 million indigenous people in Mexico (14.9% of the total national population).Citation40
Conclusion
The T allele of PiS (rs17580) is associated with the risk of developing COPD in Mexican Mestizos who are smokers and exposed to biomass-burning smoke; this association is maintained in the heterozygous state (AT). The AT genotype is a genetic risk marker most frequent in the Mexican Mestizo population of the North of the country, which has a high Caucasian component, compared with the population of the central and Southeast regions so that it would be considered a high-risk population of COPD, especially in the presence of environmental risk factors.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure
The authors declare that they have no conflicts of interest regarding this work.
Acknowledgments
The authors acknowledge the support received from physicians and technicians from the COPD clinic at INER for confirmation of diagnosis, acquisition of data on pulmonary function, and clinical care of the study participants. Partial data of this manuscript were presented in abstract form in the European Respiratory Society (ERS) 2017 international congress from September 9 to 13, 2017. Citation41
References
- BrantlyML, PaulLD, MillerBH, FalkRT, WuM, CrystalRG. Clinical features and history of the destructive lung disease associated with alpha-1-antitrypsin deficiency of adults with pulmonary symptoms. Am Rev Respir Dis. 1988;138(2):327–336. doi:10.1164/ajrccm/138.2.3273264124
- BerginDA, ReevesEP, MeleadyP, et al. Alpha-1 antitrypsin regulates human neutrophil chemotaxis induced by soluble immune complexes and IL-8. J Clin Invest. 2010;120(12):4236–4250. doi:10.1172/JCI4119621060150
- American Thoracic Society Documents. American thoracic society/European respiratory society statement: standards for the diagnosis and management of individuals with alpha-1 antitrypsin deficiency. Am J Respir Crit Care Med. 2003;168(7):818–900. doi:10.1164/rccm.168.7.818.14522813
- The National Center for Biotechnology Information. National library of medicine. Available from: http://www.ncbi.nlm.nih.gov/gene?cmd=Retrieve&dopt=Graphics&list_uids=5265. Accessed 11, 2015.
- DeMeoDL, SilvermanEK. Alpha1-antitrypsin deficiency. 2: genetic aspects of alpha(1)-antitrypsin deficiency: phenotypes and genetic modifiers of emphysema risk. Thorax. 2004;59(3):259–264. doi:10.1136/thx.2003.00650214985567
- FlicekP, AmodeMR, BarrellD, et al. Ensembl 2014. Nucleic Acids Res. 2014;42(Database issue):D749–55. doi:10.1093/nar/gkt119624316576
- GooptuB, EkeowaUI, LomasDA. Mechanisms of emphysema in alpha1-antitrypsin deficiency: molecular and cellular insights. Eur Respir J. 2009;34(2):475–488. doi:10.1183/09031936.0009650819648523
- VidalR, BlancoI, CasasF, JardiR, MiravitllesM. Guidelines for the diagnosis and management of alpha-1 antitrypsin deficiency. Arch Bronconeumol. 2006;42(12):645–659. doi:10.1157/1309597417178069
- de SerresFJ, LuisettiM, FerrarottiI, BlancoI, Fernandez-BustilloE. Alpha-1 antitrypsin deficiency in Italy: regional differences of the PIS and PIZ deficiency alleles. Monaldi Arch Chest Dis. 2005;63(3):133–141. doi:10.4081/monaldi.2005.63016312203
- de SerresFJ. Worldwide racial and ethnic distribution of alpha1-antitrypsin deficiency: summary of an analysis of published genetic epidemiologic surveys. Chest. 2002;122(5):1818–1829. doi:10.1378/chest.122.5.181812426287
- DahlM, HershCP, LyNP, BerkeyCS, SilvermanEK, NordestgaardBG. The protease inhibitor PI*S allele and COPD: a meta-analysis. Eur Respir J. 2005;26(1):67–76. doi:10.1183/09031936.05.0013570415994391
- Pérez-RubioG, Ramírez-VenegasA, Noé DíazV, et al. Polymorphisms in HTR2A and drd4 predispose to smoking and smoking quantity. ZhangH ed. PLoS One. 2017;12(1):e0170019. doi:10.1371/journal.pone.017001928103253
- Pérez-RubioG, Pérez-RodríguezME, Fernández-LópezJC, et al. SNPs in NRXN1 and CHRNA5 are associated to smoking and regulation of GABAergic and glutamatergic pathways. Pharmacogenomics. 2016;17:10. doi:10.2217/pgs-2016-0020
- Ramírez-VenegasA, Velázquez-UncalM, Pérez-HernándezR, et al. Prevalence of COPD and respiratory symptoms associated with biomass smoke exposure in a suburban area. Int J Chron Obstruct Pulmon Dis. 2018;13:1727–1734. doi:10.2147/COPD.S15640929872290
- Perez-PadillaR, ValdiviaG, MuinoA, et al. [Spirometric reference values in 5 large Latin American cities for subjects aged 40 years or over]. Arch Bronconeumol. 2006;42(7):317–325. Spanish. doi:10.1016/S1579-2129(06)60540-516945261
- Juárez-MartínAI, González-SobrinoBZ, CamarenaA, Falfán-ValenciaR. HLA class II alleles in the Otomi population of the Mezquital Valley: a genetic approach to the history of interethnic migrations in the Mexican central plateau. Hum Biol. 2014;86(3):167–184. doi:10.13110/humanbiology.86.3.016725836745
- Pérez-RubioG, Silva-ZolezziI, Fernandez-LopezJ, et al. Genetic variants in IL6R and ADAM19 are associated with COPD severity in a Mexican Mestizo population. COPD. 2016;13(5):610–615. doi:10.3109/15412555.2016.116101727078193
- The International HapMap Consortium. Browser release #28. Available from: http://hapmap.ncbi.nlm.nih.gov/index.html. Accessed 31, 2015.
- Silva-ZolezziI, Hidalgo-MirandaA, Estrada-GilJ, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci U S A. 2009;106(21):8611–8616. doi:10.1073/pnas.090304510619433783
- FalushD, StephensM, PritchardJK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7(4):574–578. doi:10.1111/j.1471-8286.2007.01758.x18784791
- Henschke, H. De Finetti Diagram. 2008. Available from: https://web.archive.org/web/20110719102655/https://finetti.meb.uni-bonn.de/download. Accessed March 27, 2018.
- DeanAG, ArnerTG, SunkiGG, et al. 2004 Epi Info, a database and statistics program for public health professionals. Available from: https://www.cdc.gov/epiinfo/index.html.
- Public Health Information Department of the General Public Health Administration of the Health Consulate. (Directorate of Galicia), health analysis and health information systems Unit of the Pan-American Health Organization (OPS-WHO). EPIDAT, program for epidemiological analysis of tabulated data. Version 3.1 (2006).
- PurcellS, NealeB, Todd-BrownK, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi:10.1086/51979517701901
- RStudio Team. RStudio: integrated development for R. rStudio; 2015 Available from: http://www.rstudio.com/.
- Pritchard Lab. Structure 2.3.4; 2012 Available from: http://pritchardlab.stanford.edu/structure_software/release_versions/v2.3.4/html/structure.html.
- Pérez-RubioG, Jiménez-ValverdeLO, Ramírez-VenegasA, et al. Prevalence of alpha-1 antitrypsin high-risk variants in Mexican mestizo population and their association with lung function values. Arch Bronconeumol. 2015;51:2. doi:10.1016/j.arbres.2014.09.01026707419
- Ponce-GallegosMA, Pérez-RubioG, García-CarmonaA, et al. Haplotype in SERPINA1 (AAT) is associated with reduced risk for COPD in a Mexican Mestizo population. Int J Mol Sci. 2019;21(1):195. doi:10.3390/ijms21010195
- FregoneseL, StolkJ, FrantsRR, VeldhuisenB. Alpha-1 antitrypsin Null mutations and severity of emphysema. Respir Med. 2008;102(6):876–884. doi:10.1016/j.rmed.2008.01.00918353624
- Perez-RubioG, Cordoba-LanusE, CupertinoP, Cartujano-BarreraF, CamposMA, Falfan-ValenciaR. Role of genetic susceptibility in nicotine addiction and chronic obstructive pulmonary disease. Rev Investig Clin. 2019;71(1):36–54. doi:10.24875/RIC.1800261730810540
- CampPG, Ramirez-VenegasA, SansoresRH, et al. COPD phenotypes in biomass smoke- versus tobacco smoke-exposed Mexican women. Eur Respir J. 2014;43(3):725–734. doi:10.1183/09031936.0020611224114962
- HanMK. The other COPD. Eur Respir J. 2014;43(3):659–661. doi:10.1183/09031936.0016991324585858
- RiveraRM, CosioMG, GhezzoH, SalazarM, Pérez-PadillaR. Comparison of lung morphology in COPD secondary to cigarette and biomass smoke. Int J Tuberc Lung Dis. 2008;12(8):972–977.
- Del Angel-PabloAD, Juárez-MartínAI, Pérez-RubioG, et al. HLA allele and haplotype frequencies in three urban Mexican populations: genetic diversity for the approach of genomic medicine. Diagnostics. 2020;10(1):47. doi:10.3390/diagnostics10010047
- PiitulainenE, TanashHA. the clinical profile of subjects included in the Swedish national register on individuals with severe alpha 1-Antitrypsin deficiency. COPD. 2015;12(Suppl 1):36–41. doi:10.3109/15412555.2015.102190925938290
- LaraB, MiravitllesM. Spanish registry of patients with alpha-1 Antitrypsin deficiency; comparison of the characteristics of PISZ and PIZZ individuals. COPD. 2015;12(Suppl 1):27–31. doi:10.3109/15412555.2015.102191225938288
- TanashHA, NilssonPM, NilssonJ-A, PiitulainenE. Survival in severe alpha-1-antitrypsin deficiency (PiZZ). Respir Res. 2010;11:44. doi:10.1186/1465-9921-11-4420420704
- Encuesta Nacional de Adicciones 2011. Reporte de Tabaco; 2011 Available from: http://www.conadic.salud.gob.mx/pdfs/ENA_2011_TABACO.pdf.
- Ambrocio-OrtizE, Pérez-RubioG, Ramírez-VenegasA, et al. Effect of SNPs in HSP family genes, variation in the mRNA and intracellular Hsp levels in COPD secondary to tobacco smoking and biomass-burning smoke. Front Genet. 2020;10(1):11. doi:10.3389/fgene.2019.01307
- La Población Indígena en México. Instituto nacional de estadística, geografía e informática; 2004 Available from: http://internet.contenidos.inegi.org.mx/contenidos/productos//prod_serv/contenidos/espanol/bvinegi/productos/historicos/2104/702825497583/702825497583_1.pdf. Accessed 71, 2016.
- Falfan-ValenciaR, Pérez-RubioG, Ambrocio-OrtizE, et al. Heterozygous genotype rs17580 AT (PiS) in SERPINA1 is associated to COPD secondary to biomass and tobacco smoke. Eur Respir J. 2017;50(suppl61). doi:10.1183/13993003.00711-2017
