 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Recently, several genes and genetic loci associated with both asthma and chronic obstructive pulmonary disease (COPD) have been described as common susceptibility factors for the two diseases. In complex diseases such as asthma and COPD, a large number of molecular and cellular components may interact through complex networks involving gene–gene and gene–environment interactions. We sought to understand the functional and regulatory pathways that play central roles in the pathobiology of asthma and COPD and to understand the overlap between these pathways. We searched the PubMed database up to September 2012 to identify genes found to be associated with asthma, COPD, tuberculosis, or essential hypertension in at least two independent reports of candidate-gene associations or in genome-wide studies. To learn how the identified genes interact with each other and other cellular proteins, we conducted pathway-based analysis using Ingenuity Pathway Analysis software. We identified 108 genes and 58 genes that were significantly associated with asthma and COPD in at least two independent studies, respectively. These susceptibility genes were grouped into networks based on functional annotation: 12 (for asthma) and eleven (for COPD) networks were identified. Analysis of the networks for overlap between the two diseases revealed that the networks form a single complex network with 229 overlapping molecules. These overlapping molecules are significantly involved in canonical pathways including the “aryl hydrocarbon receptor signaling,” “role of cytokines in mediating communication between immune cells,” “glucocorticoid receptor signaling,” and “IL-12 signaling and production in macrophages” pathways. The Jaccard similarity index for the comparison between asthma and COPD was 0.81 for the network-level comparison, and the odds ratio was 3.62 (P < 0.0001) for the asthma/COPD pair in comparison with the tuberculosis/ essential hypertension pair. In conclusion, although the identification of asthma and COPD networks is still far from complete, these networks may be used as frameworks for integrating other genome-scale information including expression profiling and phenotypic analysis. Network overlap between asthma and COPD may indicate significant overlap between the pathobiology of these two diseases, which are thought to be genetically related.
Introduction
Both asthma and chronic obstructive pulmonary disease (COPD) are characterized by chronic inflammation and remodeling of the airways.Citation1,Citation2 A common pathogenetic basis for asthma and COPD is implied based on overlapping clinical characteristics, epidemiologic studies, and the association of genes common to both asthma and COPD. Genetics provides a unique tool for studying the pathophysiology of asthma and COPD. Traditional candidate gene studies may focus on a single gene or on a few genes in combination, with these genes identified on the basis of prior knowledge or on suspected pathogenetic mechanisms. In contrast, genome-wide association studies (GWAS) and linkage studies allow for the comprehensive evaluation of the entire genome without prior assumptions regarding the pathobiology. Nonetheless, the many genetic variations discovered can explain only a small fraction of the genetic risks associated with such complex diseases;Citation3 complex biological systems and cellular networks may underlie most genotype–phenotype relationships. The high polygenicity of asthma and COPD, therefore, could suggest that genetic variants confer risk by functioning together within the same network, and that the functional unit conferring a disease risk may not be a single gene but rather the network itself.Citation4
The Dutch hypothesis maintains that asthma and airway hyperresponsiveness predispose patients to developing COPD later in life and that asthma and COPD are different expressions of a single disease,Citation5 which is based on the timing of environmental and epigenetic influences with a common genetic background. Host factors such as airway hyperre-sponsiveness, family history of asthma, and low lung function are common risk factors for asthma and COPD, as are environmental stimuli such as environmental tobacco smoke and air pollution.Citation6,Citation7 In recent decades, substantial progress has been made in characterizing the susceptible genes involved in asthma or COPD. Genes that have been implicated in both asthma and COPD include ADRB2, GSTM1, GSTP1, IL13, TGFB1, TNF, ADAM33, CCL5, and IL17F.Citation8–Citation13 Several common genetic predispositions, therefore, may contribute to the development of asthma and COPD, including predisposition to abnormal lung growth, resulting in lower lung function and delayed immune maturation; predisposition to lower respiratory viral infections and early allergic sensitization; and predisposition to bronchial hyperresponsiveness. By interacting with each other, genes and their products form complex cellular networks. Therefore, the Dutch hypothesis implies a polygenetic variation affecting the same pathway and the alteration of a functional network as a common root for increased risk for asthma and COPD.
The purpose of the current study was to identify the biological pathways and processes critical to asthma and COPD, and to analyze the genetic similarities between the two diseases using genes associated with asthma or COPD, as well as constructing interaction networks among those genes and their products. We used Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA, USA) to evaluate whether loci across the genome associated with asthma or COPD were enriched for connectivity among genes representing particular pathways or molecular processes. Assessing relatedness between diseases involves exploring the mechanisms that influence susceptibility and phenotype expression. Thus, the overlap of genes and their products could provide comparative insight into the patho-genetic mechanisms of asthma and COPD. The results from the current study will serve as a first step towards a better understanding of these chronic inflammatory lung diseases and the associated phenotypes with similar symptoms or modes of treatment.
Materials and methods
Database searches
We systematically searched the results of gene association studies on asthma or COPD phenotypes that were published up to September 2012. We also searched disease-associated genes for two other diseases, tuberculosis (TB) and essential hypertension (E-HTN), as disease controls. We chose TB as a disease control because, like COPD and asthma, it too is characterized by chronic pulmonary inflammation. In contrast, we chose E-HTN as a negative disease control; it is highly unlikely that E-HTN shares any common pathogenesis with asthma, COPD, or TB. Both TB and E-HTN are also common complex diseases in which multiple genetic and environmental factors are involved in the etiology. In addition, for both diseases, genetic factors have been extensively studied, including in GWAS.
We selected genes associated with each disease that were reported in two or more independent association studies or demonstrated by GWAS. We identified the genes by searching the PubMed database using the keywords “association” and “SNP or polymorphism” with each of the following terms: “asthma,” “COPD or chronic bronchitis or emphysema,” “tuberculosis,” and “essential hypertension.” Search results were checked manually for relevance. The search was restricted to English-language and human studies. We also excluded pharmacogenetic studies from the search.
Ingenuity pathway analysis
We sought to identify asthma- and COPD-associated gene networks from these candidate genes. We used IPA software, which links specific genes to a database of gene functions gleaned from the biomedical research literature. IPA core analysis allows us to find interactions between genes and proteins, related networks, functions, and canonical pathways in the context of biological processes. Briefly, a set of genes identified by the PubMed search was uploaded into the web-delivered application and each gene identifier was mapped to its corresponding gene object in the Ingenuity Knowledge Base (Ingenuity Systems). Genes associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. The significance of the association between the gene dataset and the canonical pathway was measured in two ways: (1) a ratio of the number of genes from the dataset that map to the pathway divided by the total number of molecules that exist in the canonical pathway; and (2) the Benjamini–Hochberg procedure for multiple testing correction, which allows us to calculate the false discovery rate for each of the probability values to determine whether the association between the genes in the dataset and the canonical pathway is explained by chance alone.
IPA network generation
Molecules of interest that interact with other molecules in the Ingenuity Knowledge Base are designated as network eligible molecules. IPA considers all network-eligible molecules on our gene list to be of equal importance when generating networks for molecule lists. Network-eligible molecules are combined into networks that maximize their specific connectivity, which is their interconnectedness with each other relative to all of the molecules with which they are connected in the Ingenuity Knowledge Base. Additional molecules from the Ingenuity Knowledge Base are used to specifically connect two or more smaller networks by merging them into a larger network. Networks are limited to 35 molecules each to keep them to a usable size. Networks are scored on the basis of the number of network-eligible molecules they contain. The score takes into account the number of network-eligible molecules in the network and its size, as well as the total number of network-eligible molecules analyzed, and the total number of molecules in the Ingenuity Knowledge Base that could potentially be included in the networks. In fact, the higher the score, the lower the probability of finding the observed number of network-eligible molecules in a given network by random chance. Focus molecules simply indicate the number of network-eligible molecules per network. The three most significant functions for each network are listed.
Canonical pathways are distinct from networks in that they are generated prior to data input, are based on the literature, and do not change upon data input, whereas networks are generated de novo on the basis of the researcher’s own input data. Biological understanding of the function of genes in pathways, and the currently available lists of ‘‘canonical’’ pathways are evolving rapidly. In this study, five gene datasets were used to identify the most significantly associated canonical pathways: 108 genes for asthma, 58 genes for COPD, 37 genes for TB, 55 genes for E-HTN, and 229 genes common to both asthma and COPD.
Relatedness between asthma and COPD
A pairwise comparison of asthma and COPD and of TB and E-HTN was performed at the network level. The JaccardCitation14 similarity index was used to measure the degree of association between the two diseases. This index considers the similarity between two diseases as the number of genes shared divided by the total number of genes present in either of them. It may be expressed as follows:
where A is the number of genes present in a given disease A; B is the number of genes present in disease B; and C is the number of genes present in both disease A and disease B. The number of genes present in either of the diseases is given by A + B – C. To assess the significance of the relatedness in network-level comparisons, we compared the Jaccard similarity index for asthma and COPD with that for TB and E-HTN as the reference because it is highly unlikely that TB and E-HTN are genetically related. Using a 2 × 2 contingency table for the two groups of disease comparisons, each was sorted according to the genes that were common or unique to the diseases; the odds ratios (ORs) and associated significance levels were calculated.
Results
The PubMed search identified 108 asthma-, 58 COPD-, 37 TB-, and 55 E-HTN-associated genes (). Using information on these genes, the IPA program created 12, eleven, seven, and five networks for these diseases, respectively (–). The networks for each disease consisted of 419, 320, 244, and 175 genes or their products for asthma, COPD, TB, and E-HTN, respectively.
Table 1 genes associated with BA, COPD, TB, or E-HTN
Table 2 Networks associated with asthma
Table 3 Networks associated with COPD
Table 4 Networks associated with tuberculosis
Table 5 Networks associated with essential hypertension
Biological pathway analysis (IPA)
For asthma, the main canonical pathways identified were the “T-helper cell differentiation,” “altered T-cell and B-cell signaling in rheumatoid arthritis,” “role of cytokines in mediating communication between immune cells,” and “communication between innate and adaptive immune cells” pathways (). For COPD, the main pathways identified were the “hepatic fibrosis/hepatic stellate cell activation,” “aryl hydrocarbon receptor signaling,” “glucocorticoid receptor signaling,” and “differential regulation of cytokine production in macrophages and T-helper cells by IL-17A and IL-17F” pathways ().
Table 6 Top ten canonical pathways significantly associated with each group of disease susceptibility genes
The canonical pathway “aryl hydrocarbon receptor signaling” was most significantly associated with the overlap of gene datasets between asthma and COPD, followed by multiple canonical pathways that showed highly significant associations such as the “role of cytokines in mediating communication between immune cells,” “glucocorticoid receptor signaling,” “IL-12 signaling and production in macrophages,” and “hepatic frosts/hepatic stellate cell activation” pathways ().
Table 7 Canonical pathways significantly associated with genes common to both BA and COPD
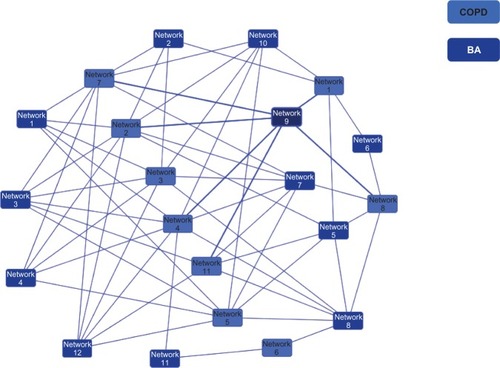
Twelve asthma and eleven COPD networks generated a large single network (); 229 genes were common to both diseases, and 190 and 91 genes were unique to asthma and COPD, respectively. The numbers of network genes common or unique to each disease comparison are shown in . The Jaccard similarity index for the network level comparison between asthma and COPD was 0.81. The OR was 3.62 (P < 0.0001) for the asthma/COPD pair in comparison with the TB/E-HTN pair (). The Jaccard similarity index for the comparisons between asthma and TB and between COPD and TB were 0.67 (OR 2.98; P < 0.0001) and 0.63 (OR 2.79; P < 0.0001), respectively ().
Table 8 Numbers of genes and related molecules that are unique or common to given disease pairs
Table 9 The Jaccard similarity index calculated for each disease pair
Figure 1 Overlapping networks between asthma and COPD.
Abbreviations: COPD, chronic obstructive pulmonary disease; BA, bronchial asthma; IPA, Ingenuity Pathway Analysis.
Discussion
IPA is a software program that helps researchers model, analyze, and understand complex systems by integrating data from a variety of experimental platforms and providing insight into molecular and chemical interactions, cellular phenotypes, and disease processes. To expand upon the understanding of the genetic architecture and molecular basis of asthma and COPD, we used the IPA program to evaluate whether loci across the genomes previously associated with asthma or COPD were enriched for connectivity among genes representing particular pathways or molecular processes. We observed a significant overlap between asthma- and COPD-associated genetic loci, which may refect the significant epidemiologic and clinical overlap between the two diseases. The common and distinct functional and regulatory pathways identified in this study may play central roles in the pathophysiology of asthma and COPD, which could help us to understand the primary pathogenesis underlying these diseases.
The literature provides good evidence that the canonical pathways highlighted in the current study could lead to the development of both asthma and COPD. The aryl hydrocarbon receptor (AhR) is a ligand-dependent transcription factor whose activity is modulated by xenobiotics as well as by physiologic ligands. The AhR is an attenuator of pulmonary inflammation caused by tobacco smoke, regulating pathogenic processes implicated in COPD etiology and progression, inflammation, and cell death.Citation15 It also modulates allergic inflammatory responses. Mast cells, located at the boundaries between tissues and the external environment, are produce IL-17, a critical player in chronic inflammation and a potential target of AhR ligands. Murine and human mast autoimmunity, suggesting a novel pathway for mast cell cells constitutively express AhR, and its activation by the activation in the pathogenesis of asthma and COPD.Citation16 high-affinity ligand 6-formylindolo[3,2-b]carbazole induces The “differential regulation of cytokine production a boost in degranulation. Moreover, AhR-activated mast cells in macrophages and T-helper cells and epithelial cells by IL-17A and IL-17F” pathway (see ) has also been identified as a gene dataset common to both asthma and COPD. Neutrophilic airway inflammation is a common feature of COPD and is recognized in asthma, particularly in severe disease. The Th17 cytokines IL-17A and IL-17F have been implicated in the development of neutrophilic airway inflammation, and increased expressions of these cytokines have been implicated in asthma and COPD.Citation17 Notably, tobacco smoke is a selective adjuvant that augments in vitro and in vivo Th17 cell differentiation via the AhR, suggesting that tobacco smoke is a potent Th17 adjuvant, and that IL-17RA signaling is required for the chemokine expression necessary for MMP12 induction and tissue emphysema.Citation18
In addition, identification of the canonical pathways for a common gene dataset such as the “role of cytokines in mediating communication between immune cells,” “role of pattern recognition receptors in recognition of bacteria and viruses,” and “IL-12 signaling and production in macrophages” pathways (see ) also suggest that asthma and COPD share common genetic associations related to impaired innate and adaptive immunity. These key pathways may be disrupted via many different causes – genetic, epigenetic, and environmental – in patients with asthma or COPD. Even if the disease arises from a different specific cause in different individuals, the disease in each of these individuals could nonetheless share disruption of these related key biological processes.
In the obtained disease networks, asthma and COPD belong to a single interconnected main giant component (), which is consistent with the idea that asthma and COPD are much more connected to each other than hitherto believed. Therefore, asthma and COPD could be viewed as perturbations of highly interlinked cellular networks. For chronic inflammatory lung disease, polygenetic variation alters the behaviors of a biological pathway in response to environmental exposures including allergens, infections (bacterial or viral), tobacco smoke, and ambient or indoor air pollution. Functional alterations of any single node through function-modifying mutation either may have no effect at all, owing to the emergent network property of robustness, or, for network hubs, may yield a phenotype such as asthma or COPD. The heterogeneity between asthma and COPD could be due, at least in part, to mutations in different genes having similar phenotypic effects owing to their acting on the same functional pathway. The functional pathways identified for the genes common to asthma and COPD, therefore, could play a role in the proposed association between asthma and COPD.
Although patients with asthma or COPD are believed to have no increased risk for TB, TB showed more similarities to asthma and COPD than did E-HTN. This may reflect the fact that these three inflammatory disorders are characterized by chronic inflammation in the lung, sharing common immune and inflammatory networks for their pathogenesis. In this study, many canonical pathways related to innate and adaptive immune responses were commonly identified for the networks identified in asthma, COPD, and TB, suggesting the important role of nonspecific inflammatory responses in asthma and COPD as well as in TB, which are mediated by cytokines and chemokines induced by the interaction of innate receptors expressed in macrophages and dendritic cells.
We acknowledge that there are inherent biases in such literature reviews, given the reduced likelihood of negative studies being published. Moreover, if the first published study yields a negative result, other investigators are less likely to study that gene. However, because even the most replicated genes have one or more negative studies, a gene with an initial negative result could still be a true susceptibility gene in other populations, or with respect to phenotypes or variants other than those studied in the initial report. In addition, we did not take into account the size of the study samples, the effect size, the overall quality of the analysis, or whether the genotype data were in Hardy–Weinberg proportions. Rather, we reported positive findings when the investigators interpreted their results as supporting an association, and we reported negative findings when the investigators interpreted their results as not supporting an association.
Conclusion
The current study suggests that the genetic contribution to chronic inflammatory lung diseases operates through multiple genes interacting in different functional pathways, providing insight into similarities in the underlying pathogenetic mechanisms between asthma and COPD. Environmental factors including allergens, infections, and smoking may alter the expression and regulation of common networks that mediate risk for asthma and COPD. Identification of networks shared by asthma and COPD may prove useful in the diagnosis and treatment of these two diseases. Although the link between these two diseases warrants further study, we anticipate that these genetic insights will transform the landscape of common complex diseases such as asthma and COPD.
Acknowledgments
We thank Mrs Flaminia Miyamasu, an associate professor at the Faculty of Medicine, University of Tsukuba, Tsukuba, Japan, who proofread and commented on this paper. The study was partly supported by a Grant-in-Aid for Scientific Research (B), No 24390206, from the Japan Society for the Promotion of Science.
Disclosure
The authors report no conflicts of interest in this work.
References
- BarnesPJImmunology of asthma and chronic obstructive pulmonary diseaseNat Rev Immunol20088318319218274560
- GelbAFZamelNKrishnanAPhysiologic similarities and differences between asthma and chronic obstructive pulmonary diseaseCurr Opin Pulm Med2008141243018043272
- VercelliDDiscovering susceptibility genes for asthma and allergyNat Rev Immunol20088316918218301422
- SullivanPFPuzzling over schizophrenia: schizophrenia as a pathway diseaseNat Med201218221021122310687
- OrieNGThe Dutch hypothesisChest20001175 Suppl 1299S10843963
- BrutscheMHDownsSHSchindlerCSAPALDIA TeamBronchial hyperresponsiveness and the development of asthma and COPD in asymptomatic individuals: SAPALDIA cohort studyThorax200661867167716670173
- SvanesCSunyerJPlanaEEarly life origins of chronic obstructive pulmonary diseaseThorax2010651142019729360
- HizawaNYamaguchiEKonnoSTaninoYJinushiENishimuraMA functional polymorphism in the RANTES gene promoter is associated with the development of late-onset asthmaAm J Respir Crit Care Med2002166568669012204866
- HizawaNKawaguchiMHuangSKNishimuraMRole of interleukin-17F in chronic inflammatory and allergic lung diseaseClin Expl Allergy200636911091114
- Van EerdeweghPLittleRDDupuisJAssociation of the ADAM33 gene with asthma and bronchial hyperresponsivenessNature2002418689642643012110844
- van DiemenCCPostmaDSVonkJMBruinenbergMSchoutenJPBoezenHMA disintegrin and metalloprotease 33 polymorphisms and lung function decline in the general populationAm J Respir Crit Care Med2005172332933315879414
- HizawaNMakitaHNasuharaYHokkaido COPD Cohort Study Group. Functional single nucleotide polymorphisms of the CCL5 gene and nonemphysematous phenotype in COPD patientsEur Respir J200832237237818385174
- PostmaDSKerkhofMBoezenHMKoppelmanGHAsthma and chronic obstructive pulmonary disease: common genes, common environments?Am J Respir Crit Care Med2011183121588159421297068
- JaccardPThe distribution of the flora in the alpine zoneNew Phytol19121123750
- Rico de SouzaAZagoMPollockSJSimePJPhippsR PBagloleCJGenetic ablation of the aryl hydrocarbon receptor causes cigarette smoke-induced mitochondrial dysfunction and apoptosisJ Biol Chem201128650432144322821984831
- SibilanoRFrossiBCalvarusoMThe aryl hydrocarbon receptor modulates acute and late mast cell responsesJ Immunol2012189112012722649193
- DoeCBafadhelMSiddiquiSExpression of the T helper 17-associated cytokines IL-17A and IL-17F in asthma and COPDChest201013851140114720538817
- ChenKPociaskDAMcAleerJPIL-17RA is required for CCL2 expression, macrophage recruitment, and emphysema in response to cigarette smokePLoS One201165e2033321647421