Abstract
Recent candidate gene studies using a human liver bank and in vivo validation in healthy volunteers identified polymorphisms in cytochrome P450 (CYP) 3A4 gene (CYP3A4*22), Ah-receptor nuclear translocator (ARNT), and peroxisome proliferator-activated receptor-α (PPAR-α) genes that are associated with the CYP3A4 phenotype. We hypothesized that the variants identified in these genes may be associated with altered clopidogrel response, since generation of clopidogrel active metabolite is, partially mediated by CYP3A activity. Blood samples from 211 subjects, of mixed racial background, with established coronary artery disease, who had received clopidogrel, were analyzed. Platelet aggregation was determined using light transmittance aggregometry (LTA). Genotyping for CYP2C19*2, CYP3A4*22, PPAR-α (rs4253728, rs4823613), and ARNT (rs2134688) variant alleles was performed using Taqman® assays. CYP2C19*2 genotype was associated with increased on-treatment platelet aggregation (adenosine diphosphate 20 μM; P=0.025). No significant difference in on-treatment platelet aggregation, as measured by LTA during therapy with clopidogrel, was demonstrated among the different genotypes of CYP3A4*22, PPAR-α, and ARNT. These findings suggest that clopidogrel platelet inhibition is not influenced by the genetic variants that have previously been associated with reduced CYP3A4 activity.
Introduction
Clopidogrel is a thienopyridine antiplatelet agent that bioactivates in a two-step process, with varying contributions to these steps from cytochrome P450 (CYP) 2C19, CYP1A2, CYP2C9, CYP3A4, CYP3A5, and CYP2B6 isoenzymes.Citation1 Inactivation of the precursor, and metabolites, is dependent, predominantly, on hepatic carboxylesterase-1 enzyme.Citation2,Citation3 Diminished metabolic status of CYP2C19, due to drug interactions or variants in the CYP2C19 gene, results in lower active clopidogrel metabolite generation, and higher on-treatment platelet reactivity.Citation4–Citation7 However, CYP2C19 metabolic status alone was found to account for only 12% of the variability in clopidogrel response.Citation8 Several lines of evidence suggest that the CYP3A4 and CYP3A5 enzymes may play a role in clopidogrel activation, although data are not uniform.Citation1,Citation5,Citation9,Citation10CYP3A5*3 polymorphism has not been shown to significantly predict clopidogrel pharmacokinetics.Citation8,Citation11,Citation12
In humans, CYP3A isoenzymes comprise the majority of hepatic CYP450 proteins. CYP3A4 is the most abundant, and metabolizes approximately 50% of drugs which are cleared via metabolism.Citation13,Citation14 A high intersubject variability has been documented in the activity of CYP3A4, an enzyme that metabolizes a wide range of clinically important substrates.Citation14–Citation16 Despite estimates that suggested that up to 90% of functional CYP3A4 variability is heritable, the genetic basis of CYP3A4 variable expression and activity is poorly understood.Citation17 While functional variants of many CYP genes have been established, the few common exonic variants of the CYP3A4 gene that have been reported so far only marginally explain variations in CYP3A4 activity.
Recently, Klein et al, using a candidate gene approach, in a human liver bank, identified several single nucleotide polymorphisms (SNPs) that are associated with CYP3A4 activity phenotype.Citation15 In that study, 40 candidate genes, including 334 SNPs, were examined for association with CYP3A4 protein expression and CYP3A4 activity, measured by atorvastatin 2-hydroxylation in hepatocyte supernatants from 159 liver samples. Validation was performed by measurement of atorvastatin hydroxylation in 56 healthy volunteers, after single-dose atorvastatin administration.Citation15 Among the SNPs identified, CYP3A4*22 is a recently-identified intronic variant associated with reduced CYP3A4 activity.Citation14,Citation16,Citation18–Citation20 The CYP3A4*22 allele is characterized by a C>T substitution (NG_008421.1:g.20493C>T), and reduced CYP3A4 messenger (m)RNA expression has been linked to the minor T-allele.Citation14 Other variants newly identified are located in the peroxisome proliferator-activated receptor-α (PPAR-α) (rs4253728, rs4823613) and Ah-receptor nuclear translocator (ARNT) (rs2134688) genes.Citation15 Functional validation of these SNPs by Klein et al, measuring atorvastatin 2-hydroxylation in atorvastatin-treated volunteers, supported the in vivo contribution of PPAR-α (rs4253728) and CYP3A4*22 variants toward reduced functional activity of CYP3A4.Citation15 The ARNT (rs2134688) polymorphism was associated with decreased CYP3A4 expression and activity in vitro, but was not significantly associated with atorvastatin hydroxylation in vivo.Citation15 In the case of CYP3A4*22, previous studies have shown improved simvastatin cholesterol lowering effects and increased tacrolimus levels in carriers of the variants.Citation14,Citation16,Citation18,Citation19 Atorvastatin hydroxylation, measured in vivo by the ratio of 2-OH-atorvastatin to atorvastatin, was reduced by 35% in carriers of CYP3A4*22.Citation15 Clopidogrel bioactivation is, at least partially, mediated by CYP3A4, and some pharmacologic studies have suggested a contribution of CYP3A4 inhibition – by statins or calcium channel blockers – to clopidogrel response.Citation5,Citation9 Therefore, the variants described by Klein et al may also be associated with altered pharmacodynamic response to clopidogrel, due to diminished generation of active clopidogrel metabolite. In comparison, generation of active clopidogrel thiol metabolite, in CYP2C19-poor metabolizers, is reduced by 43%, as compared to extensive metabolizers.Citation6
We hypothesized that adenosine diphosphate-(ADP) induced platelet reactivity – a well-established pharmacodynamic measure of clopidogrel response – is increased in carriers of CYP3A4*22 and variants of PPAR-α and ARNT that previous studies have associated with reduced functional activity of CYP3A4.
Methods
Patients
The study’s protocols were approved by the Indiana University Institutional Review Board for research. Written informed consent was obtained from all subjects. Subjects were eligible to be enrolled if they had established coronary disease, and were on dual antiplatelet therapy with clopidogrel and aspirin (81–325 mg per day). Subjects were included in this analysis if either they had been taking clopidogrel (75 mg) for at least 5 days prior to enrollment, or had received a 600 mg loading dose of clopidogrel during a percutaneous coronary intervention (PCI), at least 6 hours prior to blood sampling. Subjects were excluded if their platelet counts were lower than 100,000 per mm3, if they were taking warfarin, or if a glycoprotein IIb/IIIa antagonist was used during PCI.
Blood samples
To determine on-treatment platelet aggregation, peripheral venous blood samples were obtained from subjects after at least 5 days of clopidogrel treatment (75 mg daily), prior to the next dose of clopidogrel, or at least 6 hours after administration of a clopidogrel loading dose. The time points were chosen to allow for steady-state platelet inhibition during maintenance therapy and to achieve maximal inhibition for subjects who had been administered loading dose clopidogrel.Citation21,Citation22 All blood samples were directly transferred into vacutainer tubes containing 3.2% sodium citrate, and were analyzed within 2 hours.
Platelet aggregation studies
Ex vivo platelet function was assessed by light transmittance aggregometry (LTA) at 37°C using an optical lumi-aggregometer (Model 700 with Aggro/Link 8 software; Chrono-log Corp, Havertown, PA, USA). Platelet-rich plasma (PRP) and platelet-poor plasma (PPP) were obtained by differential centrifugation, as previously described.Citation23,Citation24 Platelet aggregation in PRP was induced using arachidonic acid (AA), at 1 mM, and ADP at 5 μM, 10 μM, and 20 μM.
Genotyping
Genomic DNA was isolated from whole blood using Qiagen’s QIAamp® DNA Blood Midi Kit (Germantown, MD, USA). Subjects were genotyped for CYP2C19*2 (681G>A; rs4244285), CYP3A4*22 (rs35599367 C>T), PPAR-α (rs4253728 G>A and rs4823613 A>G), and ARNT (rs2134688 A>G) using a real-time polymerase chain reaction (PCR) system from Bio-Rad Laboratories (Hercules, CA, USA). Sequence-specific primers were used to amplify the alleles of interest, along with two allele-specific TaqMan® probes (Life Technologies, Carlsbad, CA, USA). Allelic discrimination was used to determine individual genotypes (iCycler 3.1 optical system software; Bio-Rad Laboratories).
Statistical analysis
Categorical variables were compared using the χ2 test. Unpaired two-sided Student’s t-test was used to compare normally distributed continuous data between two groups, and for analysis of variance between multiple groups. An additive dose model was used in univariate linear regression analysis, which was performed using platelet aggregation as the outcome variable, and genetic variants of CYP3A4*22 (rs35599367 C>T), PPAR-α (rs4253728 G>A and rs4823613 A>G), ARNT (rs2134688 A>G), and CYP2C19*2 (681G>A; rs4244285) as covariates. Smoking and diabetes mellitus were identified as significant clinical covariates of platelet reactivity in univariate analysis (P<0.1). Multivariate linear regression analysis of individual CYP3A4*22, PPAR-α, and ARNT genotypes, with adjustments for smoking status, diabetes mellitus, and CYP2C19*2 genotype, was performed using the additive model. The data analysis was conducted using SPSS statistical software version 21 (IBM Corp, Armonk, NY, USA). Statistical significance was defined as P<0.05.
Results
Baseline characteristics of the study subjects are described in . A total of 106 subjects on maintenance clopidogrel therapy for >5 days, and 105 subjects who had received 600 mg clopidogrel loading dose prior to PCI, were enrolled. The majority of subjects were Caucasian (72%), while the remainder was African American (27%) and a small minority of Asian descent (1%).
Table 1 Demographics and clinical variables of subjects
Genotype frequencies for the 211 patients included in this study are shown in . For all genotype distributions, no significant deviations from Hardy–Weinberg equilibrium were observed (P>0.05). We found that two out of 12 carriers of CYP3A4*22 alleles were African American (4% of all African Americans in our study).
Table 2 Frequency of observed and expected genotypes (according to Hardy–Weinberg equilibrium) for CYP3A4*22, PPAR-α, ARNT, and CYP2C19*2
Platelet aggregation induced by ADP demonstrated wide interindividual variability during therapy with clopidogrel, as previously documented ().Citation8,Citation25 Only two out of 211 subjects were aspirin non-responders, as defined by >20% maximal platelet aggregation stimulated by AA.Citation26 There was no significant difference in on-treatment platelet aggregation between subjects on maintenance dose clopidogrel and those who had received a 600 mg loading dose (ADP 5 μM: 31%±14% versus [vs] 34%±18%; P=0.33). No significant difference in on-treatment platelet aggregation was demonstrated between Caucasian subjects and African Americans (ADP 10 μM: 40%±16% vs 39%±19%; P=0.8).
Figure 1 Platelet aggregation.
Abbreviations: ADP, adenosine diphosphate, CYP, cytochrome P450; ARNT, Ah-receptor nuclear translocator; PPAR, peroxisome proliferator-activated receptor.
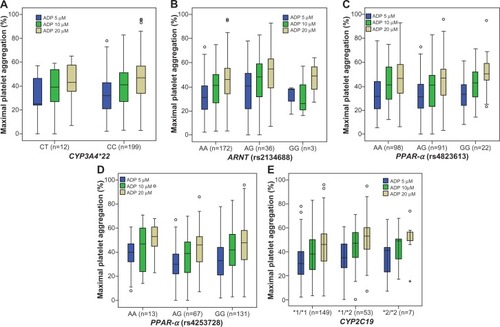
Carriers of CYP2C19*2 alleles had higher platelet aggregation (ie, lower platelet inhibition) than non-carriers (ADP 5 μM: 36%±15% vs 31%±15%; P=0.09) (ADP 10 μM: 44%±16% vs 39%±17%; P=0.034) (ADP 20 μM: 50%±16% vs 44%±17%; P=0.02) (). Carriers of CYP3A4*22 and PPAR-α (rs4253728, rs4823613) variants showed a nonsignificant trend toward lower platelet aggregation (ie, higher platelet inhibition) than non-carriers (). Carriers of the ARNT variant showed a nonsignificant trend toward higher platelet aggregation (ie, lower platelet inhibition) than non-carriers (). The nonsignificant association of the ARNT variant with platelet aggregation was not gene dose dependent, with lower platelet aggregation in homozygous individuals, compared with heterozygous carriers of the variant (ADP 10 μM; P=0.6) ( and ).
Table 3 Comparison of MPA (measured by LTA), grouped according to non-carrier and carrier status of variants of CYP3A4*22, PPAR-α, ARNT, and CYP2C19*2
Table 4 Linear regression analysis of platelet aggregation and genetic polymorphisms
Among carriers of CYP3A4*22 variants, four out of 12 subjects were also heterozygous for CYP2C19*2, with no significant additive effect on maximal ADP-induced platelet aggregation, compared with CYP2C19*2 carriers alone (ADP 10 μM: 33%±23% vs 45%±15%; P=0.16). After exclusion of carriers of CYP2C19*2 variants, on-treatment platelet aggregation in carriers of CYP3A4*22 variants remained nonsignificant, compared with wild type individuals (ADP 10 μM: 40%±17% vs 39%±15%; P=0.6). In the univariate linear regression analysis, only CYP2C19*2 genotype (ADP 20 μM; P=0.025) (), current smoking (ADP 10 μM; P=0.01), and diabetes mellitus (ADP 10 μM; P=0.004) were significantly associated with ADP-induced platelet aggregation. These covariates with P<0.1 were included, and adjusted for, in multivariate regression analysis. Proton pump inhibitor and calcium channel blocker use were not significantly associated with platelet reactivity in this analysis (ADP 10 μM; P=0.38 and P=0.3, respectively).
CYP3A4*22, PPAR-α, and ARNT genotypes were not associated with significant differences in ADP-induced platelet aggregation, neither in the univariate nor in the multivariate analyses ( and ). The use of a recessive, instead of an additive model did not significantly change the results of the regression analysis, with no significant association found between genotype and platelet aggregation for the ARNT and PPAR-α variants studied (P>0.05, for all concentrations of ADP). When analysis was restricted to Caucasian subjects and African Americans separately, no significant differences were found between CYP3A4*22 carriers and non-carriers (Caucasian ADP 10 μM: 37%±20% vs 40%±16%; P=0.6) (African American ADP 10 μM: 35%±6% vs 40%±20%; P=0.72).
Among Caucasian subjects, no significant differences were found between wild type, heterozygous, and homozygous genotypes of PPAR-α (rs4253728 ADP 10 μM: 42%±15% vs 37%±16% vs 41%±19%; P=0.16) (rs4823613 ADP 10 μM: 42%±15% vs 38%±16% vs 40%±19%; P=0.24), and ARNT variants (ADP 10 μM: 40%±16% vs 42%±18% vs 40%±16%; P=0.68).
Among African Americans, no significant differences were found between wild type, heterozygous, and homozygous genotypes of PPAR-α (rs4253728 ADP 10 μM: 39%±19% vs 39%±25% vs 60%±[CI not available]; P=0.6) (rs4823613 ADP 10 μM: 37%±17% vs 39%±22% vs 44%±14%; P=0.67). Among African Americans, platelet aggregation was significantly different between wild type, heterozygous, and homozygous ARNT genotypes (ADP 10 μM: 35%±18% vs 51%±18% vs 33%±21%; P=0.014). However, after adjustment for confounding variables (diabetes mellitus, smoking status, and CYP2C19*2 genotype) in multivariate regression analysis, no significant association remained for ARNT genotypes and platelet aggregation among African Americans (ADP 10 μM: X1=6; P=0.22).
Discussion
The prevalence and frequency of the variants in our study are broadly comparable to findings from previous studies. We demonstrated the presence of the CYP3A4*22 allele in African Americans, which had previously only been documented in subjects of European descent.Citation14 This may be due to the relatively large number of African Americans included in our study, as well as to the mixed ancestral background of African Americans living in the United States.
Multiple enzymes participate in clopidogrel activation, which includes CYP3A4.Citation1 The intronic variant CYP3A4*22 is associated with reduced protein expression, by unknown mechanism, and thus, reduced activity.Citation27 Therefore, it is expected that the proportion of clopidogrel activation via CYP3A4 is diminished. This led us to speculate that shunting of clopidogrel activation occurs in favor of other enzymes, such as CYP2C19, CYP2B6, and CYP1A2, when CYP3A4 activity is reduced. We measured ADP-induced platelet aggregation – a well-established pharmacodynamic measure of clopidogrel response that has been found to correlate well with active clopidogrel metabolite concentrations.Citation12 In contrast to our hypothesis, our data suggest that the CYP3A4*22 allele is not associated with altered clopidogrel response. While our study included only a limited number of carriers of the CYP3A4*22 variant, it is unlikely that a significantly reduced clopidogrel response could be found in a larger cohort of carriers, since the trend observed is opposite to what would be expected with reduced CYP3A4 clopidogrel activation. Similarly to CYP3A4*22, none of the variants of PPAR-α and ARNT genes were associated with increased platelet reactivity, in patients treated with clopidogrel. There may be several reasons for the lack of influence on clopidogrel response of the CYP4A4*22, PPAR-α, and ARNT variants which have been demonstrated to be associated with CYP3A4 expression and activity. First, it has previously been shown that inhibition of CYP3A4 by competing drugs, such as atorvastatin, is not sufficient to reliably reduce clopidogrel platelet inhibition. This is possibly because CYP3A5 and CYP1A2 are alternative pathways for the first step in clopidogrel activation, and are sufficiently active to overcome a reduction in CYP3A4 activity associated with the variants.Citation28 Second, it has not been established whether the variants in ARNT and PPAR-α, which are ligand-dependent nuclear receptors, could be associated with differential expression of other hepatic isoenzymes, which could balance the effects of reduced active clopidogrel metabolite formation by CYP3A4. PPAR-α is a well-known nuclear receptor with a key role in regulation of lipid homeostasis, as well as in immunomodulatory function and regulation of gene transcription.Citation29,Citation30 The ARNT gene encodes the aryl hydrocarbon receptor nuclear translocator protein, which is required for aryl hydrocarbon receptor function, and has been shown to be involved in enzyme induction related to xenobiotic metabolism. Interaction of this receptor with omeprazole metabolites has been documented.Citation31 The ARNT variant (rs2134688) has also been previously linked to increased CYP1A2 expression, which potentially could compensate for reductions in CYP3A4 activity and clopidogrel metabolite generation.Citation32 While the unadjusted regression effect of the ARNT variant is of nearly similar size to that seen with CYP2C19*2 (in our study), it appears not to be gene dose dependent, with better inhibition in homozygous individuals, and worse inhibition in heterozygous individuals. The effect size of the ARNT variant was significantly reduced after multivariate adjustment, compared to the effect size of CYP2C19. Further studies of the effect of the ARNT variant and clopidogrel response may be warranted. In comparison to the small, nonsignificant effect sizes documented in our study for the novel variants in CYP3A4*22 and PPAR-α, the CYP2C19*2 genotype was associated with a 15% difference in platelet inhibition between homozygous and wild type individuals in the study by Shuldiner et al.Citation8 Thus, it is unlikely that the small nonsignificant differences in platelet aggregation observed with the CYP3A4*22 and PPAR-α variants in our study would be associated with altered clinical outcomes similar to those observed, in larger patient cohorts, for CYP2C19*2. In a study by Klein et al, C-reactive protein, sex, and bilirubin levels were significant covariates contributing to CYP3A4 activity, and were adjusted for in multivariate analysis. Elevated C-reactive protein, and sex, are not independent predictors of clopidogrel response, and therefore were not included as covariates in our analysis.Citation33 The influence of bilirubin levels on clopidogrel response is unknown. However, none of the patients included in our study suffered from acute liver failure, and bilirubin levels were not routinely measured. While differences in clopidogrel dosing between individuals could have contributed to interindividual differences in platelet reactivity, the clopidogrel dosing protocols and inclusion criteria are comparable to previous studies.Citation8 Also, dosing was not a significant predictor of clopidogrel response in our study. In addition, maximal platelet aggregation was generally achieved within 6 hours after a clopidogrel 600 mg loading dose and was comparable to on-treatment platelet aggregation during maintenance therapy.Citation21,Citation22 Limitations of our study include the relatively small number of subjects with variant alleles, and lack of pharmacokinetic analysis to detect differences in active metabolite concentrations.
Conclusion
In conclusion, our study suggests that variants of CYP3A4*22, PPAR-α, and ARNT that have been associated with reduced CYP3A4 function are not associated with altered platelet inhibition by clopidogrel. However, given the low frequency of these variants, these findings may require further validation in larger patient cohorts.
Acknowledgments
This study was supported, in part, by the Indiana Clinical and Translational Sciences Institute, funded, in part, by grant number (RR025761) from the National Institutes of Health, National Center for Research Resources, Clinical and Translational Sciences Award. The study was supported, in part, by the Indiana University Health Value Grant, the Department of Medicine, Indiana University School of Medicine, and the Indiana University Health - Indiana University School of Medicine Strategic Research Initiative.
Disclosure
Enrollment of subjects occurred, in part, during the employment of Yan Jin by Indiana University School of Medicine. Yan Jin is currently an employee of Eli Lilly and Co, Indianapolis, IN, USA. The other authors have no conflicts of interest to declare.
References
- KazuiMNishiyaYIshizukaTIdentification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metaboliteDrug Metab Dispos2010381929919812348
- ScottSASangkuhlKGardnerEEClinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-452C19 (CYP2C19) genotype and clopidogrel therapyClin Pharmacol Ther201190232833221716271
- LewisJPHorensteinRBRyanKThe functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel responsePharmacogenet Genomics20132311823111421
- AngiolilloDJGibsonCMChengSDifferential effects of omeprazole and pantoprazole on the pharmacodynamics and pharmacokinetics of clopidogrel in healthy subjects: randomized, placebo-controlled, crossover comparison studiesClin Pharmacol Ther2011891657420844485
- BatesERLauWCAngiolilloDJClopidogrel-drug interactionsJ Am Coll Cardiol201157111251126321392639
- UmemuraKFurutaTKondoKThe common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjectsJ Thromb Haemost2008681439144118532997
- KreutzRPNystromPKreutzYInfluence of paraoxonase-1 Q192R and cytochrome P450 2C19 polymorphisms on clopidogrel responseClin Pharmacol20124132022427735
- ShuldinerARO’ConnellJRBlidenKPAssociation of cytochrome P450 2C19 genotype with the antiplatelet effect and clincal efficacy of clopidogrelJAMA2009302884985719706858
- LauWCGurbelPAWatkinsPBContribution of hepatic cytochrome P450 3 A4 metabolic activity to the phenomenon of clopidogrel resistanceCirculation2004109216617114707025
- LauWCWelchTDShieldsTRubenfireMTantryUSGurbelPAThe effect of St John’s Wort on the pharmacodynamic response of clopidogrel in hyporesponsive volunteers and patients: increased platelet inhibition by enhancement of CYP3A4 metabolic activityJ Cardiovasc Pharmacol2011571869320980920
- MegaJLCloseSLWiviottSDCytochrome p-450 polymorphisms and response to clopidogrelN Engl J Med2009360435436219106084
- FrelingerAL3rdBhattDLLeeRDClopidogrel pharmacokinetics and pharmacodynamics vary widely despite exclusion or control of polymorphisms (CYP2C19, ABCB1, PON1), noncompliance, diet, smoking, co-medications (including proton pump inhibitors), and pre-existent variability in platelet functionJ Am Coll Cardiol201361887287923333143
- DanielsonPBThe cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humansCurr Drug Metab20023656159712369887
- ElensLvan GelderTHesselinkDAHaufroidVvan SchaikRHCYP3A4*22: promising newly identified CYP3A4 variant allele for personalizing pharmacotherapyPharmacogenomics2013141476223252948
- KleinKThomasMWinterSPPARA: a novel genetic determinant of CYP3A4 in vitro and in vivoClin Pharmacol Ther20129161044105222510778
- WangDGuoYWrightonSACookeGESadeeWIntronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugsPharmacogenomics J201111427428620386561
- OzdemirVKalowWTangBKEvaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration methodPharmacogenetics200010537338810898107
- ElensLBouamarRHesselinkDAA new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipientsClin Chem201157111574158321903774
- ElensLBeckerMLHaufroidVNovel CYP3A4 intron 6 single nucleotide polymorphism is associated with simvastatin-mediated cholesterol reduction in the Rotterdam StudyPharmacogenet Genomics2011211286186621946898
- ElensLBouamarRHesselinkDAHaufroidVvan GelderTvan SchaikRHThe new CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with an increased risk of delayed graft function and worse renal function in cyclosporine-treated kidney transplant patientsPharmacogenet Genomics201222537338022388796
- JakubowskiJAPayneCDLiYGA comparison of the antiplatelet effects of prasugrel and high-dose clopidogrel as assessed by VASP-phosphorylation and light transmission aggregometryThromb Haemost200899121522218217157
- GurbelPABlidenKPButlerKRandomized double-blind assessment of the ONSET and OFFSET of the antiplatelet effects of ticagrelor versus clopidogrel in patients with stable coronary artery disease: the ONSET/OFFSET studyCirculation2009120252577258519923168
- KreutzRPTantryUSBlidenKPGurbelPAInflammatory changes during the ‘common cold’ are associated with platelet activation and increased reactivity of platelets to agonistsBlood Coagul Fibrinolysis200718871371817982310
- KreutzRPAllooshMMansourKMorbid obesity and metabolic syndrome in Ossabaw miniature swine are associated with increased platelet reactivityDiabetes Metab Syndr Obes201149910521660293
- KreutzRPBreallJAKreutzYProtease activated receptor-1 (PAR-1) mediated platelet aggregation is dependent on clopidogrel responseThromb Res2012130219820222459907
- TantryUSBlidenKPGurbelPAOverestimation of platelet aspirin resistance detection by thrombelastograph platelet mapping and validation by conventional aggregometry using arachidonic acid stimulationJ Am Coll Cardiol20054691705170916256872
- OkuboMMurayamaNShimizuMShimadaTGuengerichFPYamazakiHThe CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with reduced CYP3A4 protein level and function in human liver microsomesJ Toxicol Sci201338334935423665933
- FaridNASmallDSPayneCDEffect of atorvastatin on the pharmacokinetics and pharmacodynamics of prasugrel and clopidogrel in healthy subjectsPharmacotherapy200828121483149419025429
- MichalikLWahliWPPARs mediate lipid signaling in inflammation and cancerPPAR Res2008200813405919125181
- RakhshandehrooMHooiveldGMüllerMKerstenSComparative analysis of gene regulation by the transcription factor PPARalpha between mouse and humanPLoS One200948e679619710929
- PascussiJMGerbal-ChaloinSDuretCDaujat-ChavanieuMVilaremMJMaurelPThe tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequencesAnnu Rev Pharmacol Toxicol20084813217608617
- KleinKWinterSTurpeinenMSchwabMZangerUMPathway-targeted pharmacogenomics of CYP1 A2 in human liverFront Pharmacol2010112921918647
- ParkDWLeeSWYunSCA point-of-care platelet function assay and C-reactive protein for prediction of major cardiovascular events after drug-eluting stent implantationJ Am Coll Cardiol201158252630263922152948