Abstract
Background
Psoriasis is a widespread chronic, immune-mediated skin disease with frequent recurrences, and is extremely harmful to the physical and mental health of patients, causing enormous suffering and exerting considerable economic burdens on the health care system as a whole. In more than a decade of clinical use, the optimized formula of Yinxieling (PSORI-CM01) has consistently demonstrated its effectiveness for treating psoriasis. However, its underlying mechanism remains largely unexplored.
Methods
The network pharmacology analysis was conducted to predict the mechanism and protective effect of PSORI-CM01 in treating psoriasis. Subsequently, we collected blood samples from 21 patients with psoriasis as part of a randomized, double-blind, and double-dummy clinical trial for microRNA expression profiling. Finally, it was experimentally confirmed that PSORI-CM01 improved psoriasis by regulating miR-20a-3p and miR-3184-3p expression.
Results
As a result of the network pharmacology analysis, PSORI-CM01 improved psoriasis through the regulation of autophagy, cellular apoptosis, cellular proliferation, and anti-inflammatory processes. In the target-miRNA regulatory network, these key targets were mainly associated with the regulation of hsa-miR-20a-3p, hsa-miR-155-5p, has-miR-3184-3p, hsa-miR-328-3p and hsa-miR-124-3p. Based on the microRNA expression profiling results, the PSORI-CM01 treatment group exhibited five up-regulated genes and 16 down-regulated genes compared with the healthy control group. In particular, miR-20a-3p and miR-3184-3p were the primary differentially expressed microRNAs, and they were significantly enriched in the signaling pathways involving autophagy, apoptosis, proliferation, and anti-inflammation. Further experiments confirmed that PSORI-CM01 effectively regulates miR-20a-3p and miR-3184-3p, resulting in increased autophagy.
Conclusion
We demonstrated by combining network pharmacology and clinical studies of miRNA expression profiles in PBMCs that PSORI-CM01 effectively modulated miR-20a-3p and miR-3184-3p, leading to an increase in autophagy and a decrease in keratinocyte proliferation.
Introduction
Psoriasis is a widespread chronic, immune-mediated skin condition characterized by symmetrical, erythematous, scaling papules and plaques.Citation1 It is known that there are five main types of psoriasis, based on their morphological characteristics: psoriasis vulgaris, guttate psoriasis, inverse psoriasis, pustular psoriasis, and erythrodermic psoriasis. Psoriasis vulgaris, being the most prevalent type, accounts for approximately 90% of cases.Citation2 The disease is characterized by frequent recurrences, significantly impacting the physical and psychological well-being of those affected, causing considerable suffering, and placing a substantial economic burden on the public health system.Citation3 Despite the unclear pathogenesis of psoriasis, it is closely associated with factors such as immune system abnormalities, genetic factors, abnormal proliferation of skin cells, and environmental influences. In particular, the abnormal proliferation of skin cells plays a crucial role in the onset and progression of psoriasis.Citation4
Autophagy is a lysosomal-mediated catabolic system that is commonly found in eukaryotic cells and plays a crucial role in cell growth and metabolism, proliferation and differentiation, and inflammatory regulation.Citation5 Dysregulated autophagy has been associated with a variety of diseases due to its fundamental role in maintaining cellular homeostasis, including cancer, metabolic disorders, neurodegenerative conditions, cardiovascular and hepatic diseases, infections, and autoimmune disorders.Citation6 Moreover, in recent years, an increasing number of studies have identified the role of autophagy in the pathogenesis of skin diseases.Citation7,Citation8 The immune cells in psoriatic lesions produce excessive amounts of pro-inflammatory factors, which contribute to chronic inflammation and KC proliferation. Autophagy-related protein ATG16L1 deficiency affects the upstream autophagy mechanism, leading to the production of cytokines, which further results in the accumulation of damaged proteins and organelles, triggering cell death, tissue damage, and inflammation.Citation9 However, the specific mechanisms regulating autophagy require further investigation.
A growing body of evidence indicates that micro-RNAs (miRNAs) play a significant role in autophagy regulation.Citation10 It plays a significant role in regulating a variety of aspects of disease progression, including proliferation and apoptosis of disease-related cells, the release of inflammatory factors, oxidative stress, and signaling pathways.Citation11 Under stressful conditions, certain miRNAs can modulate autophagy by altering the levels of critical autophagy-related proteins within the cells. MiR-20 is known to be significantly expressed in numerous cancer cells, including small cell lung cancer, prostate cancer, and gastric cancer. Notably, recent studies have demonstrated its involvement in platinum resistance in ovarian cancerCitation12 and its abnormal expression in Hashimoto’s thyroiditisCitation13 and β-thalassemia in elderly breast cancer patients.Citation14 Exploring the mechanism of action and function of specific miRNAs related to psoriasis may provide new clues for the treatment of psoriasis.Citation11
At present, the main drugs for patients with psoriasis are corticosteroids, vitamin D analogues, calcineurin inhibitors, and keratolytic. In addition, biological therapy is typically reserved for severe cases due to its high cost and may not be suitable for long-term treatment. Additionally, dermatologists and patients are also looking for safer and more cost-effective treatment options for psoriasis.Citation15 Psoriasis Optimization Formula (PSORI-CM01) is derived from the “Yinxieling” prescription for the treatment of psoriasis, which has been clinically validated and was developed by Professor Guowei Xuan, a national TCM master. Over 10 years ago, Professor Chuanjian Lu’s team improved PSORI-CM01, and it has since been validated through clinical trials and reliable experimentation.Citation16,Citation17 It is made up of seven herbs, including Rhizoma Curcumae (E Zhu, Chinese), Sarcandra glabra (Zhong-Jie Feng, Chinese), Mume Fructus (Wu Mei, Chinese), Rhizoma Smilacis Glabrae (Tu-Fu Ling, Chinese), Radix Arnebiae (Zi Cao, Chinese), Radix Paeoniae Rubra (Chi Shao, Chinese), and Glycyrrhiza uralensis Fisch (Gan Cao, Chinese), and their ratio is 3:5:5:5:2:3:2.Citation16,Citation18 These herbs have the ability to nourish blood, moisten dryness, detoxifying and resolving blood stasis.Citation19 In our previous research on the PSORI-CM01 intervention for psoriasis patients, the PASI score was improved by 50% in 92.7% of patients. The combination of all the herbs together can promote blood circulation, dispel blood stasis and eliminate blemishes. The results of the study speculate that PSORI-CM01 may improve its pathological manifestations by activating blood circulation.Citation20
Our study utilized a network pharmacology approach to investigate the specific mechanism of PSORI-CM01 in psoriasis treatment and found differences in miRNA expression by miRNA microarray analysis. Additionally, we examined the miRNA expression profiles of peripheral blood mononuclear cells (PBMCs) from patients with psoriasis, as well as PBMCs from a control group before and after treatment with PSORI-CM01.
Materials and Methods
Materials and Reagents
PSORI-CM01 was obtained from the Tianjiang Pharmaceutical Co.Ltd. (Jiangsu province, China, Batch No. 20140815). Ficoll was purchased from Absin Science Co., Ltd (shanghai, China). TRIzol RNA reagent was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco Laboratories (Grand Island, NY, US). 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) was purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA).
Collecting Compounds of PSORI-CM01 and Their Targets
The Traditional Chinese Medicine Systems Pharmacology Database (TCMSP; http://lsp.nwu.edu.cn/) was used to identify the chemical components of PSORI-CM01; the candidate compounds with oral bioavailability (OB) 30% and drug-likeness (DL) 0.18 were identified as the active compounds, indicating the compounds had chemical stability and solubility.
Collecting Targets of Psoriasis and Venn Analysis
The targets of psoriasis were discovered using the Genecards Database (https://www.genecards.org/) and DisGeNET database (https://www.disgenet.org/), and a relevance score of at least 5 was necessary. Then, a Venn diagram was used to screen the intersection targets between psoriasis targets and targets of PSORI-CM01 compounds. The places where PSORI-CM01 chemicals might have an effect on psoriasis pharmacologically were identified as the intersecting targets.
Herbs-Compounds-Targets Networks Construction
In order to generate a network map that displays the distribution characteristics of PSORI-CM01 active compounds and their targets, the PSORI-CM01 herbs, active compounds, and pertinent targets were uploaded to the Cytoscape 3.1.1 program. The important active compounds were then screened by network topology analysis according to the connection degree value.
Protein-Protein Interaction Analysis
The Search Tool for the Retrieval of Interacting Genes/Proteins database (STRING, http://string-db.org; version 11.0) was used to obtain the Protein-Protein Interaction (PPI) network for the species “Homo sapiens”, and Cytoscape 3.1.1 software was then used for network topology analysis to screen the significant targets (Hub-Target).
Key Target-miRNA Regulatory Network Analysis
The core target-miRNA regulatory network was constructed using data imported into Cytoscape 3.1.1 software, with regulated miRNAs chosen based on the highest node degree. The MiRNet database (https://www.mirnet.ca/), which contains information about miRNAs and regulated genes, was used to collect the key target-miRNA regulatory network.
Gene Ontology Annotation and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis
DAVID Database (https://david.ncifcrf.gov) was used to undertake a Gene Ontology Annotation Enrichment Analysis (GO) of the screened hub targets, including cellular component (CC), molecular function (MF), and biological process (BP). The “org.Hs.eg.db” package in R software (Version 3.6.3) was used to perform the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The OMICShare tool web platform (http://www.omicshare.com/tools) was used to create enrichment bubbles.
Molecular Docking on the Interaction Between Key Compounds and the Hub Targets
The protein structures of the screened targets were preprocessed using the Discovery studio software (Version 2.5) after being retrieved from the PDB Database (https://www.rcsb.org/). The structural data of the main active substances were downloaded and stored in *SDF format from the PubChem Database (https://pubchem.ncbi.nlm.nih.gov/). Hydrogens were added to the aforementioned structures, and Discovery studio 2.5 software estimated the charges. Finally, after docking molecules and proteins using Discovery studio 2.5 software, the model with the greatest binding score was chosen, and Discovery studio 2.5 software carried out structural visualizations of the chosen model.
Preparation of PSORI-CM01
This study’s PSORI-CM01 included seven ingredients of Chinese herbs (). All plant materials were obtained from the Tianjiang Pharmaceutical Co.Ltd. (Jiangyin, Jiangsu province, China). The Guangdong Provincial Laboratory approved the quality control of the PSORI-CM01 granule by coupling liquid chromatography with an LTQ Orbitrap mass spectrometer.Citation18
Table 1 Constituents of PSORI-CM01
Clinical Study
Blood samples were taken from psoriasis patients and healthy participants at the Guangdong Provincial Hospital of Chinese Medicine’s Department of Dermatology. A clinical trial that was double-blind, randomized, and double dummy was used for the investigation. The previous study’s findings were compatible with the diagnostic criteria for the psoriasis. Psoriatic patients’ demographics and the PASI (Psoriasis Area and Severity Index) were covered in detail in . The human investigation was conducted in compliance with the Code of Ethics of the World Medical Association (Trial registration ChiCTR-TRC-14005185 was registered on August 8, 2014). The Clinical Research Ethics Committee of the Guangdong Provincial Hospital of Traditional Chinese Medicine’s ethical guidelines were followed in all procedures involving human subjects (Ethical approval No. B2014-026-01). An informed consent form was initially needed of all patients who were enrolled. The aim of the study and available treatments were then discussed by the researchers. Following that, the samples were gathered.
Table 2 Clinical Serum Samples for miRNA Analysis
Group Intervention
The previously stated treatments and procedures were described in detail.Citation20 Participants in the PSORI-CM01 treatment group consumed 5.5 g of PSORI-CM01 granules twice daily after meals, whereas those in the control group consumed 5.5 g of placebo granules three times daily for a period of 12 weeks.
Sample Collection and Processing
10 mL of blood in heparin-anticoagulant tubes were drawn from each subject’s cubital vein. Lymphocyte separation solution was used to isolate PBMCs using the Ficoll density gradient centrifugation method.
RNA Extraction
The total RNA was extracted using the TRIzol reagent per the manufacturer’s instructions, and then purified using the miRNeasy Micro Kit and RNase-Free DNase Set. At each RNA extraction, total RNA sample was used as the positive control, and RNase-free water was used as the negative control. We evaluated the integrity of extracted total RNA using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
MicroRNA Chip Scanning
12 psoriasis patients and 9 healthy controls had their PBMCs samples taken. Prior to and following the PSORI-CM01 intervention, 6 blood samples were collected.21 of the PBMCs samples were then collected and put through an Agilent microarray chip analysis (Human miRNA, Release 21.0). Total RNA was quantified via a spectrophotometer (NanoDrop ND-2000, Thermo Scientific, MA, USA). Following reverse transcription to cDNA, the samples were purified and labeled with cyanine 3-CTP. In the microarray chip, the samples were then hybridized. Finally, the Agilent Scanner G2505C was used to scan the microarray expression (Agilent Technologies, Santa Clara, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
According to the manufacturer’s instructions, SuperScript IV Reverse Transcriptase (Thermo Fisher, Dalian, China) reverse transcribed the RNA samples into cDNA. The following PCR amplification settings were used: 40 cycles of 94 °C for 20s and 60 °C for 34s during the initial 15 min of denaturation at 95 °C. SYBR Green PCR Kits were used to look at the expression of microRNAs.
Cell Culture
The Cell Culture Unit of Shanghai Science Academy provided the HaCaT cell line with its source (Shanghai, China). The cells were cultivated in DMEM (Gibco, USA) supplemented with 10% FBS (Gibco, USA) at 37 °C in a humidified environment with 5% CO2.
Cell Transfection
The mimics and inhibitor of MiR-20a-3p, besides their negative controls were designed and synthesized by RiboBio Inc. (Ribobio, Guangzhou, China). The sequences of miR-20a-3p mimics: sense strand: 5′ ACUGCAUUAUGAGCACUUAAAG 3′ and antisense strand: 5′ CUUUAAGUGCUCAUAAUGCAGU 3′, miR-20a-3p inhibitor is single-strand: 5′ CUUUAAGUGCUCAUAAUGCAGU 3′. The above-mentioned steps for cell transfection were carried out using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. HaCaT cells were transfected with miR-20a-3p mimics, with the final concentration of its negative control being at 100 nM, or miR-20a-3p inhibitor, with the final concentration of its negative control being at 200 nM.
MTT Assay
The MTT assay was used to measure cell viability. Tumor necrosis factor (TNF) was added at a concentration of 20 ng/mL after the cells were seeded into 96-well plates with 10% FBS in MEM at a density of 5000 cells per well. MiR-20a-3p mimics (control mimics) or inhibitors (control inhibitors) were added to the wells after 12 hours of incubation, and then the wells were left to sit for 24 hours. The 96-well plates were incubated for 4 hours after the MTT solution had been applied in 10 L to each well. Finally, 0.04 N HCl in isopropyl alcohol was used to lyse the cells, and the absorbance in 96-well plates was measured at 570 nm.
Statistical Analysis
SPSS 19.0 software (Chicago, IL, USA) was applied to statistical analysis, which was visualized using GraphPad Prism 5.0 (La Jolla, CA, USA). Results were expressed as means ± standard deviation (SD). For data comparisons between groups, one-way ANOVA was employed, then Duncan’s test, P< 0.05 was deemed statistically significant.
Results
Collecting of Chemical Ingredients and Their Targets of PSORI-CM01
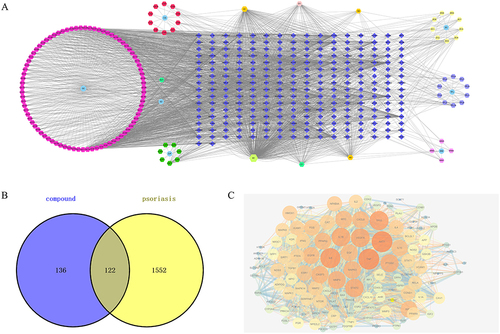
According to the TCMSP Database, 169 chemical ingredients of PSORI-CM01 were collected according to OB ≥ 30% and DL ≥ 0.18, including 29 in Radix Padoniae Rubra, 10 in Herba Sarcandrae, 3 in Curcuma phaeocaulis Valeton, 8 in Dark Plum Fruit, 12 in Lithospermum erythrorhizon Sieb. et Zucc, 15 in Rhizoma Smilacis Glabrae, and 92 in Glycyrrhiza uralensis Fisch. Then, 154 active compounds with 258 targets were selected after removing duplicate targets. In order to screen the hub active components of PSORI-CM01, a network of herb-compounds-targets was constructed. This network, which has 420 nodes and 2194 edges with an average degree of 10.45, is depicted in . Nine hub components in all were discovered, and their node degrees were three times higher than the typical node degree, including 139 in MOL000098, 65 in MOL000422, 40 in MOL004328, 40 in MOL003896, 39 in MOL000358, 35 in MOL004373, 34 in MOL002565 and 33 in MOL000392. In the network’s findings, these 9 hub components were identified as potential compounds of PSORI-CM01 that could be used for the treatment of psoriasis.
Figure 1 Activated Chemical Composition, targets of PSORI-CM01 and target of psoriasis. (A) Herb-compounds-targets network of PSORI-CM01; (B) Venn analysis of targets between targets of PSORI-CM01 compounds and target of psoriasis; (C) protein–protein interaction (PPI) network of intersection targets.
Identifying Targets of PSORI-CM01 Acting on Psoriasis
1308 targets of psoriasis and 906 targets of psoriasis were collected from DisGeNET database and GeneCards database with relevance scores no less than 5, respectively. A total of 1674 psoriasis diseases targets were selected after duplication. Subsequently, using Venn analysis, 122 intersection targets between PSORI-CM01 candidate targets and psoriasis targets were screened (). The intersecting targets were believed to be the targets operating on psoriasis from the PSORI-CM01 candidate targets.
Protein–Protein Interaction Analysis
As shown in , 122 intersection targets were analyzed by PPI network analysis to illustrate the interaction between them. There were 122 nodes and 2347 edges in this network. The shared targets had an average degree node of 38.46. Nine hub targets in all were found, and they had node degrees that were twice as high as the average node degree in this network (94 in TNF, 94 in AKT1, 90 in IL6, 85 in TP53, 83 in IL1B, 83 in VEGFA, 81 in STAT3, 79 in PTGS2 and 78 in MMP9).
GO and KEGG Enrichment Analysis of PSORI-CM01
The DAVID Database was used to conduct GO enrichment analysis to look at the enrichment of the related targets in biological processes (BP), cellular components (CC), and molecular function (MF). The primary functional modules in BP, as depicted in , included cell proliferation, apoptosis, aging, immunological response, autophagy, and ERK1 and ERK2 cascade, among other processes. The related targets enriched in CC comprised the cell surface, extracellular space, mitochondria, plasma membrane, cytosol, nucleus, and cytoplasm (). The key components of MF enrichment were protein binding, enzyme binding, DNA binding, and receptor binding (). Previous studies have validated that cell proliferation, apoptotic process, aging, immune response and autophagy participates the pathological process of psoriasis, but there is no literature exploring whether PSORI-CM01 exert pharmacological effect via these biological processes.
Figure 2 Go function and KEGG pathway enrichment analysis of PSORI-CM01 in the treatment of psoriasis. (A) Biological processes (BP); (B) cellular component (CC); (C) molecular function (MF); (D) KEGG pathway enrichment of the intersection targets; (E) Categorization of KEGG pathway enrichment; (F) gene-concept network analysis on KEGG enrichment; (G) enrichment map analysis on KEGG enrichment.
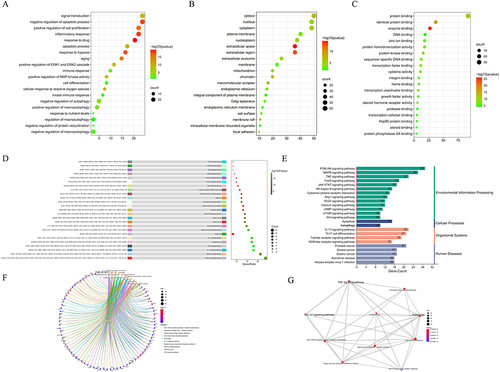
To investigate the molecular mechanism of PSORI-CM01 involving psoriasis therapy, KEGG enrichment analysis was employed the mechanisms mainly included PI3K-Akt signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, TNF signaling pathway, and Autophagy etc (). suggested that intersection targets mainly participated in the environmental information process, which was in consistent with the pathology of psoriasis. A study of the gene-concept network () and enrichment map () revealed that the PSORI-CM01 may have been caused by the autophagy signaling pathway.
Molecular Docking Results
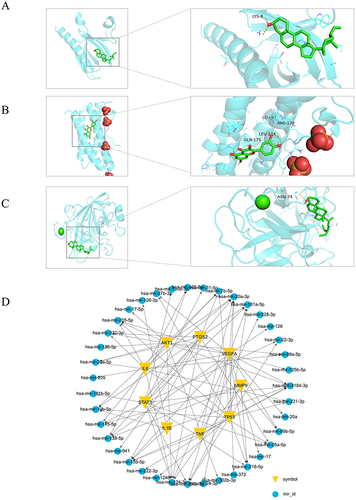
According to network topology analysis, MOL000098, MOL000422, MOL004328, MOL003896, MOL000358, MOL004373, MOL002565 and MOL000392 were identified as the key active components of PSORI-CM01. The major targets of PSORI-CM01 for treating psoriasis include TNF, AKT1, IL6, TP53, IL1B, VEGFA, STAT3, PTGS2, and MMP9. To better understand the interactions between the key targets and active ingredient, these targets were docked with the discovered component. According to the findings (), the three medicines MOL000098, MOL000422, and MOL004328 may have a pharmacological effect on psoriasis by targeting IL6, TP53, and STAT3 respectively.
Key Target-miRNA Regulatory Network
As TNF, AKT1, IL6, TP53, IL1B, VEGFA, STAT3, PTGS2 and MMP9 were identified as the key target of PSORI-CM01 involving the treatment of psoriasis. Target-miRNA regulatory network showed that these key targets were mainly associated with regulation of hsa-mir-20a-3p, hsa-mir-155-5p, has-miR-3184-3p, hsa-mir-328-3p and hsa-mir-124-3p (Degree≥6), as showed in .
Differentially Expressed miRNAs Between Groups
Before treatment, compared with the control group by an FDR_BH < 0.05 and fold change > 2, we found that 184 miRNAs were differentially expressed in the PSORI-CM01 group (0 week), including 81 up-regulated and 103 down-regulated. After PSORI-CM01 treatment (12 weeks), 284 differentially expressed miRNAs were observed in comparison with healthy control. There were 96 up-regulated and 188 down-regulated. However, only 21 miRNAs were differentially expressed before and after treatment with PSORI-CM01, of which 5 were up-regulated, 16 were down-regulated ().
Table 3 Analysis of Differentially Expressed miRNAs Between Groups
Differences in miRNAs Expression Profiles in the PSORI-CM01 and the Control Groups
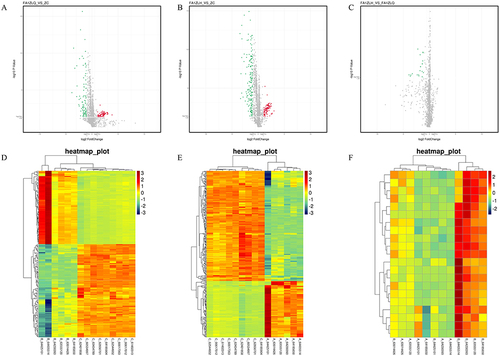
The differences in miRNA expression between the PSORI-CM01 group and the control group were shown in heat maps and volcanic maps () before and after treatment. In the volcano plot, red represented up-regulated genes, and green was the downregulation. The larger the Fold Change value, the more significant the difference in miRNA expression between groups. In the heatmap, each small square meant a miRNA, and its color indicated the level of miRNA expression. The darker the color, the higher the expression level (red for up-regulation, green for down-regulation). Each column represented the expression level of all miRNAs in each sample, whereas each row represented the expression level of each miRNA in various samples.
Figure 4 Unsupervised hierarchical cluster analysis of differentially expressed miRNAs before and after PSORI-CM01 treatment. (A) Volcanic map of PSORI-CM01 group before treatment versus the control group; (B) Volcanic map of PSORI-CM01 group after treatment versus the control group; (C) Volcanic map of PSORI-CM01 group after treatment versus PSORI-CM01 group before treatment; (D) Heat map of PSORI-CM01 group before treatment versus the control group; (E) Heat map of PSORI-CM01 group after treatment versus the control group; (F) Heat map of PSORI-CM01 group after treatment versus PSORI-CM01 group before treatment.
Pathway Enrichment Analysis for Differentially Expressed miRNAs
In order to identify the biological function or signalling pathway which may be influenced by the differentially expressed miRNAs, we set up a KEGG enrichment analysis through the David Online tools. We unearthed that the top 10 critical signal pathways targeted by PSORI-CM01 were as follows: pluripotency of stem cells signalling, axon guidance, MARK pathway, proteoglycans in cancer, breast cancer, PI3K-Akt signalling, Wnt pathway, focal adhesion, mTOR signalling and gastric cancer ( and ).
Figure 5 Pathway enrichment analysis for differentially expressed miRNAs and Verification of selected microRNAs. (A and B) Pathway enrichment analysis for differentially expressed miRNAs. (C) The miRNAs validation of miR-20a-3p, miR-3184-3p, miR-487a-3p, miR-4793-3p, miR-3653-3p, miR-5681b and miR-26a-5p by qRT-PCR. (*P < 0.05, **P < 0.01 vs PSORI-CM01 group 0-week group; #P < 0.05, ##P < 0.01 vs PSORI-CM01 group 12 weeks group).
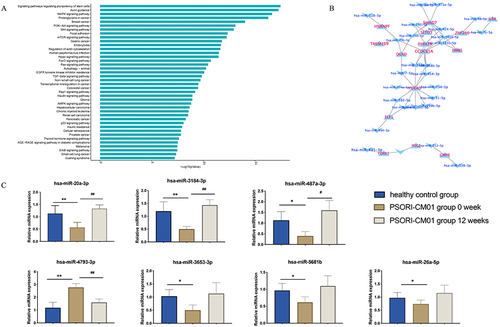
Verification of Selected microRNAs
Seven differentially expressed miRNAs including miR-20a-3p, miR-3184-3p, miR-487a-3p, miR-4793-3p, miR-3653-3p, miR-5681b and miR-26a-5p were found through microarray analysis, which were validated further by qRT-PCR (). In this study, we observed changes in the expression levels of specific microRNAs following a 12-week treatment with PSORI-CM01. Among these microRNAs, miR-20a-3p, miR-3184-3p, miR-487a-3p, miR-3653-3p, miR-5681b, and miR-26a-5p exhibited an upregulation, while miR-4793-3p showed a downregulation.
MiR-20a-3p Inhibits TNF-ɑ Induced Keratinocyte Proliferation
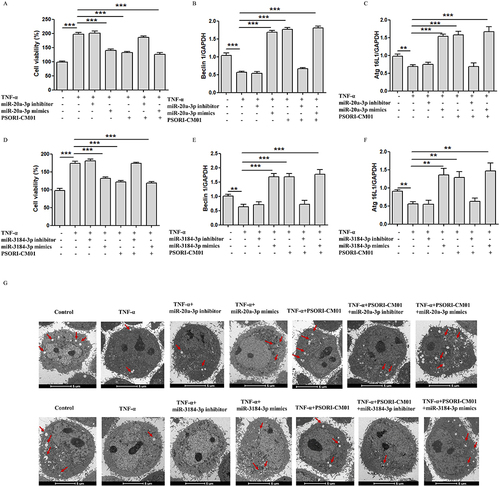
In this study, we further investigated the induction of autophagy and the inhibition of cell proliferation by PSORI-CM01. To validate this effect, we conducted intervention experiments using inhibitors and mimics of miR-20a-3p and miR-3184-3p. The Results demonstrated that under the stimulation of miR-20a-3p and miR-3184-3p inhibitors, the autophagic process was suppressed, but when PSORI-CM01 was administered, no significant change in autophagy levels was observed. However, under the stimulation of miR-20a-3p and miR-3184-3p mimics and with the administration of PSORI-CM01, the autophagic process was promoted, and cell proliferation was effectively inhibited (). Therefore, we propose that PSORI-CM01 treatment in psoriasis involves regulating the expression of miR-20a-3p and miR-3184-3p to promote the autophagic process while inhibiting cell proliferation.
Figure 6 miR-20a-3p and miR-3184-3p induces autophagy and inhibits proliferation of TNF-ɑ induced keratinocytes. (A) Inhibitors and mimics of miR-20a-3p on HaCaT cells were detected by MTT; (B and C) Expression of Beclin1 and ATG16L1 evaluated by qRT-PCR after inhibitors and mimics of miR-20a-3p; (D) Inhibitors and mimics of miR-3184-3p on HaCaT cells were detected by MTT; (E and F) Expression of Beclin1 and ATG16L1 evaluated by qRT-PCR after inhibitors and mimics of miR-3184-3p; (G) Autophagosomes in HacaT was observed by TEM (Scale bar = 5 μm; red arrow: autophagosome); (**P < 0.01, ***P < 0.001 vs TNF-α).
Discussion
Psoriasis is a chronic autoimmune inflammatory disease caused by various factors that result in the excessive proliferation and abnormal differentiation of epidermal keratinocytes (KCs) and abnormal T-cell function. KCs have the ability to release a variety of cytokines that regulate growth, differentiation, and inflammation, thereby activating and regulating the skin’s immune response.Citation21 Although current treatment options such as topical agents, oral agents, biological agents, light therapy, and combination therapy can induce remission in most psoriasis patients, these therapeutic drugs cannot fully cure psoriasis, and the condition remains prone to relapse.Citation22 Additionally, prolonged use of these drugs often leads to significant adverse reactions in patients. Currently, there is no curative treatment available for psoriasis, and the primary aim of treatment is to control the disease, minimize skin damage, prevent disease recurrence, and enhance the quality of life for patients. Therefore, it is urgent to develop a new therapeutic strategy with higher efficacy and fewer side effects to treat psoriasis, and Traditional Chinese medicine plays an important and active role in the treatment of the condition.Citation23
The PSORI-CM01 is a clinical treatment for psoriasis developed by Professor Xuan Guowei, a renowned medical expert. The preliminary findings of our research have also demonstrated the potential of PSORI-CM01 in ameliorating the symptoms of psoriasis, which includes significant reductions in PASI scores, BSA, VAS, and DLQI.Citation24 To gain deeper insights into the specific mechanisms through which PSORI-CM01 treats psoriasis, we employed network pharmacology, and our analysis revealed that PSORI-CM01 exerts its therapeutic effects by modulating autophagy, cellular apoptosis, cellular proliferation, and anti-inflammatory processes. Target-miRNA regulatory network showed that these key targets were mainly associated with regulation of hsa-miR-20a-3p, hsa-miR-155-5p, has-miR-3184-3p, hsa-miR-328-3p and hsa-miR-124-3p. Forty-three patients with psoriasis provided randomized blood samples, and subsequently, a double-blind and double-dummy clinical trial was conducted to analyze microRNA expression profiles. The microRNA expression profile results indicated that, in comparison to the healthy control group, the PSORI-CM01 treatment group exhibited up-regulation of 5 genes and down-regulation of 16 genes. Among these genes, miR-20a-3p and miR-3184-3p were the primary differentially expressed microRNAs. Additionally, they were significantly enriched in the signaling pathways associated with autophagy, cellular apoptosis, cellular proliferation, and anti-inflammation processes.
Previous studies have shown that miR-20a is crucial for the development of several cancers, including lung cancer cells,Citation25 breast cancer cells,Citation26 and prostate cancer cells.Citation27 MiR-20a-3p is another mature microRNA product produced from the precursor miR-20a stem-loop structure. Traditionally, researchers believed it to be non-functional. However, growing evidence suggests that miR-20a-3p can indeed play functional roles in normal cellular behavior.Citation28 In psoriasis and IL-22-induced keratinocyte proliferation, Li R et alCitation12 investigated at the potential functional involvement of miR-20a-3p. They found that miR-20a-3p was down-regulated in both human keratinocyte cell line HaCaT and psoriatic lesions. MiR-20a-3p is essential for psoriasis, as shown by the possibility that keratinocyte hyperproliferation and aberrant apoptosis result from miR-20a-3p deletion in psoriasis.
Currently, majority of research have shown that miR-3184-3p is closely related to tumorCitation29,Citation30 and cardiovascular related aspects.Citation31 Xu H et alCitation29 observed that miR-3184-3p was enriched in cerebrospinal fluid (CSF) exosomes of glioma patients but downregulated after tumor resection. They showed that miR-3184-3p inhibits apoptosis in glioma cells while directly promoting proliferation, migration, and invasion. These findings uncover a new mechanism and underscore the significant role of miR-3184-3p in glioma progression. Moreover, Liu et alCitation31 explored the molecular mechanism underlying the therapeutic effects of total flavonoids from Dracocephalum moldavica L. (TFDM) in vascular dementia (VaD) through bioinformatics analysis and experimental validation. Their study revealed that TFDM and its active compounds, including tilianin, luteolin, and apigenin, significantly upregulated miR-3184-3p and downregulated miR-6875-5p in oxygen-glucose deprivation (OGD)-injured cells, leading to improved cell viability.
This study employed TNF-α stimulation on HaCaT cells for in vitro research, resulting in the stimulation of inflammatory cytokine production, thereby triggering the accumulation of inflammatory cells in the dermal and epidermal layers, leading to psoriasis lesions. Psoriasis is an inflammatory skin disease characterized by abnormal epidermal keratinocyte proliferation and infiltration of inflammatory cells.Citation1 Recent research has increasingly demonstrated the critical role of autophagy in regulating the pathological processes of psoriasis.Citation8 Autophagy primarily functions by degrading excessive or aberrant keratinocytes and inflammatory mediators, which helps alleviate psoriatic skin lesions and inflammation.Citation7 Consistent with our findings, our study also revealed that PSORI-CM01 significantly promotes autophagy to inhibit cell proliferation. To further validate the results of network pharmacology and miRNA microarray analysis, we employed mimics and inhibitors of miR-20a-3p and miR-3184-3p in our research, confirming that PSORI-CM01 exerts its therapeutic effects through the mediation of cell autophagy via miR-20a-3p and miR-3184-3p.
Although the number of available drugs is continuously growing, there still exist additional limitational such as predicting side-effect profile, immunogenicity, and primary lack or secondary loss of efficacy; hence, long-term safety and efficacy are sometimes impaired. Chinese medicine has demonstrated its role in treating psoriasis by regulating various miRNAs, making it highly promising to explore its genetic perspective for psoriasis treatment.Citation32 Although the efficacy of PSORI-CM01 in the treatment of psoriasis is clear, it has the characteristics of multi-component, multi-target, multi-pathway and multi-function.Citation16 By elucidating the intricate mechanisms through which PSORI-CM01 exerts its therapeutic effects, our research contributes to the development of personalized treatment strategies tailored to the genetic makeup and specific needs of individual patients. This personalized approach holds immense promise in optimizing treatment outcomes, minimizing adverse effects, and enhancing the overall quality of life for patients battling with psoriasis.Citation33 Moreover, our findings underscore the importance of integrating traditional Chinese medicine practices into mainstream treatment protocols, offering a comprehensive and integrative approach to managing psoriasis that addresses its multifactorial nature.Citation34
Our study acknowledges limitations, primarily due to the small sample size, constrained by the availability of clinical samples. While our findings provide valuable insights, further investigation with larger sample sizes is necessary to strengthen the robustness and generalizability of our conclusions. Additionally, it’s noteworthy that autophagy, a pivotal cellular self-repair process, is governed by a network of signaling pathways, such as PI3K/AKT/mTOR, AMPK/mTOR,Citation35 and ROS pathways.Citation36 Although our study mainly focuses on the regulatory impact of PSORI-CM01 on miRNA in autophagy, there’s a recognition of the necessity to broaden our exploration to include other autophagy-regulating signaling pathways. To comprehensively understand the regulatory mechanisms behind PSORI-CM01’s therapeutic effects, future investigations should analyze its effects on these additional pathways, enabling the establishment of a more comprehensive theoretical framework and enhancing the clinical applicability of PSORI-CM01.
Conclusion
In this study, we thoroughly elucidated the therapeutic mechanism of PSORI-CM01 in treating psoriasis by employing a comprehensive approach involving network pharmacology, miRNA microarray analysis. Furthermore, we investigated the miRNA expression profiles of PBMCs in patients with psoriasis before and after treatment with PSORI-CM01 preparations, as well as in a control group. We experimentally demonstrated that PSORI-CM01 effectively regulates miR-20a-3p and miR-3184-3p, leading to increased autophagy and reduced keratinocyte proliferation. These molecular changes significantly contribute to the therapeutic effects of PSORI-CM01 against psoriasis. Overall, this study offers valuable insights into the therapeutic mechanism of PSORI-CM01 for psoriasis treatment, providing a solid foundation for further advancements in psoriasis therapy.
Abbreviations
BP, biological process; CC, including cellular component; DL, drug-likeness; OB, oral bioavailability; GO, Gene Ontology; KCs, keratinocytes; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function; miRNAs, micro-RNAs; PBMCs, peripheral blood monocytes cells; PPI, Protein-Protein Interaction; PSORI-CM01, Optimized formula of Yinxielin; PASI, psoriasis area and severity index; TEM, Transmission Electron Microscopy. Tumor necrosis factor α, (TNF)-α.
Ethics Statement
All procedures were performed in accordance with the guidelines of the Guangdong Provincial Hospital of Traditional Chinese Medicine ethics committee (B2014-026-01).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
Data Sharing Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Additional information
Funding
References
- Reid C, Griffiths CEM. Psoriasis and treatment: past, present and future aspects. Acta Derm Venereol. 2020;100(3):adv00032. doi:10.2340/00015555-3386
- Zhang P, Wu MX. A clinical review of phototherapy for psoriasis. Lasers Med Sci. 2018;33(1):173–180. doi:10.1007/s10103-017-2360-1
- Kim WB, Jerome D, Yeung J. Diagnosis and management of psoriasis. Can Fam Physician. 2017;63(4):278–285.
- Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker J. Psoriasis. Lancet. 2021;397(10281):1301–1315. doi:10.1016/S0140-6736(20)32549-6
- Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14(2):207–215. doi:10.1080/15548627.2017.1378838
- Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO j. 2021;40(19):e108863. doi:10.15252/embj.2021108863
- Guo Y, Zhang X, Wu T, Hu X, Su J, Chen X. Autophagy in Skin Diseases. Dermatology. 2019;235(5):380–389. doi:10.1159/000500470
- Hailfinger S, Schulze-Osthoff K. Impaired autophagy in psoriasis and atopic dermatitis: a new therapeutic target? J Invest Dermatol. 2021;141(12):2775–2777. doi:10.1016/j.jid.2021.06.006
- Hamaoui D, Subtil A. ATG16L1 functions in cell homeostasis beyond autophagy. Febs j. 2022;289(7):1779–1800. doi:10.1111/febs.15833
- Hawkes JE, Nguyen GH, Fujita M, et al. microRNAs in Psoriasis. J Invest Dermatol. 2016;136(2):365–371. doi:10.1038/JID.2015.409
- Mirzaei R, Zamani F, Hajibaba M, et al. The pathogenic, therapeutic and diagnostic role of exosomal microRNA in the autoimmune diseases. J Neuroimmunol. 2021;358:577640. doi:10.1016/j.jneuroim.2021.577640
- Pink RC, Samuel P, Massa D, Caley DP, Brooks SA, Carter DR. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol Oncol. 2015;137(1):143–151. doi:10.1016/j.ygyno.2014.12.042
- Zhao L, Zhou X, Shan X. Differential expression levels of plasma microRNA in Hashimoto’s disease. Gene. 2018;642:152–158. doi:10.1016/j.gene.2017.10.053
- Lai K, Jia S, Yu S, Luo J, He Y. Genome-wide analysis of aberrantly expressed lncRNAs and miRNAs with associated co-expression and ceRNA networks in β-thalassemia and hereditary persistence of fetal hemoglobin. Oncotarget. 2017;8(30):49931–49943. doi:10.18632/oncotarget.18263
- Bujoreanu FC, Bezman L, Radaschin DS, et al. Nevi, biologics for psoriasis and the risk for skin cancer: a real concern? Case presentation and short review. Exp Ther Med. 2021;22(6):1354. doi:10.3892/etm.2021.10789
- Wei JA, Han L, Lu CJ, et al. Formula PSORI-CM01 eliminates psoriasis by inhibiting the expression of keratinocyte cyclin B2. BMC Complement Altern Med. 2016;16(1):255. doi:10.1186/s12906-016-1234-6
- Han L, Sun J, Lu CJ, et al. Formula PSORI-CM01 inhibits the inflammatory cytokine and chemokine release in keratinocytes via NF-κB expression. Int Immunopharmacol. 2017;44:226–233. doi:10.1016/j.intimp.2017.01.023
- Chen SD, Lu CJ, Zhao RZ. Identification and quantitative characterization of PSORI-CM01, a Chinese medicine formula for psoriasis therapy, by liquid chromatography coupled with an LTQ Orbitrap mass spectrometer. Molecules. 2015;20(1):1594–1609. doi:10.3390/molecules20011594
- Parker S, Zhang AL, Zhang CS. Add-on effect of PSORI-CM01 to topical calcipotriol for moderate psoriasis vulgaris: a multi-center, randomized, double-blind pilot study. Clin Transl Med. 2021;11(1):e286. doi:10.1002/ctm2.286
- Deng J, Yao D, Lu C, et al. Oral Chinese herbal medicine for psoriasis vulgaris: protocol for a randomised, double-blind, double-dummy, multicentre clinical trial. BMJ Open. 2017;7(11):e014475. doi:10.1136/bmjopen-2016-014475
- Varshney P, Saini N. PI3K/AKT/mTOR activation and autophagy inhibition plays a key role in increased cholesterol during IL-17A mediated inflammatory response in psoriasis. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5 Pt A):1795–1803. doi:10.1016/j.bbadis.2018.02.003
- Tokuyama M, Mabuchi T. New treatment addressing the pathogenesis of psoriasis. Int J Mol Sci. 2020;21(20):7488. doi:10.3390/ijms21207488
- Li T, Gao S, Han W, et al. Potential effects and mechanisms of Chinese herbal medicine in the treatment of psoriasis. J Ethnopharmacol. 2022;294:115275. doi:10.1016/j.jep.2022.115275
- Yao DN, Lu CJ, Wen ZH, et al. Comparison of PSORI-CM01 granules and Yinxieling tablets for patients with chronic plaque psoriasis: a pilot study for a randomized, double-blinded, double-dummy, multicentre trial. Ann Palliat Med. 2021;10(2):2036–2047. doi:10.21037/apm-20-2575
- Shi L, Zhu W, Huang Y, et al. Cancer-associated fibroblast-derived exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin Transl Med. 2022;12(7):e989. doi:10.1002/ctm2.989
- Gao X, Qin T, Mao J, et al. PTENP1/miR-20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J Exp Clin Cancer Res. 2019;38(1):256. doi:10.1186/s13046-019-1260-6
- Stoen MJ, Andersen S, Rakaee M, et al. Overexpression of miR-20a-5p in tumor epithelium is an independent negative prognostic indicator in prostate cancer-a multi-institutional study. Cancers. 2021;13(16):4096. doi:10.3390/cancers13164096
- Li R, Qiao M, Zhao X, Yan J, Wang X, Sun Q. MiR-20a-3p regulates TGF-β1/Survivin pathway to affect keratinocytes proliferation and apoptosis by targeting SFMBT1 in vitro. Cell Signal. 2018;49:95–104. doi:10.1016/j.cellsig.2018.06.003
- Xu H, Li M, Pan Z, et al. miR-3184-3p enriched in cerebrospinal fluid exosomes contributes to progression of glioma and promotes M2-like macrophage polarization. Cancer Sci. 2022;113(8):2668–2680. doi:10.1111/cas.15372
- Shang Y, Wang L, Zhu Z, et al. Downregulation of miR-423-5p Contributes to the Radioresistance in Colorectal Cancer Cells. Front Oncol. 2020;10:582239. doi:10.3389/fonc.2020.582239
- Liu M, Shan G, Jiang H, et al. Identification of miRNA and their regulatory effects induced by total flavonoids from dracocephalum moldavica in the treatment of vascular dementia. Front Pharmacol. 2021;12:796628. doi:10.3389/fphar.2021.796628
- Lu Y, Qi Y, Yan Y, et al. Analysis of microRNA expression in peripheral blood monocytes of three Traditional Chinese Medicine (TCM) syndrome types in psoriasis patients. Chin Med. 2020;15(1):39. doi:10.1186/s13020-020-00308-y
- Camela E, Potestio L, Fabbrocini G, Pallotta S, Megna M. The holistic approach to psoriasis patients with comorbidities: the role of investigational drugs. Expert Opin Investig Drugs. 2023;32(6):537–552. doi:10.1080/13543784.2023.2219387
- Camela E, Potestio L, Fabbrocini G, Ruggiero A, Megna M. New frontiers in personalized medicine in psoriasis. Expert Opin Biol Ther. 2022;22(12):1431–1433. doi:10.1080/14712598.2022.2113872
- Zheng X, Li W, Xu H, et al. Sinomenine ester derivative inhibits glioblastoma by inducing mitochondria-dependent apoptosis and autophagy by PI3K/AKT/mTOR and AMPK/mTOR pathway. Acta Pharm Sin B. 2021;11(11):3465–3480. doi:10.1016/j.apsb.2021.05.027
- Zhou J, Li XY, Liu YJ, et al. Full-coverage regulations of autophagy by ROS: from induction to maturation. Autophagy. 2022;18(6):1240–1255. doi:10.1080/15548627.2021.1984656