Abstract
Background
Parkinson’s disease (PD) is the most prevalent movement disorder. Curcumin, a polyphenol with hydrophobic properties, has been proved against Parkinson. Our previous study suggested that curcumin’s effectiveness in treating Parkinson’s disease may be linked to the gut-brain axis, although the specific mechanism by which curcumin exerts neuroprotective effects in the brain remains unknown.
Methods
The therapeutic efficacy of curcumin was evaluated using behavioral tests, immunofluorescence of tyrosine hydroxylase (TH). Network pharmacology and transcriptomics predicted the mechanisms of curcumin in PD. Activation of the phosphatidylinositol 3-kinase PI3K/AKT pathway was confirmed by quantitative polymerase chain reaction (qPCR) and immunofluorescence.
Results
Curcumin restored the dyskinesia and dopaminergic neurons damage of MPTP-induced mice. Curcumin against Parkinson’s disease by regulating inflammation, oxidative stress, and aging. The mechanisms of these were associated with activation of PI3K / AKT pathway.
Conclusion
In conclusion, the neuroprotective mechanisms of curcumin activate PI3K / AKT pathway in Parkinson’s disease was revealed by our study.
Introduction
Parkinson’s disease (PD) is the most prevalent movement disorder. Driven principally by aging, the number of people with Parkinson disease is projected to over 12 million by 2040.Citation1 Gene mutations contribute to only a small portion of all cases, with non-genetic factors playing a significant role in combination with susceptibility genes.Citation2 Several non-genetic factors have been identified in Parkinson’s disease (PD), including viral or bacterial exposure, pesticide exposure, and changes in gut microbiota, suggesting a potential link to disease development.Citation3,Citation4 It is hypothesized that complex interactions between genes and the environment, along with the aging process, contribute to the progression of PD. Currently, there is a lack of disease-modifying therapies available, in addition to levodopa supplementation and anticholinergic therapy.Citation5 However, there is no conclusive evidence supporting the effectiveness of any specific agents in the disease-modifying.Citation5 The absence of disease-modifying treatments may be attributed to both inadequate therapeutic candidates and insufficient attention to the mechanisms of these candidates.Citation6
Curcumin, a polyphenol with hydrophobic properties, is derived from the rhizome of a plant.Citation7 It is widely consumed in Asia, either directly as a complementary treatment or as an ingredient in various food recipes.Citation8 Recently, there has been increasing interest in using curcumin to prevent Parkinson’s disease.Citation9 The mechanisms of curcumin to anti-PD may include antioxidant, Immune modulation, and the clearance of α-syn.Citation10–12 Our previous study suggested that curcumin’s effectiveness in treating Parkinson’s disease was linked to the gut-brain axis, although the specific mechanism by which curcumin exerts neuroprotective effects in the brain remains unknown.Citation13,Citation14
The PI3K/AKT was associated with some biological processes such as apoptosis, cell proliferation, and metabolism.Citation15 There is growing evidence that the activation of the PI3K/AKT signaling pathway exerts anti-Parkinson’s disease effectsCitation15,Citation16. At the same time, it is believed that curcumin can activate the PI3K/AKT.Citation17–19 However, there were no studies to confirm that curcumin exerts neuroprotective effects in Parkinson’s disease by activating PI3K/AKT in the brain.
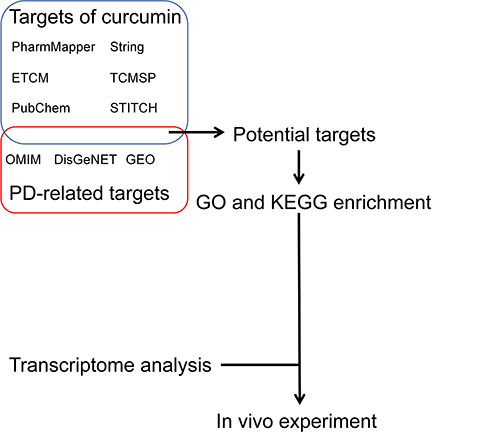
Network pharmacology is an emerging field that utilizes systems biology principles, analyzes biological system networks, and designs drug molecules that target multiple specific signaling nodes. By employing network pharmacology, effective therapeutic interventions can be predicted without the need for traditional methods, thereby expediting clinical translation.Citation20,Citation21 Transcriptomics, a highly advanced field in the post-genomic era, enables the study of mRNA expression in tissues or cells at a comprehensive level.Citation22 Our research combines these two approaches to predict how curcumin works against Parkinson’s disease. We have also confirmed in vivo that curcumin activates PI3K/AKT and has protective effects on the brain (). This study, along with our previous research, establishes a theoretical foundation for using curcumin as a disease-modifying treatment for Parkinson’s disease.Citation13,Citation14
Materials and Methods
Animals and Treatments
C57BL/6J mice (8 weeks old, male, n=30) were obtained from Hunan Slaughter Jingda Laboratory Animal Co., Ltd. Prior to the commencement of the experiments, the mice were housed in a standard environment with unlimited access to food and water. They were kept in a pathogen-free facility for one week. The animal procedures were approved by the Ethics Committee of Hainan Medical University. Carbon dioxide asphyxiation was used as a humane method to euthanize all the mice. The mice were divided into MPTP (Sigma-Aldrich, United States) + Curcumin (Macklin, Shanghai, China) group, MPTP group, and control group. To induce Parkinson’s disease in the mouse model, MPTP (2mg/mL) dissolved in sterile distilled water with 5% ethanol solution was administered through intraperitoneal injections at a dose of 20 mg/kg/3.5days for 5 weeks. Before MPTP injection 2 weeks, curcumin (100 mg/mL in DMSO to store) was given at a dose of 60 mg/kg/d for 35 days in MPTP + Curcumin group.
Behavioral Tests
All behavioral tests were conducted in a blinded fashion.
In the open field test, placing mice in a field measuring 45 × 45×50 cm3 and the mice were observed for 5 minutes using a camera. The SMART 3.0 system was used to analyze and record the results. To ensure cleanliness, the field was cleaned with alcohol before use and dried between tests.
In the pole test, the mice were given 3 days to acclimate to their surroundings before the experiment. While the test, the mice were put on a rough pole with 1 cm in diameter and 0.5 m in height with a head-up position. The time taken by the mice to climb down the pole was recorded. After 5 days of training, all mice were formally tested, and the results were analyzed to evaluate their motor function.Citation23 Ten mice of each group (n=10) were observed.
Network Pharmacology
Using the “curcumin” as a search term, Obtain the targets of curcumin in PubChem (https://pubchem.ncbi.nlm.nih.gov/), PharmMapper (http://lilabecust.cn/pharmmapper/), Targetnet (http://targetnet.scbdd.com/), TCMSP (https://old.tcmspe.com/tcmsp.php), STITCH (https://ngdc.cncb.ac.cn/databasecommons/database/id/208), ETCM (www.tcmip.cn/ETCM).Citation24 The gene names of the obtained targets were determined by querying the UniProt database (https://www.uniprot.org/). Subsequently, the target genes were combined and all duplicates were removed, resulting in a list of potential curcumin targets. To identify disease targets related to Parkinson’s disease, we utilized the disease databases DisGeNET (http://www.disgenet.org), GEO (https://www.ncbi.nlm.nih.gov/geo), and OMIM (https://www.omim.org) with the search term “Parkinson’s disease”. The overlapping targets between curcumin and Parkinson’s disease were considered as potential therapeutic target genes for curcumin in treating PD. The PPI network of the targets was achieved by STRING database (https://cn.string-db.org/). The files were downloaded, and imported into Cytoscape for visualization.
Transcriptome
Five mice of each group were sacrificed for Transcriptome. RNA extraction was performed using the TRIzol reagent. The quality and quantity of the extracted RNA were assessed using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). The integrity of the RNA was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Subsequently, libraries were prepared using the VAHTS Universal V6 RNA-seq Library Prep Kit according to the manufacturer’s protocol. The libraries were sequenced on an Illumina Novaseq 6000 platform, generating 150 bp paired-end reads. The raw reads in fastq format were processed using fastp24 to remove low quality reads, resulting in clean reads. Approximately 50 clean reads were retained for each sample for further analysis. The clean reads were then aligned to the reference genome using HISAT225. The FPKM26 of each gene was calculated and the read counts of each gene were obtained using HTSeq-count27. Principal component analysis (PCA) was performed using R (v 3.2.0) to assess the biological duplication of samples. Differential expression analysis was conducted using DESeq228, with a threshold of Q value < 0.05 and foldchange > 2 to determine significantly differentially expressed genes (DEGs). Hierarchical cluster analysis of DEGs was performed using R (v 3.2.0) to visualize the expression patterns of genes in different groups and samples. We would like to acknowledge the assistance of OE Biotech, Inc., (Shanghai, China) in sequencing and bioinformatics analysis.
GO and KEGG Analyses
In order to explore the mechanisms involved in the treatment of PD with curcumin, the targets in the network pharmacology or transcriptomics were analyzed using the DAVID (https://david.ncifcrf.gov/) for visualization, integration and annotation. This involved functional annotation, including gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis.Citation25,Citation26 The analysis was limited to the species “Homo sapiens” and a significance level of P < 0.05. The results of the GO and KEGG enrichment analysis were then visualized using bioinformatics tools.
Immunofluorescence
One-half of the brain of 5 mice per group were subjected to Immunofluorescence (n=5). The brain was dehydrated, OCT embedded, and sectioned (25 µm). The sections were blocked, incubated overnight at 4°C with primary antibodies, and then Sections were rinsed in PBS and incubated in Cy3 or FITC conjugated donkey anti-rabbit IgG. Then, sections were again rinsed by PBS, posed on slides, coverslipped, and observed by confocal microscopy. The distribution of immunoreactive cells was analyzed using digitized images. The other antibodies from Servicebio were used: anti-TH antibody, anti-PI3K antibody, anti-AKT antibody, FITC conjugated donkey anti-rabbit IgG, Cy3-conjugated donkey anti-rabbit IgG, and DAPI staining reagent.
Quantitative Real-Time PCR
The other half of the brain was used for PCR (n=5). We utilized the Eastep Super Total RNA Extraction Kit (Promega, Shanghai, China) to extract RNA from mouse brain tissue. The manufacturer’s instructions were followed for this process. Subsequently, 2µg of RNA was reverse transcribed into cDNA using the YEASEN Biotechnology kit (Shanghai, China). The target genes were amplified using the qPCR system (Quantagene q225-0304, Kubo Tech Co, Zurich, Switzerland) with the cycling conditions: pre-denaturation at 95 °C for 5 min, denaturation at 95 °C for 10s, annealing at 60 °C for 30s, and 40 cycles. The 2−(∆∆Ct) method was employed to analyze the relative mRNA expression. Primers were designed from PubMed and synthesized by Beijing Prime Biotechnology Co. (Beijing, China). Refer to for the list of primers used.
Table 1 Primer Sequences
Statistical Analyses
The data were expressed as mean values ± standard deviation (SD). Behavioral data are non-parametric and were analyzed using a Kruskal Wallis. Statistical comparisons between the groups were conducted using Student’s t-test or one- or two-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. A significance level of p < 0.05 was used to determine the statistical significance. All statistical analyses were performed using SPSS software.
Results
The Neuroprotective of Curcumin in MPTP-Induced Mouse Model
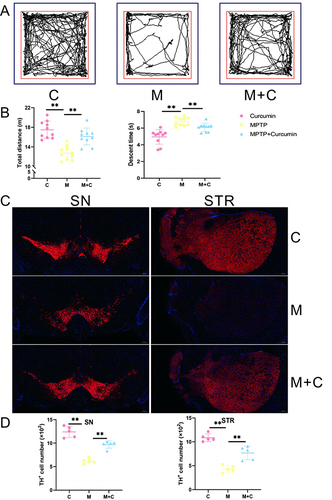
We evaluated the neuroprotective effects of curcumin in the MPTP-induced mouse model of PD. Curcumin restored the behavioral performance of MPTP-induced mice. The representative tracks in the open field test are shown in . Quantitative analyses further confirmed that the total distance was increased in the open field test of curcumin treated PD mice (). In pole test, a significant longer time to down was spent by the MPTP group vs the MPTP+Curcumin group and control group (). The number of tyrosine hydroxylase+ cells, a marker for dopaminergic neurons, was reduced in the SNc and striatum of PD mouse compared with control group (). Based on tyrosine hydroxylase staining, the quantitative analysis supported that the tyrosine hydroxylase+ cells in MPTP+Curcumin was considerably higher than that in the MPTP group ().
Figure 2 The neuroprotective of curcumin on MPTP-induced mouse model. (A) Representative traces of the open field test among control, MPTP, and MPTP + Curcumin groups (n = 10); (B) Quantification of the open field test and pole test among control, MPTP, and MPTP + Curcumin groups (n = 10); (C) Representative immunofluorescence images of TH-positive cells in the Substantia nigra and striatum of the three groups; (D) Quantification analysis of TH-positive cell number in Substantia nigra and Striatum. The analysis was performed using ImageJ (n = 5). Red staining represents immunolabeling for tyrosine hydroxylase. The significance is expressed as **p<0.01.
PPI Network and Key Targets for Curcumin Treatment of PD
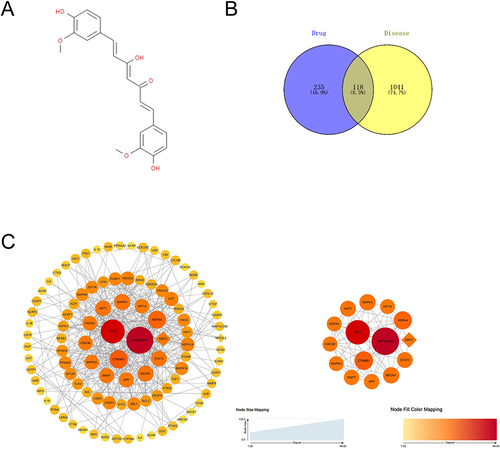
The chemical structure of curcumin was shown in . Targets of curcumin were predicted using TCMSP, STITCH, PharmMapper, ETCM, PubChem and TargetNet database. Three hundred and fifty-three genes were obtained after integrating and deleting the reduplication term (Supplementary Table 1). Parkinson Relevant targets were searched using the disease name “Parkinson’s disease”, OMIM database obtained 45 target genes, The DisGeNET database obtained 952 target genes, The GEO database obtained 162 target genes, A total of 1159 genes were obtained after integrated deduplication (Supplementary Table 2). The overlapping targets, shown by Venn diagram, were recognized as potential targets for curcumin treatment of PD (). Subsequently, PPI network for curcumin treatment of PD (), which contains 118 nodes and 393 edges. The top 15 key targets were shown as HSP90AA1, TP53, CTNNB1, HSPA4, HIF1A, MAPK1, AKT1, HSPA5, GSK3B, MAPK3, MAPT, APP, BECN1, STAT3, SIRT1. In previous studies, we confirmed the key role of SIRT1 in curcumin in inhibiting intestinal inflammation and thus against Parkinson’s disease, demonstrating the reliability of network pharmacological analysis.Citation13
Figure 3 The network pharmacology analysis of curcumin in the treatment of PD. (A) The chemical structure of curcumin. (B) Venn diagram of overlapping genes associated with curcumin in PD.(C) PPI of potential targets of curcumin to treat PD. The top 15 key targets were shown as core PPI network. Nodes are potential targets for curcumin in the treatment of PD. The larger the node, the deeper the color, the greater the Degree. The line between two nodes represents the interaction. The thicker the edge, the higher the interaction between targets.
Enrichment Analysis of PPI Network
To gain insights into the function and signaling pathways of potential targets of curcumin in treating PD, we conducted enrichment analysis of the PPI network. We utilized the online analysis system DAVID to analyze the 118 overlapping targets. Our analysis revealed that curcumin regulates a total of 120 biological processes (BP), including gene expression regulation, neuron apoptosis regulation, and phosphatidylinositol 3 kinase signaling positive regulation. Additionally, we identified 29 cellular components (CC), such as cytoplasm, extracellular components, and nucleus. Furthermore, we determined 51 molecular functions (MF), including protein binding, enzyme binding, and kinase binding. The top 20 BP, CC, and MF are depicted in .
Figure 4 Enrichment analysis of network pharmacology. (A) GO functional enrichment analysis. (B) KEGG pathway enrichment analysis. The closer the color to red, the smaller the P value, the more pronounced the enrichment.
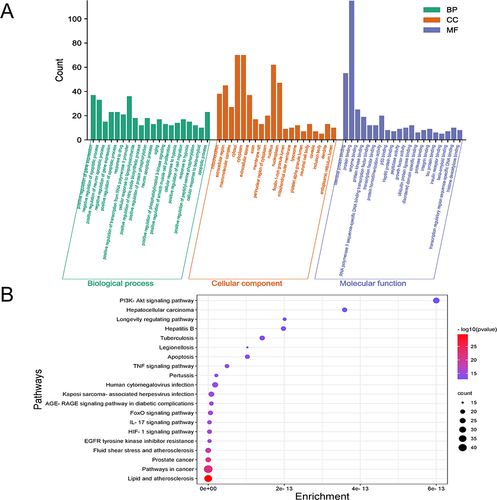
Moreover, we performed KEGG pathway enrichment analysis on the targets in the PPI network, resulting in the identification of 41 pathways. We visualized the top 20 pathways in a bubble diagram (). Our findings indicated that the PI3K/AKT pathway, apoptosis, Longevity regulating pathways, and Renin-angiotensin system are the main pathways involved. Among these, the PI3K/AKT pathway emerged as the most significant in curcumin’s therapeutic effects on PD. Therefore, the activation of the PI3K/AKT pathway may serve as a critical mechanism underlying curcumin’s efficacy in treating PD.
Transcriptome Analysis of Curcumin Treatment in MPTP-Induced Mouse Model
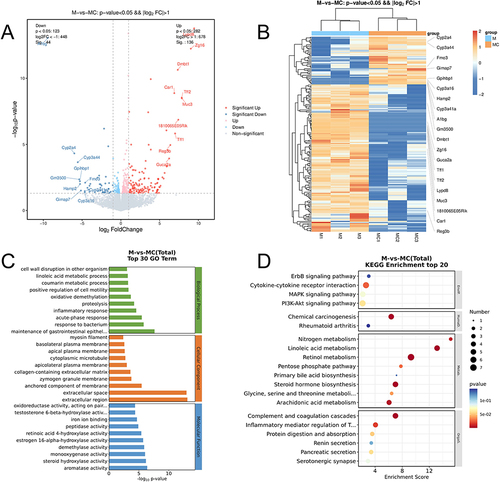
In order to validate the potential mechanisms of curcumin in treating Parkinson’s disease (PD) as obtained through network pharmacology analysis, we conducted transcriptome analysis using brain tissue from a mouse model of each groups (n=5). As a result, we identified 180 differentially expressed genes (DEGs) by comparing the MPTP+Curcumin and MPTP groups. Among these DEGs, 44 were down-regulated and 136 were up-regulated. To visually represent the overall distribution of these DEGs, we plotted volcano and heat maps ( and ). For functional enrichment analysis of the DEGs, GO and KEGG analyses were also carried out ( and ). Specifically, the insufficient data of network pharmacology and the insufficient sample used in the transcriptome, which may be responsible for the difference between the two enrichment analyses. However, the activation of PI3K/AKT pathway were consistent with the Results of network pharmacology. Thus, these transcriptome results provide support for the conclusions drawn from the network pharmacology analysis.
Figure 5 Transcriptome analysis. (A) The volcano map showed DEGs in MPTP vs MPTP+Curcumin groups. Red nodes represented up-regulated DEGs, and blue nodes represented down-regulated DEGs.(B) The heat map showed DEGs in MPTP vs MPTP+Curcumin groups.(C) GO functional enrichment analysis of DEGs. (D) KEGG pathway enrichment analysis of DEGs.
In vivo Verification
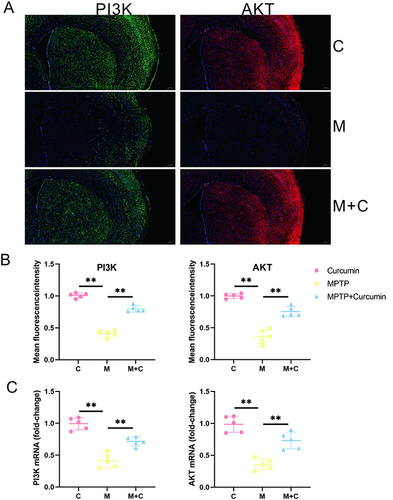
The results of network pharmacology analysis and transcriptome analysis suggested that curcumin could activate PI3K/AKT pathway and further treat PD by regulating various biological processes. The qPCR were consonant with the transcriptome, suggesting that expression levels of the PI3K/AKT pathway increased significantly after curcumin treatment in the PD mouse model ( and ). PI3K and AKT expression was confirmed by immunofluorescence stain ( and ). An immunofluorescence indicated that expression of PI3K/AKT pathway was significantly increased in MPTP+Curcumin group vs model group. The results of the quantitative analysis were consistent with the results of immunofluorescence ( and ). These results from in vivo, Transcriptome, and network pharmacology collectively support the conclusion that the role of neuroprotection of curcumin maybe achieved by activation of PI3K / AKT pathway in PD mouse model.
Figure 6 Curcumin activate PI3K/AKT pathway in the Substantia nigra. (A) Representative immunofluorescence images of pi3k-positive and akt-positive in the SN of the control, MPTP, and MPTP + Curcumin groups; (B) Quantitative analysis of mean fluorescence intensity of PI3K-positive and AKT-positive regions in the SN; (C) PI3K, and AKT mRNA levels in the SN. The significance is expressed as *p<0.05,**p<0.01. The analysis was performed using ImageJ (n = 5).
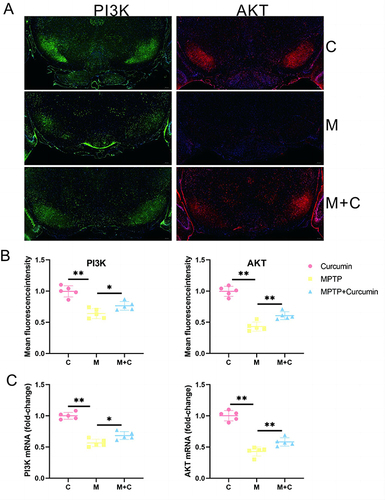
Figure 7 Curcumin activate PI3K/AKT pathway in the striatum. (A) Representative immunofluorescence images of pi3k-positive and akt-positive in the STR of the control, MPTP, and MPTP + Curcumin groups; (B) Quantitative analysis of mean fluorescence intensity of PI3K-positive and AKT-positive regions in the STR; (C) PI3K, and AKT mRNA levels in the STR. The significance is expressed as **p<0.01. The analysis was performed using ImageJ (n = 5).
Discussion
Parkinson’s disease, a severe threat to human health in aging society, is commonly treated with L-DOPA, which unfortunately has the main disadvantage of progressive loss of efficacy and L-DOPA-induced dyskinesia.Citation27,Citation28 Current treatments for Parkinson’s disease with L‐DOPA fail to stop disease progression in patients. Therefore, more therapeutic targets and treatment options need to be explored.Citation6 Studies shown that curcumin can alleviate the symptoms of Parkinson’s disease.Citation10,Citation29–32 Our previous study provides an initial understanding of curcumin in PD.Citation13,Citation14 However, further exploration was needed. To provide a basis for the application, we integrate network pharmacology, transcriptomics to reveal neuroprotective of curcumin in Parkinson’s disease.
Firstly, network pharmacology was illuminated the potential mechanism of curcumin in PD. We identified that key targets, including HSP90AA1, TP53, CTNNB1, HSPA4, HIF1A, MAPK1, AKT1, HSPA5, GSK3B, MAPK3, MAPT, APP, BECN1, STAT3, and SIRT1, were closely related to curcumin treatment of PD. GO enrichment analysis showed that the regulation of apoptosis process, response to lipopolysaccharide, positive regulation of nitric oxide biosynthetic, aging, and positive regulation of phosphatidylinositol 3-kinase signaling playing a critical role in curcumin treating PD. And KEGG enrichment analysis showed that it was involved in PI3K/AKT pathway and longevity pathway. Apoptosis, an evolutionarily conserved form of programmed cell death, is implicated in the pathophysiology of Parkinson’s disease.Citation33–35 As we know that lipopolysaccharide injection can induce symptoms similar to Parkinsonism.Citation36 This phenomenon was associated with oxidative stress.Citation37 Excessive oxidative stress was thought to play a vital role during the pathogenesis of Parkinson’s disease and much attention has been placed on lipopolysaccharide -induced nitric oxide as a key factor.Citation38,Citation39 Our analysis suggested that curcumin provided protection against LPS-induced apoptosis in Parkinson’s disease. In addition, curcumin ameliorate Parkinson’s diseases development and progression may relate with anti-aging. Notably, both GO and KEGG enrichment analyses suggested that PI3K as a key in curcumin anti-PD.
Secondly, transcriptome analysis of curcumin treatment in MPTP-induced mouse model was used to further illuminate the potential mechanism. 180 DEGs were identified, significant up DEGs in PD mouse include, Lypd8, Zg16, Dmbt1, Car1, Tiff2, Muc3, Tiff1, Reg3b, Guca2a. In the other hand, A1bg, Cyp2a4, Cyp3a44, Cyp3a16, Cyp3a41a, Fmo3, Gimap7, Gm3500, and Hamp2 was significantly down. GO enrichment analysis of transcriptome showed that maintenance of gastrointestinal epithelium, response to bacterium, and inflammatory response were involved in curcumin against PD. This finding reinforces our earlier research, indicating that curcumin depends on the gut microbiota and gut-brain axis to combat Parkinson’s disease.Citation13,Citation14 Our experimental findings validate that curcumin has the ability to protect against MPTP-induced intestinal barrier disruption in Parkinson’s disease.Citation13 There is an inextricable link between inflammation, oxidative stress, and aging, and transcriptome results further complement the underlying mechanisms of curcumin’s anti-Parkinson’s disease. Requires our attention, KEGG enrichment analysis of transcriptome also suggested that PI3K/AKT pathway may play a key role in curcumin anti-Parkinson’s disease. The PI3K/AKT signal pathway has been extensively researched and shown to have a significant impact on the central nervous system (CNS).Citation15,Citation16,Citation40 Compared with those of other signalling pathways, the components and biological functions of the PI3K/AKT pathway are complicated.Citation41 This pathway is involved in various physiological processes of the CNS, including angiogenesis, cell survival, apoptosis, autophagy, neurogenesis, neuronal proliferation and differentiation, and synaptic plasticity.Citation42 Especially in autophagy, the process of protein aggregates and damaged organelles mediated by lysosomes.Citation43,Citation44 Recent reviews have increasingly suggested that curcumin targeting the PI3K/AKT signal pathway to protect dopaminergic neurons, while also inhibiting the activation of microglia.Citation17,Citation45 As a result, curcumin has shown promising potential in the treatment and prevention of PD. Either as antioxidants or modulators of cell signalling, the influence of curcumin on oxidative and inflammation balance are key. Consistent with our enrichment analysis results, the neuroprotective of curcumin on Parkinson’s disease depend on oxidative stress, inflammation and apoptosis of nerve cells has been demonstrated.Citation45–47 However, there is no direct experimental evidence in vivo for curcumin exerts the effects of anti-oxidative stress, anti-inflammation and anti-apoptosis by activating the PI3K/AKT in PD. And AKT gene expression in the cell was the key role of anti-oxidative stress, anti-inflammation and anti-apoptosis.Citation44 Finally, our study verified the PI3K/AKT pathway activated by curcumin in Parkinson’s disease. However, to further demonstrate curcumin protects through the activation of the PI3K/AKT pathway, our future study should be incorporated an inhibitor targeting the PI3K/AKT pathway. If the inhibitor could restore this protective effect, it would be the stronger evidence.
We have employed a combination of network pharmacology, transcriptomics, and in vivo experimental evidence to uncover the neuroprotective effects of curcumin through the activation of the PI3K/AKT pathway in Parkinson’s disease. Our study not only supports the findings of recent reviews but also provides crucial evidence from bioinformatics, omics, and experiments.Citation48 This has established a solid theoretical basis for the clinical application of curcumin in treating Parkinson’s disease. Nevertheless, our research still has certain limitations, which will guide our future exploration. Firstly, our data lacks a baseline for evaluating the specific activation of the PI3K/AKT pathway solely due to curcumin administration. And our data also lacks other control conditions associated with vehicle treatment (curcumin/MPTP dilution vehicle). On the other hand, we only used MPTP-induced models, and we need more PD models to verify our conclusion, such as induced by rotenone or 6-OHDA. We will delve deeper into the role of the PI3K/AKT pathway in Parkinson’s disease, focusing on its effects against oxidative stress, inflammation, and apoptosis. Additionally, we will validate our conclusions using clinical samples and in vitro. And we also need more bioinformatics analysis techniques, to discover the underlying mechanisms, such as Reactome. In summary, our research offers innovative approaches and ideas for the identification of new therapeutic targets for PD.
Institutional Review Board Statement
This study approved by the Ethics Committee of Hainan Medical College University (2023061513). This study was retrospective, the data from public databases involved in this study does not involve additional human trials, and confirmed that the data were anonymous or confidential, ethics committee approval does not require obtaining informed consent. The welfare of the laboratory animals followed the National standard of the People’s Republic of China GB/T 35892-2018, Laboratory animal – Guideline for ethical review of animal welfare.
Disclosure
The authors declare no conflict of interest.
Data Sharing Statement
The datasets generated and analysed during the current study are available in the GEO database repository, [GSE243886].
Additional information
Funding
References
- Dorsey ER, Sherer T, Okun MS, Bloem BR. The emerging evidence of the Parkinson pandemic. J Parkinsons Dis. 2018;8(s1):S3–S8. doi:10.3233/JPD-181474
- Ye H, Robak LA, Yu M, Cykowski M, Shulman JM. Genetics and Pathogenesis of Parkinson’s Syndrome. Annu Rev Pathol. 2023;18:95–121. doi:10.1146/annurev-pathmechdis-031521-034145
- Ascherio A, Schwarzschild MA. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol. 2016;15(12):1257–1272. doi:10.1016/S1474-4422(16)30230-7
- Cryan JF, O’Riordan KJ, Sandhu K, Peterson V, Dinan TG. The gut microbiome in neurological disorders. Lancet Neurol. 2020;19(2):179–194. doi:10.1016/S1474-4422(19)30356-4
- Bloem BR, Okun MS, Klein C. Parkinson’s disease. Lancet. 2021;397(10291):2284–2303. doi:10.1016/S0140-6736(21)00218-X
- Vijiaratnam N, Simuni T, Bandmann O, Morris HR, Foltynie T. Progress towards therapies for disease modification in Parkinson’s disease. Lancet Neurol. 2021;20(7):559–572. doi:10.1016/S1474-4422(21)00061-2
- Kocaadam B, Sanlier N. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit Rev Food Sci Nutr. 2017;57(13):2889–2895. doi:10.1080/10408398.2015.1077195
- Tsuda T. Curcumin as a functional food-derived factor: degradation products, metabolites, bioactivity, and future perspectives. Food Funct. 2018;9(2):705–714. doi:10.1039/c7fo01242j
- Bhat A, Mahalakshmi AM, Ray B, et al. Benefits of curcumin in brain disorders. Biofactors. 2019;45(5):666–689. doi:10.1002/biof.1533
- Yeung AWK, Tzvetkov NT, Georgieva MG, et al. Reactive oxygen species and their impact in neurodegenerative diseases: literature landscape analysis. Antioxid Redox Signal. 2021;34(5):402–420. doi:10.1089/ars.2019.7952
- Mollazadeh H, Cicero AFG, Blesso CN, Pirro M, Majeed M, Sahebkar A. Immune modulation by curcumin: the role of interleukin-10. Crit Rev Food Sci Nutr. 2019;59(1):89–101. doi:10.1080/10408398.2017.1358139
- Pandey N, Strider J, Nolan WC, Yan SX, Galvin JE. Curcumin inhibits aggregation of alpha-synuclein. Acta Neuropathol. 2008;115(4):479–489. doi:10.1007/s00401-007-0332-4
- Zhong L, Cai B, Wang Q, Li X, Xu W, Chen T. Exploring the neuroprotective mechanism of curcumin inhibition of intestinal inflammation against Parkinson’s disease based on the gut-brain axis. Pharmaceuticals. 2022;16(1). doi:10.3390/ph16010039
- Cai B, Zhong L, Wang Q, Xu W, Li X, Chen T. Curcumin alleviates 1-methyl- 4-phenyl- 1,2,3,6-tetrahydropyridine- induced Parkinson’s disease in mice via modulating gut microbiota and short-chain fatty acids. Front Pharmacol. 2023;14:1198335. doi:10.3389/fphar.2023.1198335
- Long HZ, Cheng Y, Zhou ZW, Luo HY, Wen DD, Gao LC. PI3K/AKT signal pathway: a target of natural products in the prevention and treatment of Alzheimer’s disease and Parkinson’s disease. Front Pharmacol. 2021;12:648636. doi:10.3389/fphar.2021.648636
- Elstner M, Morris CM, Heim K, et al. Expression analysis of dopaminergic neurons in Parkinson’s disease and aging links transcriptional dysregulation of energy metabolism to cell death. Acta Neuropathol. 2011;122(1):75–86. doi:10.1007/s00401-011-0828-9
- Jin T, Zhang Y, Botchway BOA, et al. Curcumin can improve Parkinson’s disease via activating BDNF/PI3k/Akt signaling pathways. Food Chem Toxicol. 2022;164:113091. doi:10.1016/j.fct.2022.113091
- Wu J, Li M, Huang N, et al. Curcumin alleviates high-fat diet-induced nonalcoholic steatohepatitis via improving hepatic endothelial function with microbial biotransformation in rats. J Agric Food Chem. 2023;71(27):10338–10348. doi:10.1021/acs.jafc.3c01067
- Shi HH, Chen LP, Wang CC, et al. Docosahexaenoic acid-acylated curcumin diester alleviates cisplatin-induced acute kidney injury by regulating the effect of gut microbiota on the lipopolysaccharide- and trimethylamine-N-oxide-mediated PI3K/Akt/NF-kappaB signaling pathway in mice. Food Funct. 2022;13(11):6103–6117. doi:10.1039/d1fo04178a
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi:10.1038/nchembio.118
- Nogales C, Mamdouh ZM, List M, Kiel C, Casas AI, Schmidt H. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol Sci. 2022;43(2):136–150. doi:10.1016/j.tips.2021.11.004
- Dong Z, Chen Y. Transcriptomics: advances and approaches. Sci China Life Sci. 2013;56(10):960–967. doi:10.1007/s11427-013-4557-2
- Prasad EM, Hung SY. Behavioral tests in neurotoxin-induced animal models of Parkinson’s disease. Antioxidants. 2020;9(10). doi:10.3390/antiox9101007
- Kumar A, Chetia H, Sharma S, Kabiraj D, Talukdar NC, Bora U. Curcumin resource database. Database. 2015;2015:bav070. doi:10.1093/database/bav070
- The Gene Ontology, C. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi:10.1093/nar/gky1055
- Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–484. doi:10.1093/nar/gkm882
- LeWitt PA. Levodopa therapy for Parkinson’s disease: pharmacokinetics and pharmacodynamics. Mov Disord. 2015;30(1):64–72. doi:10.1002/mds.26082
- Nonnekes J, Timmer MH, de Vries NM, Rascol O, Helmich RC, Bloem BR. Unmasking levodopa resistance in Parkinson’s disease. Mov Disord. 2016;31(11):1602–1609. doi:10.1002/mds.26712
- Peng H, Li Y, Ji W, et al. Intranasal administration of self-oriented nanocarriers based on therapeutic exosomes for synergistic treatment of Parkinson’s disease. ACS Nano. 2022;16(1):869–884. doi:10.1021/acsnano.1c08473
- Liu J, Liu C, Zhang J, et al. A self-assembled alpha-synuclein nanoscavenger for Parkinson’s disease. ACS Nano. 2020;14(2):1533–1549. doi:10.1021/acsnano.9b06453
- Perez-Pi I, Evans DA, Horrocks MH, et al. alpha-Synuclein-Confocal Nanoscanning (ASYN-CONA), a bead-based assay for detecting early-stage alpha-synuclein aggregation. Anal Chem. 2019;91(9):5582–5590. doi:10.1021/acs.analchem.8b03842
- Zhang N, Yan F, Liang X, et al. Localized delivery of curcumin into brain with polysorbate 80-modified cerasomes by ultrasound-targeted microbubble destruction for improved Parkinson’s disease therapy. Theranostics. 2018;8(8):2264–2277. doi:10.7150/thno.23734
- Moujalled D, Strasser A, Liddell JR. Molecular mechanisms of cell death in neurological diseases. Cell Death Differ. 2021;28(7):2029–2044. doi:10.1038/s41418-021-00814-y
- Ghavami S, Shojaei S, Yeganeh B, et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;112:24–49. doi:10.1016/j.pneurobio.2013.10.004
- Yuan J, Amin P, Ofengeim D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat Rev Neurosci. 2019;20(1):19–33. doi:10.1038/s41583-018-0093-1
- Gu C, Wang F, Zhang YT, et al. Microglial MT1 activation inhibits LPS-induced neuroinflammation via regulation of metabolic reprogramming. Aging Cell. 2021;20(6):e13375. doi:10.1111/acel.13375
- Zhao YF, Qiong Z, Zhang JF, et al. The synergy of aging and LPS exposure in a mouse model of Parkinson’s disease. Aging Dis. 2018;9(5):785–797. doi:10.14336/AD.2017.1028
- Kumar A, Chen SH, Kadiiska MB, et al. Inducible nitric oxide synthase is key to peroxynitrite-mediated, LPS-induced protein radical formation in murine microglial BV2 cells. Free Radic Biol Med. 2014;73:51–59. doi:10.1016/j.freeradbiomed.2014.04.014
- Singh S, Das T, Ravindran A, et al. Involvement of nitric oxide in neurodegeneration: a study on the experimental models of Parkinson’s disease. Redox Rep. 2005;10(2):103–109. doi:10.1179/135100005X38842
- Wang G, Pan J, Chen SD. Kinases and kinase signaling pathways: potential therapeutic targets in Parkinson’s disease. Prog Neurobiol. 2012;98(2):207–221. doi:10.1016/j.pneurobio.2012.06.003
- Xu F, Na L, Li Y, Chen L. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci. 2020;10(1):54. doi:10.1186/s13578-020-00416-0
- Miao L, Yang L, Huang H, Liang F, Ling C, Hu Y. mTORC1 is necessary but mTORC2 and GSK3beta are inhibitory for AKT3-induced axon regeneration in the central nervous system. Elife. 2016;5:e14908. doi:10.7554/eLife.14908
- Xiang H, Zhang J, Lin C, Zhang L, Liu B, Ouyang L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm Sin B. 2020;10(4):569–581. doi:10.1016/j.apsb.2019.10.003
- Gupta R, Sahu M, Srivastava D, Tiwari S, Ambasta RK, Kumar P. Post-translational modifications: regulators of neurodegenerative proteinopathies. Ageing Res Rev. 2021;68:101336. doi:10.1016/j.arr.2021.101336
- Patel SS, Acharya A, Ray RS, Agrawal R, Raghuwanshi R, Jain P. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit Rev Food Sci Nutr. 2020;60(6):887–939. doi:10.1080/10408398.2018.1552244
- Hatami M, Abdolahi M, Soveyd N, Djalali M, Togha M, Honarvar NM. Molecular mechanisms of curcumin in neuroinflammatory disorders: a mini review of current evidences. Endocr Metab Immune Disord Drug Targets. 2019;19(3):247–258. doi:10.2174/1871530319666181129103056
- van der Merwe C, van Dyk HC, Engelbrecht L, et al. Curcumin rescues a PINK1 knock down SH-SY5Y cellular model of Parkinson’s disease from mitochondrial dysfunction and cell death. Mol Neurobiol. 2017;54(4):2752–2762. doi:10.1007/s12035-016-9843-0
- Wang XS, Zhang ZR, Zhang MM, Sun MX, Wang WW, Xie CL. Neuroprotective properties of curcumin in toxin-base animal models of Parkinson’s disease: a systematic experiment literatures review. BMC Complement Altern Med. 2017;17(1):412. doi:10.1186/s12906-017-1922-x