Abstract
Biomarkers are of tremendous importance for the prediction, diagnosis, and observation of the therapeutic success of common complex multifactorial metabolic diseases, such as type II diabetes and obesity. However, the predictive power of the traditional biomarkers used (eg, plasma metabolites and cytokines, body parameters) is apparently not sufficient for reliable monitoring of stage-dependent pathogenesis starting with the healthy state via its initiation and development to the established disease and further progression to late clinical outcomes. Moreover, the elucidation of putative considerable differences in the underlying pathogenetic pathways (eg, related to cellular/tissue origin, epigenetic and environmental effects) within the patient population and, consequently, the differentiation between individual options for disease prevention and therapy – hallmarks of personalized medicine – plays only a minor role in the traditional biomarker concept of metabolic diseases. In contrast, multidimensional and interdependent patterns of genetic, epigenetic, and phenotypic markers presumably will add a novel quality to predictive values, provided they can be followed routinely along the complete individual disease pathway with sufficient precision. These requirements may be fulfilled by small membrane vesicles, which are so-called exosomes and microvesicles (EMVs) that are released via two distinct molecular mechanisms from a wide variety of tissue and blood cells into the circulation in response to normal and stress/pathogenic conditions and are equipped with a multitude of transmembrane, soluble and glycosylphosphatidylinositol-anchored proteins, mRNAs, and microRNAs. Based on the currently available data, EMVs seem to reflect the diverse functional and dysfunctional states of the releasing cells and tissues along the complete individual pathogenetic pathways underlying metabolic diseases. A critical step in further validation of EMVs as biomarkers will rely on the identification of unequivocal correlations between critical disease states and specific EMV signatures, which in future may be determined in rapid and convenient fashion using nanoparticle-driven biosensors.
Biomarkers
Definitions
Under the direction of the National Institute for Health (NIH), the Biomarkers and Surrogate Endpoint Working Group has agreed on key definitions as well as a classification system for biomarkers as follows: (1) a biomarker represents “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes or pharmacological responses to a therapeutic intervention”; (2) a clinical endpoint represents “a characteristic or variable that reflects how a patient feels, functions, or survives”; and (3) a surrogate endpoint represents “a biomarker intended to substitute for a clinical endpoint with the potential for predicting the clinical benefit or harm (or lack of benefit or lack of harm) on the basis of epidemiological, therapeutic, pathophysiological, or other scientific evidence.”Citation1,Citation2 Thus, the very aims of the use of biomarkers, clinical endpoints, and surrogate endpoints are: (1) the improvement of the prediction, diagnosis and prognosis, particularly of common complex multifactorial diseases; and (2) the facilitation of the drug discovery and development processes.Citation3,Citation4
According to the European Medicine Agency (EMA), clinical endpoints are distinct measurements or analyses of disease characteristics observed in a study or clinical trial that reflect the effect of the therapeutic intervention.Citation5 Consequently, clinical endpoints are regarded as the most reliable indicators for disease or therapeutic responses. The definition of the US Food and Drug Administration (FDA) is much more practical and focused, stating that surrogate endpoints or biomarkers are “used in clinical trials as a substitute for a clinically meaningful endpoint, represents a direct measure of how a patient feels, functions or survives, and is expected to predict the effect of the therapy.”Citation6
In other words, a biomarker is an indicator of change and therefore fluctuates as a function of time and biological influence. Importantly, this strict definition of a biomarker excludes single nucleotide polymorphisms. Lastly, from the perspective of the pharmaceutical industry, a pragmatic definition of the term biomarker describes it as: “A measurable property that reflects the mechanism of action of the molecule based on its pharmacology [and] the pathophysiology of the disease, and [may] be useful for internal decisionmaking within a pharmaceutical company.”Citation7
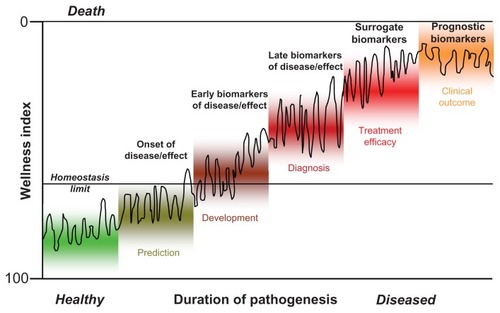
Moreover, a general classification system has been introduced for biomarkers that discriminates type 0 biomarkers, which measure the natural history of a disease and correlate over time with established clinical indicators; type I biomarkers, which indicate the intervention effect of a drug; and type II biomarkers, which are considered as surrogate endpoint biomarkers (). Both type I and type II biomarkers are ultimately aimed at monitoring the so-called wellness index, which ranges from 0 (death) to 100 (perfect homeostatic control under healthy conditions). As a function of age, environment, and genes, individuals strive to maintain a biological equilibrium of homeostatic control. Various biomarkers can be used to determine the personalized progression from homeostasis via disease initiation, disease development, and further disease progression to final disease outcome ().Citation8
Applications
Importantly, in most cases, the biomarkers for a given indication are used primarily in the pattern recognition mode on the basis of a set of unidentified markers that may be genomic, transcriptomic, proteomic, lipidomic, metabolomic, or a combination of datasets.Citation9,Citation10 A priori application of these type I-like biomarkers does not necessitate the determination of the biomarker constituent identities since the pattern or signature alone denotes the specific biological activity.Citation11 In this regard, EMVs (see “EMVs as biomarkers: General considerations”) belong to type I biomarkers. In addition, the use of type II-like biomarkers (transcripts, proteins, metabolites) for deciphering or screening the pathogenetic mechanism, prediction, diagnosis, and monitoring of common complex diseases is of critical importance and requires their structural identification and validation (for at least one of them). This approach may be supported or even substituted by the discovery of complex biomarker panels of type 0 to monitor specific disease states along the complete pathogenetic pathway for prediction, initiation, development, diagnosis, progression, regression, and treatment efficacy of the disease ().Citation1,Citation11–Citation15 The expectation of a good biomarker ranges from a molecular signature of structurally unidentified markers to a complete panel of identified biomarkers specific to the pathogenesis being studied. Importantly, the work required to establish the reliability and validity of a new biomarker should not be underestimated; it needs detailed planning for each combination of a clinical indication and a mechanism of action.Citation16 For example, type 0 biomarkers can be validated longitudinally in a well-defined patient population against a gold standard clinical assessor. In contrast, type I biomarkers should be validated in parallel with the drug candidate, and type II biomarkers must be relevant both to the mechanism of action of the drug and to the pathophysiology of the disease.
Critical questions for the discovery and development of modern drugs and the adequate use of biomarkers are how to adequately transform data into information and thence into knowledge and how to apply that to both processes. The general hope is that biomarker data will provide more predictive information and knowledge about the alterations in the biological processes induced after administration of the drug,Citation17 which should allow better predictive capability and decision-making on the part of scientists and managers involved in the drug-finding procedure.Citation18
Biomarkers for metabolic diseases
Pathogenesis
The pathogenetic mechanisms underlying metabolic diseases have previously been assumed to rely on abnormal triacylglycerol storage, which is driven by excess of energy intake and insufficient energy expenditure in the case of development of metabolic syndrome, insulin resistance, and obesity, and accompanied or followed by considerable reduction in the number and function of pancreatic β-cells in the case of development of type II diabetes.Citation19–Citation21 Chronic low-grade inflammationCitation22,Citation23 and oxidative stressCitation24,Citation25 are regarded as causally involved in both the induction and the progression of metabolic diseases. Importantly, the levels of a subset of proinflammatory adipokines, such as interleukin-2/6 (IL-2/6), tumor necrosis factor-α (TNFα), adiponectin, ferritin, and C-reactive protein (CRP) have been demonstrated by epidemiological studies to be positively correlated to the degree of insulin resistance, impaired glucose tolerance and homeostasis, and excess of adipose tissue mass.Citation26,Citation27 They have thus been regarded as the most predictive biomarkers for metabolic diseases.Citation28–Citation30 The same is true for oxidized low-density lipoprotein (LDL), which induces the monocytes in plasma for their movement into and residence in the adipose tissue upon binding to the walls of the constituting vascular endothelial cells. The elevated infiltration of the adipose tissue with immune cells seems to function as the driver for the emergence of defects in insulin signaling, ie, insulin resistance; increase in adipocyte number and size, ie, obesity; and plaque destabilization and rupture, ie, atherogenesis.Citation31–Citation33 Moreover, elevated oxidative stress as well as the redox state in the adipose tissue has also been associated with the early phases in the pathogenesis of metabolic diseases.Citation34 Interestingly, both inflammation and oxidative stress have consequences for insulin signaling, adipocyte proliferation and differentiation, and adipocyte apoptosis and angiogenesis, ultimately resulting in type II diabetes. Obesity and atherosclerosis have been linked to specific expression patterns of miRNAs (see “EMVs as biomarkers: Obesity”).Citation35–Citation37
Currently used biomarkers
Currently, the prediction within a limited period during the development of a metabolic disease prior to its diagnosis (typically 5–10 years for type II diabetes), as well as its definite diagnosis and prognosis of the future disease outcome, are based on the measurement of the levels of typical and easily accessible serum parameters, such as carbohydrate and lipid metabolites (eg, glucose, triacylglycerol, cholesterol, lipoproteins), small molecule intermediates (eg, 2-hydroxybutyrate) and metabolites (eg, creatinine) as well as surrogate entities (eg, glycated hemoglobin HbA1c).Citation38–Citation42 In routine clinical practice, these so-called traditional biomarkers in combination enable the prediction of type II diabetes with a probability of about 0.65 to 0.75 (with 0.5 representing chance in throwing a dice) (). The supplementation with information about physical body parameters (BMI, waist-to-hip ratio, sex) as well as the health state and lifestyle of the probands (eg, blood pressure, smoking, fitness, food consumption as evaluated in the Deutsche RisikoabschätzungCitation43) leads to a further increase in the probability of prediction to about 0.85 or 0.90.Citation43 In future, functional assays, currently applied on a routine basis only for the confirmatory diagnosis of a metabolic disease, such as glucose and insulin tolerance tests, as well as euglycemic clamp studies for type II diabetes and noninvasive imaging proceduresCitation44 currently in the stage of clinical testing and approval, such as magnetic resonance imaging (MRI) of the β-cell mass for type II diabetes and positron emission tomography (PET) of the brown adipose tissue mass for obesity, will supplement the portfolio of traditional biomarkers and further improve their predictive power (). However, in any case, the prerequisite and, therefore, the critical disadvantage of the traditional, functional, and imaging biomarkers is that the earlier the time points are envisaged for the prediction, potential prevention, and therapy, the less informative they are. On the other hand, they are of particular value for prediction along further disease development and advanced stages of the pathogenesis. In sum, therefore, these phenotypic biomarkers do support prediction independent of the individual life stage and style in contrast to the prediction of disease susceptibility by genotypic biomarkers ().
Figure 2 Correlation between the prediction of the risk for disease and the earliest time point feasible for its prediction.
Abbreviation: EMVs, exosomes and microvesicles.
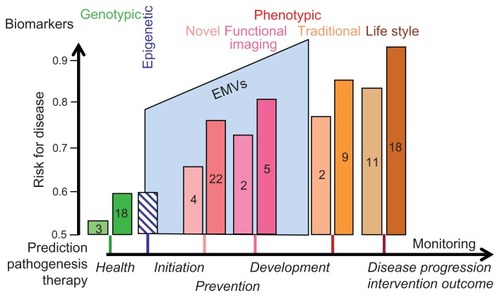
In research clinical studies, the determination of so-called novel biomarkers, predominantly peptides and proteins, such as hormones, cytokines (eg, TNF-α), adipokines (leptin, adiponectin), incretins (GLP-1), and others (eg, ferritin, cross-reactive protein), in combination but not alone, led to prediction values approaching, but hardly exceeding, those achieved with well-designed combinations of traditional biomarkers.Citation45 The highest prediction probabilities have been reported for combinations of traditional and novel biomarkers,Citation46 as for example those evaluated by the “EASD Risk Score”.Citation46 However, because of the partial overlap of the pathogenic mechanisms and pathways covered by the two classes of biomarkers, the predictive values of the combinations did not increase to a degree as calculated from the individual contributions of each of the two classes.
In contrast to phenotypic biomarkers, genotypic biomarkers, ie, determination of the complete genetic profile encompassing all relevant polymorphisms in all metabolic disease susceptibility genes, would fulfill the typical demand for life stage/style-independent prediction. However, the research clinical studies so far reported were rather disappointing: predictive values for type II diabetes ranging from 0.54 for genetic polymorphisms in three independent susceptibility genes in combinationCitation28 to 0.60 for single nucleotide polymorphisms in 18 distinct susceptibility genes ().Citation29,Citation43,Citation45 Apparently, common complex multifactorial metabolic diseases cannot be predicted with sufficient probability or precision on the basis of genotypic biomarkers that have been derived from the identification of disease genes as well as susceptibility genes altered in their amount and/or function by mutations, single nucleotide polymorphisms, copy number variants, monoallelic expression, and complex combinations. Even more discouraging was the repeated observation that combining the polymorphisms identified in known metabolic disease susceptibility genes with traditional and novel biomarkers only marginally increased the total predictive value compared to the values of the traditional and/or novel biomarkers in the absence of genotypic markers.Citation27–Citation29 It may be argued that in future the further increase in the number of susceptibility genes identified for metabolic disorders will lead to considerable improvement of the predictive power of genotypic biomarkers in polymorphism and gene combinations of increasing complexity, affecting multiple target tissues and pathogenic pathways. However, the path from the genotype to the phenotype with underlying gene–gene interactions, gene-environment interactions, genome plasticity (somatic and mitochondrial mutations), and epigenetic modifications is long and complex. For these principal considerations, which are beyond the scope of this review article, it remains questionable whether the complete genetic profiling per se, ie, the determination of all relevant (single nucleotide) polymorphisms of each susceptibility gene in all relevant combinations encompassing each complete disease mechanism (eg, insulin release and insulin signaling) in all relevant tissues contributing to metabolic diseases (eg, β-cells and liver, muscle, adipose tissues) will truly enable predictions of metabolic diseases with the required high probabilities of ≥0.90.
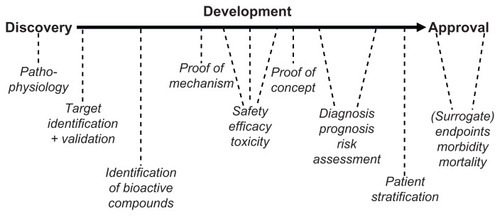
Thus, there is a critical gap between the genotypic biomarkers with their capability for very early and life stage-independent prediction of rather limited probability and the traditional as well as functional/imaging biomarkers with their capability for rather late and pathogenesis-dependent prediction of higher probability (). Apparently, this gap has not yet been filled by novel biomarkers with their intermediary predictive values and intermediary positioning between the initiation of the pathogenesis and the full development of a complex common metabolic disease (). With regard to drug discovery and development, the efficacy of new antidiabetic drugs has been evaluated traditionally in clinical trials using morbidity and mortality as the endpoints (). However, such trials may require 10,000–15,000 subjects and at least 5 years of follow-up to demonstrate significant benefits. Smaller and shorter studies based on biomarkers and surrogate endpoint effects for monitoring the various stages from drug discovery via drug development to drug approval have the potential to revolutionize the drug finding and approval process ().
Requirements for future biomarkers
In future, biomarkers should enable monitoring of the upregulation of the number and size of adipocytes, the downregulation of the number and/or functionality of pancreatic β-cells, the impairment in insulin sensitivity of peripheral tissues, such as muscle, fat, and liver, the development of impaired glucose tolerance and homeostasis, and finally the manifestation of obesity and type II diabetes and further progression to late complications of diabetes, such as retinopathy, nephropathy, neuropathy, and micro/macroangiopathy. These biomarkers should preferably circulate in the plasma and allow monitoring (1) of the overall pathogenesis or critical pathogenic steps; (2) of drug efficacy in a target-independent fashion; and (3) with limited expenditure in preclinical and clinical studies. Furthermore, they should fill the above-mentioned gap left by genotypic, traditional, and novel biomarkers ().
Although it seems reasonable to determine biomarkers in disease-relevant and affected organs, tissues and cells where they typically occur at higher concentrations, it is of huge practical advantage to measure them in the plasma. However, the huge dynamic range in the amounts of the individual protein components in plasma (>10 orders of magnitude in difference) hampers the discovery as well as routine determination of novel biomarkers, since typical plasma proteomics is biased towards the detection of primarily high-abundance proteins. This necessitates complex and tedious fractionation procedures that may facilitate the access to low-abundance proteins. However, these are not typically compatible with high-throughput analysis, which is, however, a prerequisite for the monitoring of large clinical trials that are appropriate and required for metabolic diseases.
Exosomes and microvesicles (EMVs)
In contrast, plasma proteomics based on EMVs have the distinct advantage that the information obtained may encompass a considerable reservoir of novel biomarker candidates being transferred from organs, tissues, and cells to that compartment and protected from degradation. Moreover, the plasma EMV sub-proteome is characterized by a smaller dynamic range and a higher portion of undegraded soluble and membrane proteins at considerably high concentrations, and their amount and composition is determined by specific stimuli (eg, drugs) or microenvironmental and pathogenic factors (eg, cellular stress, high glucose). EMVs are expected to fulfill and connect the above criteria of very early prediction, high prediction probability, and feasibility of measurement. This expectation is based on (1) the structure and composition of EMVs with constituent mRNAs encoding genotypic biomarkers as well as constituent cytokine biomarkers, signaling proteins, receptors, transporters, and enzymes; (2) the function of EMVs in intercellular information transfer in various pathogenic processes; (3) the sensitivity of EMVs toward environmental stimuli with regard to their release from the donor cells of almost each tissue and organ into the circulation; and (4) the accessibility and ease of detection and technological determination of EMVs in the plasma. For routine applications in the mid-term, it will be tremendously important to identify significant and physiologically relevant correlations between EMVs and metabolic diseases along subsequent stages of each of the different disease mechanisms covering (1) the healthy state; (2) the initiation of the disease process; (3) its subsequent development to the established disease; (4) the benefits and failures of therapeutic intervention; and (5) the further progression to complications linked to diabetes and obesity ().
Structure and composition
It has long been known that small membrane vesicles are released from most animal cell types,Citation47,Citation48 such as mast cells,Citation49 dendritic cells, B lymphocyte cell lines,Citation49 astrocytes, platelets, neurons, endothelial cells, and epithelial cells.Citation50,Citation51 They have been regarded as “extracellular organelles” and a family member of the “bioactive vesicles” (). They have the same topology regarding outer and inner phospholipid bilayer leaflets as the donor cell, and are of variable diameters, which are used to discriminate the larger so-called microvesicles (200–1000 nm) and the smaller so-called exosomes (50–200 nm) (). It is critically important to distinguish EMVs from the other large membrane vesicles (0.5–3 μm), the so-called apoptotic bodies, which are released from almost each cell type when they are challenged by apoptotic and death signals or mechanical stress ().Citation52 Despite a variety of morphological and structural similarities, apoptotic bodies and EMVs appear to differ considerably with regard to the cellular origin and molecular composition as well as the releasing signals and mechanisms. Interestingly, EMVs were identified more than three decades ago as being released from reticulocytes during their maturation into erythrocytes, whereby the transferrin receptor as a constituent component of those EMVs becomes downregulated in the mature erythrocytes.Citation53
Figure 4 Structure of “model” EMVs with functions of some of their components (see text for details).
Abbreviations: EPCR, endothelial protein C receptor; PECAM-1, platelet endothelial cell adhesion molecule-1; VCAM-1, vascular cell adhesion molecule-1; ICAM-1, intercellular cell adhesion molecule-1; E-selectin, endothelial selectin; S-Endo, CD146/melanoma cell adhesion molecule; VE -cadherin, vascular endothelial cadherin; eNOS, endothelial nitride oxide synthase; MMP, matrix metalloproteases; uPA, urokinase plasminogen activator; uPAR, uPA receptor; EPC, endothelial protein C; TM, thrombomodulin.
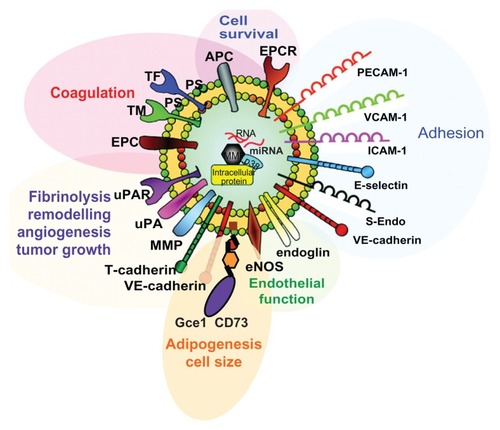
Figure 5 Cellular origin and composition of EMVs and apoptotic vesicles released from donor cells in response to inductors, such as physiological and stress signals (in green) or apoptotis and necrosis signals (in red).
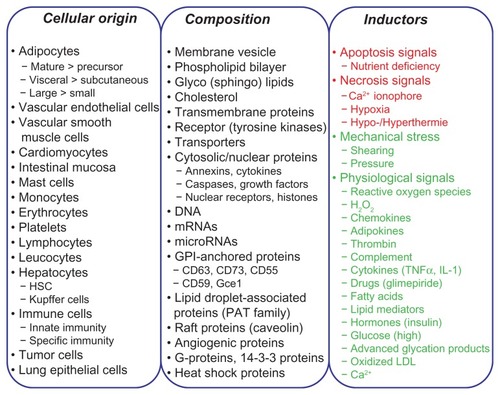
EMVs have been detected in the circulatory system (plasma) and various body fluids, such as urine, bronchoalveolar lavage, mucus, saliva, bile, ascites, cerebrospinal fluid, and breast milk.Citation54–Citation56 On the basis of the identification of tumor-derived EMVs in the plasma of patients with glioblastoma, multiform malignant glioma, ovarian carcinoma, and lung adenocarcinoma,Citation55,Citation57 it may be concluded that EMVs are transferred across the blood-brain barrier and are released from both the apical and basolateral plasma membranes of polarized epithelial and endothelial cells, respectively. EMVs harbor a wide variety of (glyco)phospholipids, mRNAs, and microRNAs (miRNAs), but not ribosomal RNAs,Citation58 in concert with soluble, peripheral, transmembrane, and glycosylphosphatidylinositol- (GPI-)anchored proteins ( and ) with overlapping yet distinct patterns between different cell types (–). It seems plausible that their composition depends on the cell type from which they originate and is therefore heterogeneous, which is revealed by proteomic analysis of EMV components in the large scale derived from various cell typesCitation59 (detailed summarized information in http://dir.nhlbi.nih.gov/papers/lkem/exosome) and from the urine.Citation60 The latter study described the presence of 1132 polypeptides and, in addition, of phosphoproteins in EMVs. Moreover, the current version (3.1) of ExoCartaCitation61 compiles 11,261 protein entries, 2375 mRNA entries, and 764 miRNA entries derived from 134 studies that deal with EMVs.Citation61 All these EMV components originate from the cytoplasm, nucleus, cytoskeleton, proteasome, plasma membranes, and intracellular membranes (eg, mitochondria, endoplasmic reticulum) of the donor cell (). Thus, the differential composition of EMVs from various body fluids is a prerequisite for their potential use as biomarkers for metabolic diseases.
Table 1 Some protein components identified in EMVs from rodent adipocytesCitation300,Citation302,Citation307,Citation322
Table 2 Some protein components identified in EMVs from rodent adipocytesCitation300,Citation302,Citation307,Citation322
Table 3 Some protein components identified in EMVs from various cell typesCitation303,Citation322,Citation361,Citation363,Citation372–Citation375
Table 4 Some protein components identified in EMVs from various cell typesCitation303,Citation322,Citation361,Citation363,Citation372–Citation375
Biogenesis
At present, the detailed mechanisms underlying the biogenesis of EMVs remain unclear. According to current models, microvesicles develop as buds directly at the plasma membrane and are subsequently released by shedding under control of the intracellular calcium concentration and reorganization of the cytoskeleton, with annexin playing a key regulatory role (). Alternatively, the membrane of intracellular endosomes buds into the luminal space. After shedding, they form multivesicular bodies (MVB), which in the course of trafficking to and fusion with the plasma membrane result in exosomes released into the extracellular space ().Citation47,Citation62 The energy-dependent formation of soluble NSF attachment protein receptor (SNARE) complexesCitation63 is thought to mediate the attachment of the exosomal membranes (via v-SNAREs) with the target plasma membranes (via t-SNAREs).Citation64 Upon initiation of complex assembly at the amino-terminal regions of the SNAREs and further movement toward the membrane-anchoring carboxy-terminal regions,Citation65 an intermediary complex constituted by SNAP-25 and syntaxin-1 is formed that interacts with synaptobrevin-2. The SNARE-dependent docking of the MVB at the plasma membrane is apparently regulated by Rab27a, Rab27b, and Rab35Citation66,Citation67 since downregulation of their expression and/or activity led to size increases/accumulation and redistribution to the perinuclear region of the MVB/endosomes.
Figure 6 Molecular mechanisms for the release of exosomes. (A) involves the budding and subsequent shedding of specialized areas of the plasma membrane, which are identical to lipid rafts (in red), from the donor cells. (B) involves exocytosis, ie, fusion with the plasma membrane, of exocytic multivesicular bodies (MVB) which originated from endocytic cisternae and escaped degradation by degradative MVB and lysosomes as well as recycling by endosomes, and of microvesicles.
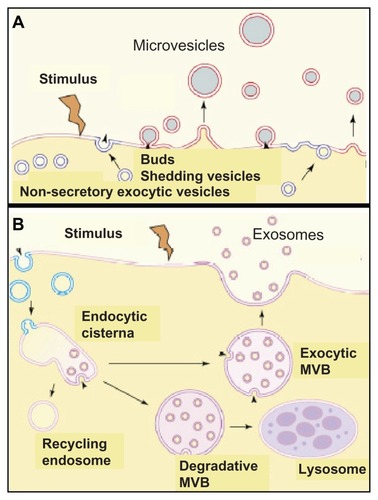
A recent study described a novel and interesting mechanism for the regulation of exosome biogenesis and release, which involves members of the tetraspanin protein family. The tetraspanin transmembrane proteins possess four membane-spanning domains and are engaged in a very wide range of specific molecular interactions that result in the formation in the plane of the membrane of tetraspanin-enriched microdomains, which are identical or similar to lipid rafts.Citation68–Citation71 Tetraspanins have been implicated in a multitude of biological processes, such as cell adhesion, migration, fusion, and signal transduction through their associated partner molecules.Citation72,Citation73 Microarray analysis showed that CD9 expression in tumor cells correlated with the down-regulation of several Wnt family genes and their targets, suggesting that CD9 may act as an upstream negative regulator and tumor suppressor in the Wnt signaling pathway.Citation74 Previous immunoelectron microscopic studies showed that several members of the tetraspanin family, including CD63 and CD82, are rich in exosomes.Citation75 At steady state, tetraspanins CD9 and CD82 are organized in a signaling complex with the membrane adaptor and β-catenin-binding protein, E-adherin, at the plasma membrane.Citation76 This signaling complex, including CD9, CD82, E-cadherin, and the transcription factor, β-catenin, becomes internalized and delivered to early endosomes where the exosome biogenesis is initiated with inward vesicle budding. After maturation of the exosome-containing endosomes into late MVB, they fuse with the plasma membrane and thereby release the β-catenin-harboring exosomes. Thus, in general, the tetraspanins CD9/CD82 seem to drive and control the release of exosomes. In the case of β-catenin-containing exosomes they apparently trigger the reduction in the cytoplasmic/nuclear pool of β-catenin with the accompanying downregulation of Wnt/β-catenin signaling and tumorigenesis.Citation76
According to their biogenesis, a principal differentiation between microvesicles and exosomes seems feasible (; ); however, their classification according to composition and cellular origin is problematic (). This is predominantly due to the lack of unambiguous physical properties or unique molecular markers of EMVs (–) and, consequently, the difficulties in knowing their cellular origin and biogenetic pathway. Moreover, a number of experimental observations are difficult to reconcile with a strict separation between microvesicle and exosome biogenesis (). For instance, (glyco)sphingolipids and GPI-anchored proteins – eg, the prion proteinCitation77 – already embedded in the outer leaflet of the plasma membrane can also be efficiently released via exosomes.Citation78 This process can be quite extensive, as demonstrated in reticulocytes, which release approximately 50% of GPI-anchored acetylcholinest-erase from the plasma membrane via exosomes during their differentiation into erythrocytes.Citation79 Thus, exosomal proteins and phospholipids are able to bud directly from the plasma membranes of the donor cells.Citation80 In addition, exosomes are also detectable as deep invaginations of plasma membranes and thus possibly share the site of biogenesis with that of microvesicles.Citation81 Consequently, it has been speculated that exosomes and microvesicles use similar or overlapping pathways in their biogenesis. Surprisingly, recent findings suggested that exosome biogenesis does not depend on the formation of MVB since the latter but not the former process requires functional endosomal sorting complexes required for transport (ESCRT) machinery.Citation80,Citation82 This argues for identical MVB-independent biogenetic pathways for both microvesicles and exosomes. Given these uncertainties, it has recently been suggested to use the collective acronym, EMVs, for exosomes plus microvesiclesCitation83 for any released nonapoptotic small membrane vesicle ( and ) that is also used in this review article.
Table 5 Comparison of composition and biogenesis between microvesicles and exosomesCitation376–Citation381
Releasing signals
Multiple signaling mechanisms are presumably involved in the cell- or agonist-specific release of both types of EMVs (). It is important to point out that EMV release is not a random process, such as the degradation of plasma membranes of dying necrotic cells, but a highly controlled process triggered by a multitude of structurally diverse stimuli (). For instance, in platelets, EMV release is induced by the following stimuli with increasing potency: epinephrine, adenosine diphosphate, thrombin, collagen, thrombin plus collagen, and Ca2+ ionophore A23187.Citation84 Furthermore, the complement membrane attack complex C5b-9 or antiplatelet antibodies represent other stimuli that cause EMV release in platelets. In addition to biological and chemical stimuli, mechanical factors, such as shear stress, induce EMV release in platelets and many other cell types. Significantly, many stimuli can trigger the release in additive or even synergistic fashion to other stimuli.
miRNA components
miRNAs are small, noncoding RNA sequences of about 22 nucleotidesCitation85 expressed in animals, plants, and viruses.Citation86 All precursor miRNAs have stem loop structures that are cleaved by the Drosha and Dicer protein complexes to form mature functional miRNA. Through the formation of RNA-induced silencing complexes, miRNAs can either cleave mature mRNA molecules or inhibit their translation, thus representing an additional posttranscriptional layer of gene regulation. Interestingly, miRNAs were found incorporated into EMVs released from primary or cultured cells in vitro or circulating in the plasma. For instance, a large number of miRNA species has been detected associated with EMVs in the plasma of normal subjects, which are predicted to control homeostasis and metabolism of hematopoietic cells.Citation87 Importantly, the miRNA species (as revealed by advanced array technologies) of those EMVs did not reflect the total miRNA profiles of the donor cells,Citation57,Citation58,Citation87–Citation89 which is an argument for rather selective miRNA packaging into EMVs. Nevertheless, EMVs released from T-cells, B-cells, and dendritic cells have been demonstrated to harbor miRNA species unique to the expression pattern in their donor cells.Citation90 The molecular mechanisms underlying the selective packaging of miRNAs into EMVs remain unknown so far.
Importantly, miRNAs have been demonstrated to be more stable than their cellular counterpartsCitation91 and to resist degradation during prolonged storage and repeated freezing/thawing cycles.Citation92 This apparent stability of EMV contents underscores the attractiveness of EMVs as biomarkers. Moreover, during the development of many diseases and in a multitude of pathological states, EMVs prepared from patients were found to harbor specific miRNAs that were detected at lower levels or not at all in EMVs from normal healthy subjects.Citation57,Citation88,Citation93,Citation94 For instance, levels of miR-133a, which regulates the NFATc4 protein known to accompany or even induce the development of cardiac hypertrophy, have been found increased in circulating EMVs isolated from injured myocardium of patients with cardiometabolic diseases.Citation95 In agreement, the administration of miR-133a antagomirs that specifically inhibit miR-133a function was reported to decrease the extent of cardiac hypertrophy.Citation95
The administration and targeted cellular delivery of specific miRNAs via EMV-like vehicles prepared from natural sources and subsequently chemically modified or reconstituted in vitro is under intense investigation and discussion.Citation96 Antagomirs represent a new class of chemicals, which have been successfully used to downregulate the endogenous expression of miRNAs.Citation97 They represent chemically modified small synthetic RNA oligonucleotides perfectly complementary to the selected miRNA target but mispairing at the cleavage site of argonaute 2 or harboring some modified bases to interfere with argonaute 2 cleavage. Furthermore, antagomirs must be protected from rapid degradation, which is usually accomplished by the introduction of 2′ methoxy groups or phosphothioates. The molecular mechanism by which antagomirs block the miRNA function remains to be elucidated, but it may involve irreversible binding to the targeted miRNA.Citation97,Citation98 Alternatively, miRNAs can be inactivated, even in vivo, upon administration of so-called “locked” nucleic acids.Citation99–Citation101 On the contrary, the endogenous function of a selected miRNA can be recapitulated upon introduction into the target cells of synthetic miRNA mimics consisting of a “guide strand,” which is identical to that of the selected miRNA and a “passenger strand,” which is chemically linked to a “carrier” molecule, such as cholesterol, for facilitating cellular uptake. Both strands have to be chemically modified to increase their stability to a considerable extent.
In summary, there is increasing evidence for the potential use of EMVs-associated miRNA signatures in body fluids, particularly peripheral blood, as biomarkers for the prediction of metabolic diseases.Citation102 In addition, the technologies for the up- and downregulation of miRNA activity introduced for eluciduation of its (patho)physiology will open new avenues for the miRNA/EMV-based therapy of metabolic diseases.
GPI-anchored protein components
GPI-anchored proteins are widely distributed among all eukaryotic organisms.Citation103–Citation106 They harbor lipidic tail structures that tether them to the outer phospholipid leaflet of plasma membranes and EMVs, respectively, as well as relatively large hydrophilic proteinaceous domains that protrude into extracellular/vesicular spaces (). The evolutionary purpose of membrane anchorage via GPI linkage for surface protein expression remains unclear so far. GPI- anchored enzymes, receptors, binding proteins, and structural proteins are known to be involved in a number of different physiological processes and functions, such as catalysis, signal transduction, cell adhesion, and complement regulation ().Citation107–Citation123 GPI-anchored proteins also have a great role in embryogenesis, since abrogation in their biosynthesis results in embryonic lethality. Recently, it was shown that interaction of GPI-anchored EphrinA with its receptor is crucial for closure of the neural tube.Citation122
Table 6 Some representatives for GPI-anchored proteinsCitation382–Citation387
First indications that proteins might be attached to plasma membranes by lipidic anchors were reported in 1963 with the finding that bacterial phospholipase can release alkaline phosphatase from cells.Citation124 The existence of inositol-containing phospholipid protein anchors was suggested in 1976Citation125 and was finally accepted in 1985, when detailed compositional data about Torpedo electric organ and human acetylcholinesterase,Citation126,Citation127 rat brain and thymocyte Thy-1,Citation128 and Trypanosoma brucei variant surface glycoproteinCitation129,Citation130 became available. Today, hundreds of GPI-anchored proteins are known, and it is estimated that approximately 0.5% of all proteins in lower and higher eukaryotes are modified in this manner.Citation131
All GPI anchors share a common glycolipidic core structure.Citation105,Citation106 Phosphatidylinositol is glycosidically linked through carbon-6 of the myoinositol ring to the reducing end of a nonacetylated glucosamine moiety. Three mannosyl residues, linked to α1-4, α1-6, and α1-2, respectively, are attached to this glucosamine. The terminal α1-2 linked mannose is linked to phosphoethanolamine via a phosphodiester linkage. The GPI anchor becomes attached to the carboxyl terminus of the protein by an amide linkage to the amino group of phosphoethanolamine. This common core structure can be further modified in a way that depends on both the organism and the cell type in which it is synthesized.Citation104
The whole process of GPI biosynthesis is carried out in the endoplasmic reticulum,Citation106 and nearly 20 enzymes are involved in this pathway. Corresponding genes have been cloned from mammals, yeast, and protozoa.Citation132,Citation133 The initial step, the attachment of N-acetylglucosamine to phosphatidylinositol, depends on the product of the X-chromosomal gene, phosphatidylinositolglycan class A (PIG-A) in humans.Citation134 A deficiency in PIG-A results in a rare human disease called paroxysmal nocturnal hemoglobinuria (PNH).Citation134–Citation137 Patients with PNH have abnormal cells of various hematopoietic lineages that are defective in the biosynthesis of GPI-anchored proteins. These include the complement-regulatory proteins, CD55 and CD59, whose absence results in enhanced complement-mediated lysis.Citation138,Citation139 Since deficiency of GPI biosynthesis is embryonically lethal,Citation119–Citation121 all PNH patients reported to date apparently have acquired a somatic mutation in PIG-A.Citation140 The exact mechanism how one or a few of the large number of pluripotent hematopoietic stem cells that bear a mutation in PIG-A achieve dominance in the bone marrow and the peripheral blood is not known so far.Citation141 Possibly, PIG-A deficient cells have lower susceptibility to TNF-α and IFN-γ, and this resistance might contribute to their clonal dominance.
Once the biosynthesis of the GPI anchor is completed, it is transferred to a specific site upstream of the carboxyterminal end of the protein in the endoplasmic reticulum lumen by the action of a transamidase complex, which simultaneously cleaves off the remaining carboxy-terminal peptide.Citation132,Citation133 A specific signal for GPI anchor attachment has been identified at the carboxy terminus of the protein moiety.Citation103,Citation106 After attachment of the prefabricated GPI anchor, the GPI-modified proteins are then transferred from the endoplasmic reticulum to the Golgi complex where they are subjected to further modifications at their GPI moiety. Finally, they are transferred to the plasma membrane via the trans-Golgi network and secretory vesicles as mature GPI-anchored proteins. In mammalian polarized cells, the GPI-anchored proteins seem to be targeted predominantly to the basolateral plasma membrane domain.Citation142
Although the polypeptide moieties attached to the GPI anchors do not apparently share common features, the presence of the GPI anchor itself appears to confer some important characteristics on them. In particular, localization to plasma membrane microdomains or lipid rafts – which are highly enriched in (glyco)sphingolipids, cholesterol, saturated fatty acids, and certain proteins, serving as platforms for a variety of cellular functions, such as vesicular traffickingCitation143,Citation144 and signal transduction () – appears to play critical roles in the transduction of signals across the plasma membrane, the translocation of GPI-anchored proteins from plasma membrane lipid rafts onto cytoplasmic lipid droplets (LDs),Citation145–Citation147 and the release of GPI-anchored proteins into EMVs.Citation148,Citation149 The clustering of GPI-anchored proteins in lipid raftsCitation150–Citation161 may be required for the initial budding, subsequent shedding, and final release of EMVs enriched in GPI-anchored proteins.
Table 7 Some selected signaling pathways mediated by lipid rafts and major components involvedCitation388–Citation394
Release and transfer of EMV-associated GPI-anchored proteins
In addition to liberation by enzymatic, ie, lipolytic or proteolytic cleavage, GPI-anchored proteins can be released from the outer leaflet of the plasma membrane with their GPI anchors remaining intact. This release can occur via embedding the GPI-anchored proteins into small aggregates together with some membrane phospholipids.Citation162 Alternatively and more frequently, the release is provoked through EMVs. Little is known about the signals that target proteins, such as GPI- and other lipid-anchored, eg, acylated proteins to the site(s) of EMV release. Candidate signals represent amino-terminal acylation or myristoylation tags, internal phosphatidylinositol-(3,)4,5-bisphosphate-binding domains, carboxy-terminal prenylation, and palmitoylation tags as well as type-I integral plasma membrane outer leaflet targeting domains.Citation80,Citation83,Citation163,Citation164 With regard to GPI-anchored proteins, their segregation into the site(s) of EMV release may rely on the intrinsic capability of GPI anchors to spontaneously insert, accumulate, and aggregate in lipid rafts of the outer plasma membrane leaflet.Citation165
The phenomenon of the release of polypeptides from donor cells and subsequent intercellular transfer to and uptake by acceptor cells of a GPI-anchored protein were reported even before the actual discoveries of GPI anchors and the presence of GPI-anchored proteins in EMVs. While investigating phospholipid exchange between cells and artificial vesicles and liposomes, ACE and some other erythrocyte proteins were observed to be transferred from the erythrocytes to the vesicles and liposomes in reversible fashion.Citation166 The rate, direction, and extent of those apparent intermembrane transfers were found to depend on the relative phospholipid composition and fluidity of both the donor and the acceptor membranes.Citation167 In addition, the spontaneous insertion of exogenously added purified human decay accelerating factor (DAF) into the surface of sheep erythrocytes was reported,Citation168 which resulted in freely mobile and fully active DAF, as demonstrated by its inhibition of convertase complexes and mediation of resistance to complement-mediated lysis.Citation169
Since then, a number of other GPI-anchored proteins were successfully incorporated into a variety of different cell types in vitro under retention of the same characteristics and functions as their endogenously expressed counterparts.Citation162,Citation167–Citation180 For instance, CD59 was transferred from seminal plasma to erythrocytes and other cells,Citation179,Citation181,Citation182 as well as from erythrocytes to endothelial cells in mice made transgenic for this GPI-anchored protein.Citation169,Citation183 Storage of erythrocytes resulted in the loss of both CD55 and CD59 from the erythrocyte membraneCitation183 and generation of erythrocyte EMVs that are enriched in GPI-anchored proteins, including CD55 and CD59.Citation184 Interestingly, CD59 incorporated into U937 monocytic cells and allowed to equilibrate for 2 hours at 37°C showed redistribution into lipid rafts and signaling via intracellular Ca2+ fluxes.Citation185 Therefore, exogenously added GPI-anchored proteins appear to become functional within the target cell membrane once they have acquired a distribution similar to that of their endogenous counterparts during a slow process that can take even more than 24 hours.Citation186,Citation187 Incubation of rat Thy-1 antigen with murine lymphocytes showed that the rat protein was transferred to murine cells and incorporated into their plasma membranes, where the exogenous protein migrated with the same lateral mobility as endogenous murine Thy-1 protein.Citation188 Similarly, incorporation of T. brucei variant surface glycoprotein (VSG) into baby hamster kidney cells showed that the inserted VSG exhibited lateral mobility equivalent to that of endogenous VSG in T. brucei.Citation189
When erythrocytes from PNH patients who were deficient in GPI-anchored proteins were incubated with high-density lipoprotein (HDL) preparations or erythrocyte EMVs from normal blood donors, significant transfer of CD55 and CD59 to the cell surface occurred. Pretreatment of the EMVs and HDL with bacterial phosphatidylinositol-specific phospholipase C abrogated protein transfer to CD55/59-deficient cells, indicating that the elevated cell-associated CD55/59 levels were related to the insertion of an intact GPI anchor into the outer leaflet of the plasma membrane by the GPI fatty acyl chains rather than to simple adhesion.Citation184 An elegant experiment successfully demonstrated the ability of GPI-anchored proteins to transfer between cells in vivo.Citation172 PNH patients of blood group A1 were given transfusions of compatible washed group 0 blood. Patients’ group A1 cells were distinguished from the transfused group 0 cells by staining with a Dolichos biflorus lectin that specifically binds to group A1 erythrocytes. Significant transfer of GPI-anchored proteins from donor cells to patients’ erythrocytes could be demonstrated as early as one day following transfusion and persisted for several days.Citation190,Citation191
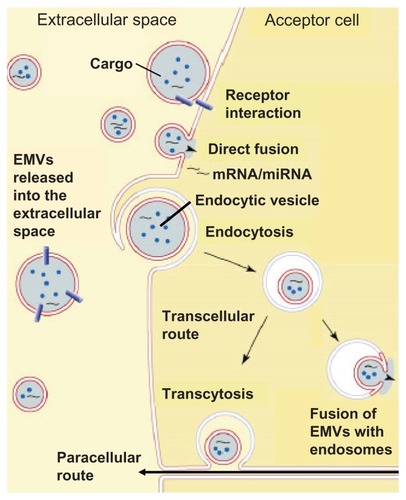
Transfer between membranes can occur without actual membrane fusion.Citation162 The GPI-anchored proteins are apparently transferred through EMVs released from the donor cells.Citation192 Moreover, GPI-anchored proteins were reported not to transfer spontaneously from erythrocytes to liposomes or between plasma membranes in vivo.Citation192 The involvement of a catalyst was supported by the observation that CD4 engineered to become biosynthetically coupled to a GPI anchor is efficiently transferred between plasma membranes in one type of cell,Citation193 while another cell line expressing CD4-GPI fusion protein failed to release it in any form.Citation194 With regard to CD59, HDL may act as its carrier and manage to transfer CD59 to erythrocytes.Citation195 The exact mechanism(s) underlying the EMV-mediated transfer of GPI-anchored proteins, which may involve receptor-receptor interactions, direct fusion or endocytosis of the EMVs by the acceptor cell, subsequent fusion with endosomes (), and identification of the hypothetical catalyst are currently under intense investigation,Citation169,Citation173,Citation185–Citation188,Citation196 but in any case, they rely on the intactness of both GPI lipid and protein moieties.Citation168,Citation186 This conclusion was drawn from the observation that transgenic mice expressing the GPI anchorless (released instead of cell surface-expressed) version of the prion protein (PrP) were infected with the pathogenic scrapie form of PrP, but they never developed manifest prion disease.Citation197 Experiments with cells expressing anchorless PrP were also resistant to scrapie infection.Citation198 Apparently, lack of the GPI anchor on PrP would prevent its transfer and could explain why cells expressing GPI anchor-less PrP were unable to sustain a scrapie infection over time.
Figure 7 Current working hypotheses for the different modes of interaction of EMVs released from donor cells into the extracellular space with target acceptor cells.
Abbreviation: EMVs, exosomes and microvesicles.
(Patho)physiological roles of EMV-associated GPI-anchored proteins
The transport of GPI-anchored proteins and miRNAs across the plasma membrane and their intracellular translocation onto cytoplasmic LDs (GPI-anchored proteins, only) or release into the circulation through EMVs (GPI-anchored proteins and miRNAs) as well as the subsequent transfer from the EMVs of the donor cell to the plasma membrane of an acceptor cell were independently discovered and rediscovered in several distinct research areas during the past decades and termed shedding, release, incorporation, painting, uptake, jumping, transfer, and translocation.Citation62,Citation159,Citation162,Citation192,Citation199–Citation201 In these studies, the differential release and transfer of EMVs originating from a multitude of cell types were implicated with the pathogenesis of a broad variety of diseases.
One prominent source of EMVs is tumor cells that use the release and transfer of certain EMV components to evade destruction by immune cells.Citation52,Citation201 Retroviruses exploit transfer via EMVs for spreading to other cells.Citation200–Citation203 However, these extensively studied mechanisms are actually examples of a misuse of EMVs and some of their components. The real reason that this process developed in the course of evolution has yet to be elucidated. One reported physiological function is the transfer of GPI-anchored proteins and gangliosides by prostasomes from prostate epithelium to spermatozoa.Citation162 Since spermatozoa do not synthesize proteins, the transfer may represent an important target cell-oriented mechanism by which spermatozoa can obtain new proteins and alter their antigenicity or acquire resistance to immune attack and other surface properties. Another rather probable function of the transfer is the modulation of the composition of lipid rafts and their function, eg, in signal transduction. Together with the GPI-anchored proteins, gangliosides are located in lipid rafts. Even though they arise from different biochemical pathways, they both have lipid anchors that tether them to the outer leaflet of plasma membranes and enable their release from the donor cell and transfer to the acceptor cell in a regulated fashion.Citation58 Since GPI-anchored proteins and gangliosides are both localized in lipid rafts, they could affect each other. Exogenous administration of gangliosides affected the distribution of GPI-anchored proteins within lipid rafts.Citation203–Citation206 It is tempting to speculate that modification of lipid rafts by removal or addition of specific gangliosides might create favorable conditions for the release of GPI-anchored proteins.
In rodent adipose tissues, the release of (GPI-anchored) proteins and mi/mRNAs from donor adipocytes and subsequent transfer to acceptor adipocytes via EMVs is known to exert multiple (patho)physiological consequences.Citation207–Citation211 Recently, the fusion of EMVs derived from large white rat adipocytes and harboring the GPI-anchored proteins, Gce1 and CD73, the mRNAs coding for the fat-specific (LD-associated) protein, FSP27, and the lipid-synthesizing enzyme, glycerol-3-phosphate acyltransferase (GPAT3), as well as the microRNAs, miR-16 and miR-222, with distinct small adipocytes was demonstrated to stimulate lipid synthesis and storage, in parallel with inhibition of lipid degradation through lipolysis.Citation207–Citation211 Importantly, the release of those EMVs from human and rat donor adipocytes was upregulated by physiological signals, such as palmitate and H2O2, as well as pharmacological signals, such as the antidiabetic sulfonylurea drug, glimepiride.Citation207–Citation212 Furthermore, there is experimental evidence that this induced release is epigenetically controlled via demethylation of histone H3 at lysine 4 and dual methylation of histone H3 at lysine 9 within the promoter of the tetraspanin protein, CD9 (Müller G et al, unpublished data). Ectopic expression of CD9 and silencing of endogenous CD9 expression in human adipocytes triggered significant upregulation and downregulation, respectively, of the release of EMVs harboring CD73 and CD9 proteins, FSP27/CideC mRNA and miR-222. This was accompanied by downregulation and upregulation of lipid degradation and oxidation, respectively. In agreement, lipid synthesis and the expression of the LD-associated proteins FSP27/CideC and perilipin A were decreased and increased upon up- and downregulation of CD9 expression, respectively. Lipolysis, LD-associated Gce1 protein and its cAMP-specific phosphodiesterase activity, the oxygen consumption rate, the extracellular acidification rate, and the expression of the mitochondrial proteins, uncoupling protein 1 (UCP1) and F1-ATPase, became elevated and diminished in response to up- and downregulated CD9 expression, respectively (Müller G et al, unpublished data). Together these findings suggest that upregulation of the release of EMVs harboring specific GPI-anchored proteins, mRNAs and miRNAs, is under the control of differential histone methylation of the CD9 promoter, which leads to fostered CD9 expression and ultimately triggers lipolysis and oxidation of the fatty acid products in (newly generated and partially uncoupled) mitochondria. This raises the possibility that upregulation of EMV release and transfer within human adipose tissue depots may represent a novel target for the therapy of obesity.Citation213,Citation214
Unfortunately, the phenomenon of transfer of GPI-anchored proteins via EMVs is difficult to discover and track for the following reasons: (1) different GPI-anchored proteins undergo different mechanisms of release and its regulation; (2) transfer efficacy is often rather limited, at least if total GPI-anchored proteins are analyzed; (3) transfer is directed to only a limited number of GPI-anchored proteins and/or specific cell types; (4) from a technological point of view, it is difficult to detect the transfer of GPI-anchored proteins in whole organisms. In conclusion, the physiological significance and function of release and transfer of EMVs are not well understood at present. It will be interesting to learn whether and, if so, how they support the integration of individual cells within and between tissues.
(Patho)physiological roles of EMV-associated mi/mRNAs
Considering the importance of miRNAs as an inevitable cornerstone of the human genetic system, the engagement of EMVs for the transfer of genetic material could be an efficient method within the human body to exchange biological information. EMVs containing miRNAs would enable intercellular and inter-organ communication in the body.Citation215 Consequently, EMVs could shuttle mRNAs and miRNAs directly from donor to acceptor cells, thereby considerably increasing the probability that the transferred genetic information would affect the function of the acceptor cells upon their successful expression and silencing functions, respectively. The release of mi/mRNA-harboring EMVs may enable the cell-to-cell communication irrespective of the distance between the cells within an organ or tissue or between cells of different, more or less remote organs and tissues. The identification of EMVs in blood and various body fluids hints at the possibility that this exchange of genetic information between organs or tissues may involve EMVs.Citation215 Moreover, the membrane of EMVs harbors donor cell-specific factors (eg, GPI-anchored proteins) that enable the EMVs to target specific acceptor cells for transfer of their mi/mRNA contents with exquisite specificity and efficacy. Those EMV-associated mi/mRNAs may reflect disease-specific causal mechanisms. Experimental evidence is emerging that the prediction, diagnosis and prognosis of certain diseases may be facilitated by measurement of the levels of specific mi/mRNAs in EMVs isolated from certain body fluids.
Initial credit for this attractive possibility was gained during cancer research.Citation216–Citation226 Certain tumors were recently demonstrated to shed EMVs, which are rich in signaling molecules and genetic material that together constitute a specific and readily identifiable signature.Citation47 Moreover, miRNAs entrapped in EMVs were detected in the serum of cancer patients by quantitative real-time polymerase chain reaction in tumor-specific fashion.Citation58,Citation221 The miRNA profiles from circulating tumor EMVs were found unique and distinct compared to those from normal controls.Citation88 Similarly, specific miRNA profiles have been reported for EMVs isolated from patients with lung cancer, glioblastoma, and heptocellular carcinoma.Citation57 In lung adenocarcinoma, the total tissue miRNA signatures differ considerably between cancer patients and normal probands, and the profiles of the EMV-associated miRNAs resemble very closely those isolated from the tumors.Citation57 Tumor-derived EMVs are known to transfer mRNAs to monocytes within the tumor microenvironment to stimulate these cells for the production of cytokines, resulting in enhanced tumor growth and dampening of the immune response.Citation220 EMV-associated miRNAs released from hepatocellular carcinoma cells were shown to induce the downregulation of the transforming growth factor-β activated kinase-1 (TAK1) signaling pathway in hepatocarcinogenesis, and thereby may cause the local spreading, intrahepatic metastases, and multifocal growth in hepatocellular carcinoma.Citation218 Tumor-released EMVs were found to be a prerequisite for the promotion of tumor metastasis in the course of proinflammatory cytokine-driven proliferation of myeloid-derived suppressor cells via the MyD88 pathway.Citation223 EMVs released from Epstein–Barr virus-infected B95–8 LCL cells and isolated from monocyte-derived dendritic cells were found to induce gene silencing in the acceptor cells.Citation222 Macrophages have been reported to increase the invasiveness of breast cancer cells by EMV-mediating transfer of oncogenic miRNAs from the macrophages to potential cancer cells.Citation225 mRNAs from endothelial-derived EMVs have been found to exert proangiogenic effects.Citation226
An additional link between EMVs and miRNAs has recently become apparent in immunology and virology research. EMVs released from human and murine mast cell lines were found to harbor over 1200 mRNA and about 121 miRNA species.Citation215 EMVs released from donor dendritic cells were reported to dock to and fuse with target dendritic cells and to transfer their contents into the target cells under accompanying specific silencing of the miRNA-targeted genes.Citation227 miRNA-harboring EMVs released from stromal cells have been shown to support quiescence of B-cells. The apparent transfer of miRNAs from bone marrow stroma to B-cells could be involved in the dormancy of bone marrow metastases.Citation224 The presence of viral miRNAs in EMVs is evidence that they may function as vesicular carriers for spreading of the disease or initiation of the infection process.Citation228,Citation229 Thus, there is increasing evidence that EMV-associated miRNAs function in the acceptor cells following their transfer. Based on the unique and specific signatures of the transferred EMV-associated miRNAs, their use as biomarkers in screening tests for the prediction and prognosis of cancer and other diseases has been proposed.
EMVs as biomarkers
General considerations
The following characteristics argue for the potential of EMVs as a promising source for new biomarkers: (1) EMVs are identifiable and isolatable on the basis of typical intrinsic and well-defined properties, such as phosphatidylserine content, size, sedimentation behavior; (2) EMVs are specific with regard to the expression of cell-lineage markers as well as the overall molecular composition and patterns of their luminal and surface contents; (3) EMV signatures critically depend on the stimulation and micro-environment of the donor cells; (4) EMVs are initial and rapid “responders” since they are released early during stimulatory or micro-environmental changes and in the pathogenic cascade of a disease; (5) EMVs are noninvasive since they are detectable in many body fluids; (6) EMVs are “translatable” since their release is not limited to one cell type or one species; and (7) EMVs act as vectors since they transport and protect biological messages, ie, mRNAs, microRNAs, proteins, and phospholipids, which are normally confined to cells/tissues.
It is well established that for each type of biomarker, its predictive value increases with the number of members of this biomarker type combined as well as with the relative proximity of the time points of biomarker measurement, ie, disease prediction and onset of the disease, ie, diagnosis (). On the basis of the specific structural and functional features of EMVs (see “EMVs as biomarkers: Type II diabetes” and “Obesity”), it seems reasonable (albeit still speculative) to assume that EMVs enable prediction with higher probability (compared to genotypic biomarkers) and at earlier time points (compared to phenotypic biomarkers) along the complete pathogenetic pathway, including the initiation, further development, diagnosis, further progression, and outcome of the disease.
Origins and sources
During the past two decades, the existence of EMVs has been demonstrated in blood and a range of other body fluids, such as urine, saliva, mucus, and breast milk. Consequently, there is an increasing interest in their potential use as tissue-borne and easily accessible diagnostic biomarkers for many diseases, including cancer, cardiovascular diseases, and metabolic diseases.Citation116,Citation117,Citation230–Citation242 As a prerequisite for the use of EMVs as multi-component biomarkers reflecting different dysfunctional or disease stages of those relevant cells/tissues from which they originate, it is important to understand the critical parameters and molecular mechanisms involved in their passage from the donor cells into the corresponding body fluid ().
Current estimates of the concentration of EMVs in peripheral blood of healthy probands are 5–50 μg/mL. Although the term “microparticles” is neither specific nor fully descriptive, it has been used for the past 30 years and unfortunately remains standard in the current literature on EMVs of the blood.Citation84,Citation242–Citation248 Using flow cytometry, it was found that the majority of the peripheral blood EMVs from normal individuals are actually derived from blood cells, particularly platelets ( and ).Citation249,Citation250 For instance, about 80% of the EMVs analyzed in human plasma samples obtained during a clinical study were determined to be of platelet origin,Citation251 and the number of nonplatelet-derived (eg, tissue-derived) EMVs were well correlated to the clinical data and disease history of the patients. Initially, platelet-derived EMVs were considered to have pathophysiological importance since they expose a multitude of procoagulant anionic phospholipids, such as phosphatidylserine, in a similar fashion as activated platelets and consequently have been the most extensively studied.Citation84,Citation244,Citation247,Citation252 In the 1940s, it was demonstrated that clotting of platelet-deprived plasma was delayed after high-speed centrifugation.Citation253 This observation suggested that procoagulant subcellular structures are present in plasma and are removed by sedimentation. Surprisingly, the release of membrane fragments from activated platelets, thereafter called “platelet dust,” was not demonstrated until 1967.Citation254
Figure 8 Schematic depiction of the origin of EMVs in the blood. © 2008, Elsevier. Adapted with permission from Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2008;19:43–51.Citation47
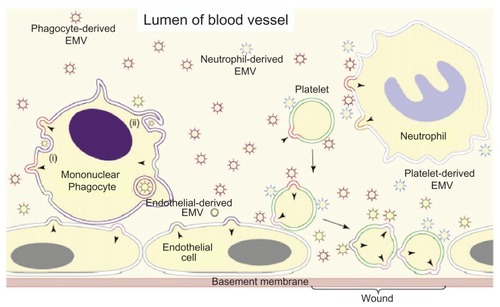
Figure 9 Analysis of the origin of peripheral blood EMVs.
Abbreviation: EMVs, exosomes and microvesicles.
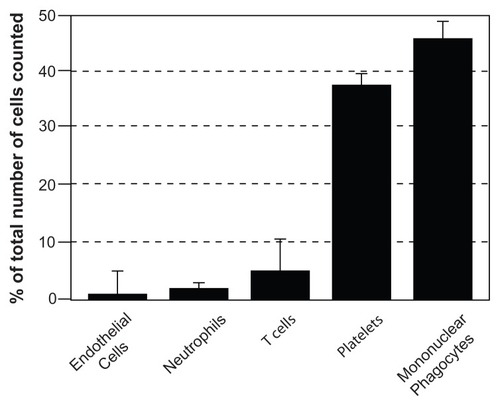
The second-largest population of blood EMVs is derived from the mononuclear phagocyte cell lineage ( and ). In contrast, only a small percentage of the peripheral blood EMVs originates from T-cells and neutrophils. A small subpopulation of blood EMVs seems to be released from endothelial cells according to the expression of corresponding cell surface antigens,Citation251 in contrast to the massive EMV production observed with human endothelial cells in vitro.Citation255–Citation257 B-cells do not seem to contribute to EMVs to any significant degree.
In addition to blood, urine is one of the most useful sources for EMVs, which are assumed to be released by tubular epithelial cells. It is of tremendous advantage that their collection is simple and noninvasive by nature. Urinary EMVs, the majority of which are thought to represent exosomes, have recently been the subject of intense proteomic analyses.Citation258–Citation260 Interestingly, aquaporin-2 has recently been identified in urinary EMVs, but not in EMVs from other sources.Citation261 Thus, aquaporin-2 positive EMVs may be useful as biomarkers for renal dysfunction and structural injury as they typically develop during diabetic nephropathy.
The transport pathway of platelet-, blood cell-, and endothelial cell-derived as well as tubular epithelial cell-derived EMVs into the circulation and urine, respectively, appears to be straightforward. In contrast, the mechanism of how EMVs released from tissue cells reach the blood is intriguing since these EMVs have to transit vascular endothelial cells during their passage from the interstitial space into the circulation. The transport of EMVs across endothelial cells may occur via (1) the transcellular route involving transport vesicles, which are generated by endocytosis of the plasma membrane and subsequently fuse with the opposite plasma membrane; or (2) the paracellular route involving the transient opening of tight junctions in course of direct interaction with certain protein components (). Recently, the operation of a third pathway has become conceivable based on the observation that small leucine-rich proteoglycans, such as biglycan and decorin, as secreted components of the extracellular matrix critically affect the communication between neighboring tissue cells in the course of their continuous formation and degradation.Citation262–Citation264 Hence, biglycan and decorin seem to regulate the active and passive transport of nutrient and hormonesCitation265,Citation266 and possibly also of EMVs. Interestingly, biglycan was found to be elevated in obese artherosclerotic mice,Citation267 in rats with metabolic syndrome, and in type II diabetes patientsCitation268,Citation269 compared to controls. Its prominent expression in stromal vascular cells of visceral adipose tissue suggests that the expression level of biglycan and other components of the extracellular matrix may modulate the efficacy of EMV transfer from donor to acceptor cells within a tissue depot (eg, from large to small adipocytes) as well as of EMV transport from donor cells into body fluids (eg, from adipocytes into plasma). These pathways and the underlying molecular mechanisms must be delineated for a better understanding of the correlation between the levels of EMVs in body fluids and the pathogenesis of metabolic diseases.
Type II diabetes
Preclinical studies aimed at finding evidence for the potential usefulness of EMVs as type II diabetes biomarkers demonstrated that: (1) db/db mice display significantly higher levels of circulating total and platelet-derived (but not tissue-derived) EMVs compared to db/+ mice; (2) male db/db mice treated for 2 weeks with antidiabetic compound and exhibiting significantly elevated body weight gain, diminished blood glucose levels, increased plasma adiponectin levels, and decreased plasma insulin levels have significantly reduced amounts of total and tissue-derived (but not platelet-derived) EMVs; (3) streptozotocin-induced mice exhibit significantly increased levels of circulating total, platelet-derived and tissue-derived EMVs prior to the onset of fully established diabetes; and (4) circulating EMVs increase in levels and change their signature between healthy volunteers and patients suffering from type II diabetesCitation270–Citation272 or metabolic syndrome.Citation272–Citation278
The development of vasculopathies and endothelial dysfunction during type II diabetes is known to involve multifactorial processes, including the pathological activation of vascular cells. The release of EMVs by activated cells was reported predominantly for diseases associated with thrombotic risk, but few data are available for type II diabetes. Platelet-derived EMVs of both CD42+ and CD41a+ phenotypes as well as monocyte-derived EMVs (CD14+) were found elevated in patients with type II diabetes.Citation279 In particular, the latter EMVs were highly increased in patients suffering from diabetic nephropathy and may be indicators of vascular complications.Citation280,Citation281 Another study demonstrated sustained, elevated amounts of circulating platelet-derived procoagulant EMVs (CD42+) after acute myocardial infarction in diabetic patients, which were higher than in nondiabetic patients with myocardial infarction.Citation282 Tissue factor-positive EMVs associated with TH cell-derived (CD4+), granulocyte-derived (CD66e+) or platelet-derived (CD61+) antigen were significantly increased in early type II diabetes when compared with healthy control subjects.Citation283 Importantly, EMV numbers did not correlate with in vivo coagulation markers, but correlations were found between various tissue factor-positive EMV subpopulations and several components of the metabolic syndrome, such as body mass index, fasting plasma glucose, insulin, TNF-α, and serum HDL cholesterol. This hints at a diabetogenic role that is distinct from the procoagulant role of the detected tissue factor antigen.
In addition, significant differences have been demonstrated in the number and the procoagulant activity of circulating EMVs between type I and II diabetic patients.Citation280,Citation284,Citation285 When compared with control subjects, type I diabetic patients presented significantly higher numbers of platelet-derived EMVs (CD41+), endothelial cell-derived EMVs (CD51+), and total phosphatidylserine-positive EMVs as well as elevated levels of EMV-associated procoagulant activity. In contrast, in type II diabetic patients, only the number in phosphatidylserine-positive EMVs was significantly higher without concomitant increase of their procoagulant activity.Citation286 Importantly, in type I diabetic patients, only procoagulant activity associated with phosphatidylserine-positive EMV was correlated with the level of glycated hemoglobin, suggesting that procoagulant activity is associated with impaired glucose tolerance and homeostasis.Citation287
Previous reports have indicated that high levels of plasma dipeptidyl peptidase-IV (DPP-IV), also known as CD26, are positively correlated with type II diabetes. DPP-IV degrades the active form of the incretin, glucagon-like peptide-1, which is released from intestinal L-cells after meal intake and enhances insulin secretion in a glucose-dependent manner (). DPP-IV inhibition causes blood glucose decrease in animal models of diabetes and type II diabetic patients. The DPP-IV inhibitors sitagliptin and vildagliptin are currently widely used as an adjunct to diet and exercise to improve glycemic control in type II diabetic patients.Citation288,Citation289 DPP-IV is a membrane-associated peptidase highly expressed in the brush border and microvillar fractions of the kidney cortex. Recent findings showed that in urine, EMV-associated DPP-IV represents the major portion of urinary DPP-IV activity.Citation290,Citation291 The excretion of this urinary EMV-associated DPP-IV was significantly higher in type II diabetic patients compared to control probands.Citation291 The urinary levels of EMV-associated DPP-IV were found to be positively correlated with the urinary albumin/creatinine ratio in type II diabetic patients.Citation292,Citation293 These results suggest that the urinary level of EMV-associated DPP-IV is associated with and may be used for the prediction of the development and outcome of diabetic nephropathy.
The potential of EMV-associated miRNAs as biomarkers for type II diabetes can be indirectly inferred from the known multiple roles of miRNAs in the regulation of lipid and glucose metabolism. These were revealed by a multi-tude of in vitro and in vivo (mice) studies, which used the delivery of miRNA precursors or antagomirsCitation97 into cells to overexpress or silence the miRNAs of interest and to evaluate their application to therapy.Citation294,Citation295 For instance, the blockade of miR-122 expression by systemic administration of an miR-122 antagomir oligonucleotide caused considerable lowering of plasma cholesterol levels as well as hepatic fatty acid and cholesterol synthesis rates in normal mice, leading to diminished levels of triglycerides and hepatic steatosis in diet-induced obese mice.Citation296 Consequently, the mice appeared to be in good health after 4 weeks of therapy with the antagomirs. Furthermore, the expression of miR-103/107 was found upregulated in obese insulin-resistant ob/ob mice.Citation297 The specific and efficient silencing of miR-103/107 in liver and fat by delivery of cholesterol-conjugated anti-miR antagomir significantly improved glucose homeostasis and insulin sensitivity in ob/ob as well as diet-induced obese, but not chow-fed wild-type mice compared to scrambled or mismatched antagomirs. Interestingly, as a consequence of the downregulation of miR-103/107, the amount of caveolin-1, a positive regulator of insulin receptor signaling, becomes elevated in parallel with improved insulin signaling, reduced adipocyte size, and upregulated insulin-stimulated glucose transport.Citation297 These findings argue for the potential use of specific miRNAs as biomarkers for the development of insulin resistance and type II diabetes, in particular, if the miRNAs are incorporated together with other typical EMV components into circulating EMVs.
In summary, the currently available experimental evidence strongly argues for the usefulness of EMVs, particularly those released into the plasma and urine, as specific biomarkers for the pathogenesis of type II diabetes. Moreover, there is an urgent need to detect those EMVs that originate from disease-relevant tissues, are released into accessible body fluids and have already increased in number in the prediabetic state.Citation298–Citation300 Putative candidates for relevant EMV donor cells and tissues are pancreatic islets, particularly β-cells, liver, skeletal muscle, adipose tissue, smooth muscle cells, vascular endothelial cells, and tissue and plasma macrophages. The future challenge will be to identify the EMV signatures that are specific to those cells and tissues causally involved in the pathogenesis of type II diabetes and/or reflecting the temporal and individual disease states.
Obesity
The increased prevalence of obesity in present day society and the current view of adipose tissue as one of the most critical regulators of energy homeostasis and metabolism have warranted a sustained interest in studying the mechanisms controlling its formation. In general, obesity can be regarded as the consequence of a long-lasting imbalance between energy intake and energy expenditure, finally leading to the storage of the surplus energy as triacylglycerol in the adipocytes, which thereby undergo both hypertrophy and hyperplasia. Clearly, both adipocyte hypertrophy and hyperplasia is now generally accepted to be involved in the dramatic increase in adipose tissue mass in obese as well as type II diabetic patients.Citation301 The underlying molecular mechanisms and defects have not yet been fully elucidated. Interestingly, they may encompass the paracrine exchange of information about the lipogenic vs lipolytic state between large and small adipocytes within adipose tissue depots via adipokines and/or EMVs.Citation302,Citation303
Alterations in the number and size of adipocytes are typically accompanied by changes in the expression patterns for subsets of miRNAs. For instance, during adipocyte differentiation, the levels of miR-130b and miR-210 were found to be decreased in both lean and obese probands. Levels of miR-222 were decreased in lean probands but increased in obese subjects.Citation304,Citation305 Levels of miR-103, miR-107, miR-143, and miR-185 were upregulated in the lean state but downregulated in the obese state.Citation304,Citation305 Interestingly, from the miRNAs analyzed so far, only miR-34a has been found to be positively correlated with the rate of adipocyte differentiation and development of the BMI.Citation305 The expression of the majority of these miRNAs is known to be controlled by certain adipokines, such as TNFα that downregulates miR-103 and miR-143 and upregulates miR-221 and miR-222.Citation304
It was recently found that EMVs released from dysfunctional and hypertrophic adipocytes manage to impair the function of vascular endothelial cells, which may have implications for the development of obesity-linked complications. For instance, EMVs released from primary rodent adipocytes were demonstrated to stimulate angiogenesis in vivo.Citation306 Strikingly, as revealed by antibody arrays and gelatin zymographic analyses, these EMVs harbored a multitude of angiogenic and antiangiogenic adipokines, growth factors and enzymes, such as leptin, TNFα, acidic fibroblast growth factor-γ and matrix metalloproteases-2/5. Upon incubation with human umbilical vein endothelial cells, these EMV cargo proteins promoted the induction of cell migration and tube formation, but only in combination after the removal of leptin, TNFα, or fibroblast growth factor-γ by neutralizing antibodies, which caused abrogation of the angiogenic effect to a major degree.Citation306 Moreover, a subset of miRNAs (let-7b, miR-103/143/146b/148/155/221) was found associated with those EMVs.Citation307 Surprisingly, let-7b, miR-143, miR-155, and miR-221 are known to play similar roles in the control of cell proliferation, apoptosis, inflammation and angiogenesis in adipose and vascular tissues.Citation308 Consequently, it may be proposed that these tissues communicate via EMVs that are released from donor adipocytes and targeted to vascular endothelial acceptor cells.Citation308 This hypothesis gained further credit upon the recent finding that EMVs isolated from plasma and peripheral blood monocytes share the expression of 71 miRNAs,Citation87 the majority of which regulate the differentiation of blood cells, immune functions, and metabolic pathways. Furthermore, a few other EMV-associated miRNAs derived from adipocytes, endothelial cells, or monocytes have been detected in the plasma and demonstrated to affect the function and phenotype of the corresponding acceptor cell type.Citation215,Citation309
However, in comparison with the approximately 400 studies that revealed differential miRNA expression profiles during different cardiovascular disease states and the limited number of medically attractive EMV-associated miRNAs left for the prediction and prevention of cardiovascular diseases so far,Citation309–Citation313 even fewer EMV-associated miRNA candidates have been reported to date as being differentially expressed in and released from adipose tissue cells in the course of obesity development. In particular, miRNAs have also been recognized as regulators of adipocyte metabolic integration, energy homeostasis, and differentiation ().Citation314–Citation315 For instance, the expression of 50 out of 70 miRNAs was found to significantly differ between human pre- or mature adipocytes from lean and obese individuals.Citation311 Moreover, EMVs released from cultured 3T3-L1 adipocytes have been reported to harbor about 140 miRNAs.Citation305 The majority of them were adipocyte-specific, abundant in correlation with their expression level in the donor cells, and considerably upregulated during adipocyte differentiation in vitro.Citation316,Citation317 Interestingly, these apparently adipocyte-derived EMVs also seem to mediate the transport of mRNAs coding for adiponectin, resistin and PPARγ into cultured macrophages and to induce angiogenesis.Citation318–Citation321 In addition, adipocyte-specific mRNAs and miRNAs have also been detected in EMVs isolated from rat serum.Citation302,Citation322,Citation323
Table 8 Functions of dysregulated miRNAs in adipose tissue
Moreover, EMV-associated miRNAs seem to play key roles in the proper functioning of adipocytes and vascular endothelial cells as well as adhesion, apoptosis, oxidative stress, hypoxia, and infiltration of inflammatory cells (). The similarity in the apparent effects of EMV-associated miRNAs on adipose and vascular tissues raises the possibility of the operation of parallel molecular mechanisms in adipocytes, vascular cells, and macrophages. Apparently, lipid storage and handling varies among adipocytes (triacylglycerols), macrophages, and smooth muscle foam cells (cholesterylesters). However, rather than the species of the accumulated lipids, changes in their amount per se may trigger phenotypic and functional alterations. This view was supported by the observation that the initial expansion of adipose tissue is accompanied by the emergence of M2-polarized adipose tissue macrophages, whereas in the course of further adipose tissue enlargement (as a consequence of both adipocyte hypertrophy and hyperplasia), the mere deposition of lipids of the same species in these macrophages induces their M1 polarization.Citation324 This conversion of the macrophage phenotype associated with obesity-induced insulin resistance, inflammation, and severe oxidative stress may be further enhanced by lowered levels of adiponectin. This adipokine is known to be preferentially released from small instead of fully lipid-loaded adipocytesCitation303,Citation325 and to improve the metabolism of macrophages through its antidiabetic and anti-inflammatory activities.Citation326 Hence, EMVs derived from adipocytes, vascular cells and macrophages, and harboring miRNA constituents seem to reflect the functional state of the adipose tissue and thereby may be useful for following the development of obesity.
The posttranscriptional cascade governing adipogenesis, ie, the differentiation of adipocytes from precursor cells and their subsequent maturation from small unilocular (ie, containing many small LDs) to large multilocular (ie, containing few large LDs) cells has been extensively studied for many years.Citation327 Novel findings suggest the involvement of EMVs in the control of maturation of rodent adipocytes within adipose tissue depots in vitro.Citation146–Citation149,Citation300,Citation302,Citation307,Citation321,Citation328 In particular, the release of EMVs from primary and cultured rat and mouse adipocytes has been demonstrated to undergo considerable upregulation upon challenge with physiological stimuli, such as excess of fatty acids (palmitate) and reactive oxygen species (H2O2), as well as pharmacological agents, such as the antidiabetic drug, glimepiride.Citation206–Citation210 These adipocyte-derived EMVs are thought to regulate lipid metabolism between large and small adipocytes within the same adipose tissue depot during the transfer of some of their constituent components (). Among them are the GPI-anchored cAMP-specific phosphodiesterase, Gce1, and 5′-nucleotidase, CD73 (), which in concerted fashion catalyze (c) AMP degradation,Citation145,Citation146,Citation328 and the mRNAs coding for GPAT3 and FSP27, which drive lipid synthesis and LD biogenesis, respectively,Citation329–Citation334 as well as the miRNAs, miR-16, and miR-222, which have been implicated in the coordination of lipid metabolic pathways.Citation292,Citation300,Citation322 The underlying mechanisms encompass (1) the release of those EMVs from preferentially large donor adipocytes; (2) the interaction, fusion or endocytosis of those EMVs () with or by, respectively, preferentially small acceptor adipocytes; (3) the translocation of Gce1 and CD73 from the (lipid rafts of the) EMV membranes to intracellular LDs of the acceptor adipocytes as well as the delivery of the GPAT3- and FSP27-encoding mRNAs and miR-16/222 from the EMV lumen to the cytoplasm of the acceptor adipocytes; (4) the degradation of (c)AMP at the LD surface zone by Gce1 and CD73 (); and, finally, (5) the upregulation of fatty acid esterification at the LDs and downregulation of fatty acid release from the LDs by concerted action of GPAT3 and FSP27 enzymic and structural activities, respectively, by decreasing of the cAMP levels and by miR-16/222 expression in the acceptor adipocytes.Citation145–Citation149,Citation207–Citation209,Citation302 These findings strongly argue that EMVs transfer lipogenic and antilipolytic information from large to small adipocytes to shift the burden of lipid loading within adipose tissue depots during certain (patho)physiological states ().Citation328 Among them are excessive fatty acid intake, antidiabetic pharmacotherapy, and exposure to cellular oxidative stress, which are well known to accompany or foster the obese state. The measurement of these adipocyte-derived EMVs in the adipose tissue depots or, more practicably, in the plasma if they became adequately released into this compartment, could provide helpful information for the prediction of obesity long before the body mass index and the waist-to-hip ratio increase considerably. In fact, EMV levels were reported to be increased in the serum of slightly overweight women compared to normal age-matched controls and suggested to account for the significantly increased risk for obesity-linked (eg, thrombotic) complications during later and advanced obesity in this population.Citation335,Citation336
Figure 10 Hypothetical model for the regulation of lipid storage and lipid oxidation in adipose tissue depots consisting of large and small white adipocytes by the release of EMVs harboring miRNAsCitation207–Citation212 under the epigenetic control of CD9 expression and their paracrine action (see text for details).
Abbreviation: EMVs, exosomes and microvesicles.
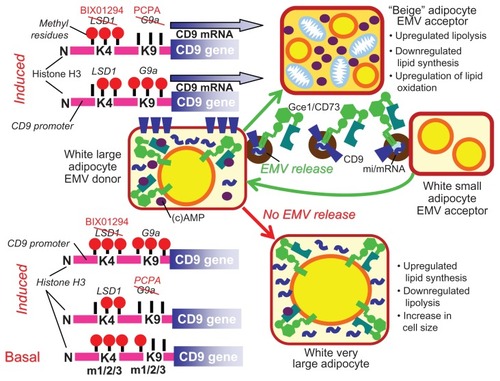
Epigenetic control
To date obesity has been studied predominantly from a thermoenergetic point of view, taking into consideration the imbalance between energy intake and energy expenditure, which is determined by complex interrelationships between gene mutations/polymorphisms and lifestyle. The putative involvement of epigenetic mechanisms – ie, of inheritable alterations in gene expression that are manifest in the absence of changes in the DNA nucleotide sequence itself in the development of obesity – has only recently begun to attract attention.Citation337,Citation338 The underlying molecular mechanisms include (but are not restricted to) covalent post-translational modifications of the histone amino-terminal tails,Citation339–Citation341 in which lysine methylation either enhances or silences the transcriptional state of a given gene, depending on the modified residue. In particular, methylation of lysine 4 of histone H3 (H3K4) usually correlates with gene activation, whereas methylation of lysine 9 of histone H3 (H3K9) is typically associated with transcriptional silencing.Citation341–Citation346
With regard to obesity, an adverse environment during in utero or lactation periods is generally assumed to affect its future development, suggesting that the mother’s nutrition or perinatal lifestyle could alter the developmental programming of the fetus and pubs.Citation337 In contrast, the role of adverse environment (eg, nutrition, physical activity, cellular stress, pharmacotherapy) during postnatal growth and adulthood for the modification of the epigenetic pattern remains a matter of debate. Thus, changes in histone methylation patterns could be the result of the interplay of various dietary and environmental factors and provide a source of interindividual differences with respect to the efficacy of adipogenesis and adipocyte differentiationCitation345–Citation349 and ultimately explain the differential susceptibility of genetically similar individuals to develop obesity.Citation337,Citation350
However, it is disappointing that despite the assignment of some miRNAs to critical functions in the epigenetic control of adipogenesis and obesityCitation307,Citation322,Citation337,Citation338 and the identification of other miRNAs in EMVs released from adipocytes and circulating in various body fluids,Citation302,Citation307,Citation322 experimental evidence for the involvement of EMVs as putative carriers of epigenetic information between somatic cells in para- or endocrine fashion has not been reported so far. However, recent findings of the regulation of cell size and lipid metabolism between large and small rat adipocytes by EMVs harboring specific GPI-anchored proteins, mRNAs, and miRNAsCitation300,Citation302 raised the possibility that the transfer of EMVs in response to certain environmental stimuli is under epigenetic control (). A recent study demonstrated that in the course of long-term incubation with inhibitors of either H3K9 methylation or H3K4 demethylation, the palmitate-, glimepiride-, and H2O2-induced release of those EMVs from large primary rat and differentiated human adipocytes is significantly reduced.Citation212 These results emphasized the putative epigenetic control of adipogenesis with emphasis on the late maturation stage of lipid storage and increased cell size.
The putative functional consequences of the apparent epigenetic regulation of the induced (by fatty acids, oxidative stress, glimepiride) EMV release for large and small adipocytes within an adipose tissue depot can be summarized in the following working model (see also ): large white donor adipocytes release EMVs harboring the GPI-anchored proteins, CD73 and Gce1, GPAT3 and FSP27 mRNAs, and miR-16 and miR-222. The loss of these EMV components from the large donor adipocytes leads to the upregulation of lipolysis (as a consequence of increased levels of cAMP at the LD surface zone) and downregulation of lipid synthesis (as a consequence of decreased levels of GPAT3 and FSP27 proteins), which trigger the degradation of LDs and eventually the induction of the brown adipose tissue-like phenotype.Citation351 The release of these EMVs from the large donor adipocytes is blocked by modulation of their histone H3 methylation state (eg, in the course of G9a or LSD1 inhibition). The retention of these EMV components in the large donor adipocytes supports the downregulation of lipolysis (as a consequence of decreased levels of cAMP at the LD surface zone) and upregulation of lipid synthesis (as a consequence of increased levels of GPAT3 and FSP27), which lead to LD biogenesis and elevation of the cell size. The EMVs fuse preferentially with the plasma membranes of small white (pre)adipocytes. Thereby, the EMVs will deliver the GPAT3/FSP27 mRNAs and miR-16/222 miRNAs into the cytoplasm and translocate the GPI-anchored proteins, CD73/Gce1, onto the LD surface of the acceptor adipocytes. Consequently, GPAT3 and FSP27 protein expression becomes upregulated in parallel with degradation of (c)AMP at the LD surface zone, resulting in the direct stimulation of lipid synthesis and inhibition of lipolysis (). The consequent promotion of LD biogenesis drives the increase in cell size of the (initially small) adipocytes and contributes to the stabilization of their white adipose tissue-like phenotype.Citation352,Citation353 Upon blockade of the release of the EMVs from large white adipocytes (as provoked experimentally by BIX01294 or tPCPA) or of their transfer to and fusion with the plasma membranes of small white adipocytes (as provoked experimentally by [c]AMP-SepharoseCitation208,Citation209 or di-annexinCitation354), the lack of upregulation of FSP27 and GPAT3 protein expression in concert with elevated levels of cAMP at the LD surface zone fosters lipolysis and impairs lipid synthesis. These regulatory events are linked to the stimulation of mitochondrial biogenesis and blockade of LD biogenesis, respectively, which ultimately lead to the induction of the brown adipose tissue-like or “beige” phenotype (). The validity of this working model has to be examined in vitro and in vivo using human adipocytes and rodent animal models that enable monitoring of adipocyte hypertrophy in response to supraphysiological concentrations of palmitate and reactive oxygen species or pharmacological concentrations of glimepiride.
Based on present and previous findings, it is already conceivable that the release of EMVs from large donor adipocytes or/and their fusion with small acceptor adipocytes within adipose tissue depots can be used as a biomarker tool for the prediction of the epigenetically controlled lipogenic/adipogenic state of this adipose tissue depot, and, in the case of EMV release from the depot into the circulation, of the whole organism. At present, only a limited number of effector molecules and tool compounds are known to modulate EMV release or fusion,Citation207–Citation209,Citation355 among them the antidiabetic sulfonylurea drug, glimepiride. The determination of adipocyte-derived EMVs in the blood of type II diabetic patients treated with glimepirideCitation356 could enable the prediction of their risk for developing obesity.
Measurement
General considerations
For the purpose of disease prediction, it is sufficient to find correlations between distinct disease states and subsets of EMVs of specific and unique composition, ie, EMV signatures. Importantly, there is no need for the elucidation of the overall molecular composition of the EMVs (ie, the identity of each constituent) and of their cellular/tissue origin. Of course, this information may turn out to be very valuable for a better understanding of the disease mechanism if the EMVs are causally associated. However, the currently available technologies for the analysis of EMVs rely on their (1) biochemical composition as predominantly revealed by (shotgun) transcriptomics, proteomics and lipidomics and the constituting basic methods of one- and two-dimensional electrophoretic separations, (LC-)MS/MS, nanoLC, Q-TOF, MALDI-TOF/TOF, and state-of-the-art data mining in combination with gene expression studies performed with the candidate donor cells; (2) size, mass and morphology; (3) cellular origin; and (4) flow cytometry.Citation318–Citation321,Citation357–Citation363 These technologies manage the biased elucidation of the exact nature of selected constituents, but only in a limited number. It is of crucial importance to considerably increase the discriminative power of future EMV analytics by the generation of multidimensional patterns reflecting the overall constituent signatures of a given EMV species as is typical for type I biomarkers (see “Biomarkers: Definitions”). This demand may be met with the introduction of so-called “nanoparticle tracking” and “chemical noses,” which are already in use for the analysis of complex macromolecules, small vesicles, cell surfaces, and microorganisms for research purposes.Citation364–Citation369
Biosensors
The principal mode of operation of a biosensor for the generation of EMV signatures based on “chemical noses” is as follows (): EMVs isolated from the body fluids of healthy control probands and patients (ie, suffering from type II diabetes) will compete with a selected reporter enzyme, such as glucose oxidase for interaction with nanoparticles. These consist of a gold core with covalently attached tentacle-like aliphatic, positively charged and aromatic substituents at their surface, and are capable of interacting with the proteinaceous and lipidic surface structures of EMVs. Moreover, it seems likely that nanoparticles equipped with similar but uncharged substituents can cross the phospholipid bilayer of the EMVs and finally interact with the protein and mi/mRNAs contents in the lumen of the EMVs. The high-avidity interactions of the differentially substituted nanoparticles with the EMV constituents are apparently based on multiple hydrogen, electrostatic, hydrophobic, and van der Waals bonds.
Figure 11 Schematic depictions of the operation of a nanoparticle-based biosensor for the identification of EMV signatures. (A) Detection principle. The sizes of the nanoparticles versus EMVs are not drawn at scale. (B) Generation of signatures specific for EMVs from different tissues from normal and type II diabetic patients (see text for details).
In the case of nanoparticles interacting with glucose oxidase that has higher avidity and specificity than EMVs have, the reporter enzyme becomes inhibited, eg, through steric hindrance of substrate (ie, glucose) access to its binding cleft/catalytic site (). Alternatively, the nanoparticles may prefer interacting with one or the other of the EMVs contained in the sample in differential fashion rather than with each species with the same avidity. Consequently, this nanoparticle species will dissociate from glucose oxidase, relieving it from inhibition. The resulting glucose oxidase activity is quantitatively evaluated by monitoring the generated protons using a biosensor chip based on ion-sensitive, field-effect transistor technology.Citation367,Citation368 Upon incorporation into microfluidic cards and lab-on-the-chips, EMV populations can be characterized in a high-throughput format. The measured currents will finally be transformed into EMV-specific signatures ().
A single nanoparticle species will usually not be sufficient for the unequivocal identification of the desired EMV species. More likely, the use of a single or a few nanoparticle species results in more or less similar or even identical signatures due to overlapping recognition patterns and similar surface structures among the various EMV species. However, the discriminative power can be drastically increased by the inclusion of a multitude of nanoparticle species that differ considerably in their surface substituents in hydrophobicity, hydrogen donors/acceptors, and charges. The combination of the different signals provided by separate cycles of biosensor read out will create signatures typical for EMVs from distinct cellular/tissue origin and reflecting the distinct stages of the various disease pathways causing type II diabetes and obesity ().
Conclusion
The physiological function of EMVs is probably intercell signaling through specific interactions with target cells and transferring of biological information encoded in transmembrane, soluble and GPI-anchored proteins, mRNAs, miRNAs, and phospholipids. Therefore, EMVs may participate in pathogenic processes, such as the development of metabolic diseases. Further insights into the molecular signatures and specificities of distinct EMV species may allow the identification of their cellular origin and contribute to the elucidation of novel drug targets for metabolic diseases. For instance, the removal from plasma of harmful EMVs may be beneficial during the development of type II diabetes and obesity, in which EMVs apparently deliver thrombogenic and inflammatory signals.
In addition, an increasing body of experimental evidence indicates the possibility that EMVs may offer predictive, diagnostic, and prognostic information for metabolic diseases, such as type II diabetes and obesity. The appearance of EMVs in body fluids makes them readily accessible. Their number, cellular origin, composition, and function seem to depend on and correlate with individual disease stages. In fact, EMV levels are positively correlated with type II diabetes and obesity and, in particular, accompany or even induce the development of diabetes- and obesity-linked complications. This finding is based on the demonstration of (1) quantitative differences in circulating EMVs in various animal disease models; (2) changes in circulating EMV levels occurring prior to the onset of atherosclerotic or diabetic symptoms; and (3) differential modulation of the EMV signature in course of pharmacological interference. Analyses of plasma (and, if feasible, tissue) samples from various cohorts of patients at very early to late stages in disease development and recruited for a multitude of clinical studies should be initiated for (1) comparisons between the different pathophysiological states along cross-sectional studies; and (2) follow-up studies for the pathogenesis along longitudinal studies for several years after the recruitment.
On the other hand, and not addressed in this review, EMVs derived from stem cells may reprogram altered functions in target cells, suggesting that they could be exploited in regenerative medicine to repair damaged tissues.Citation370 In addition, EMV-mediated transfer of genetic information could explain the observed plasticity and the functional effects of stem cells without the need for their transdifferentiation into tissue cells.Citation371,Citation372
Many issues remain to be clarified, such as (1) the stimuli and the molecular mechanisms that regulate the assembly of the EMVs and their bioactive load that they transfer from donor to acceptor cells; (2) the stimuli that trigger the release of the EMVs from donor cells; (3) the nature of the surface receptors at the acceptor cells that confer exquisite specificity; (4) the complete diagnostic potential of EMVs for the whole range of metabolic diseases; (5) the strategies to prevent the formation or to induce the removal from the circulation of potentially harmful EMVs; and (6) the therapeutic exploitation in regenerative medicine of the potency of EMVs to modify the phenotype and function of acceptor cells.
The introduction of detection methods that enable the identification of EMV signatures, in-depth characterization, and specific tracking of EMVs in plasma (and other body fluids) derived from cells and tissues, which are relevant for the pathogenesis of metabolic diseases, should be considerably intensified. It is hoped that recognition of the potential of EMVs will open novel perspectives in the prediction, diagnosis, prognosis, and therapy of metabolic diseases.
Disclosure
The author is an employee of Sanofi Deutschland GmbH and reports no conflicts of interest in this work.
References
- Frank R Hargreaves R Clinical biomarkers in drug discovery and development Nature Drug Discov 2003 2 566 580
- Biomarker Definitions Working Group Biomarkers and surrogate end-points: Preferred definitions and conceptual framework Clin Pharmacol Ther 2002 69 89 95
- Müller G Personalized strategies for the diagnosis and therapy of type II diabetes and obesity Immun Endoc Metab Agents Med Chem 2012 12 80 109
- Jorgensen JT A challenging drug development process in the era of personalized medicine Drug Discov Today 2011 16 891 897 21945860
- Manolis E Vamvakas S Isaac M New pathways for qualification of novel methodologies in the European Medicines Agency Proteomics Clin Appl 2011 5 248 255 21538915
- Temple R Are surrogate markers adequate to address cardiovascular disease drugs? JAMA 1998 282 790 795 10463719
- Lathia CD Biomarkers and surrogate endpoints: How and when they might impact drug development Disease Markers 2002 18 83 90 12364814
- Zineh I Huang SM Biomarkers in drug development and regulation: a paradigm for clinical implementation of personalized medicine Biomark Med 2011 5 705 713 22103607
- Ioannidis JP A roadmap for successful applications of clinical proteomics Proteomics Clin Appl 2011 5 241 247 21523915
- Calligaris D Villard C Lafitte D Advances in top-down proteomics for disease biomarker discovery J Proteomics 2011 74 920 934 21477672
- Colburn WA Biomarkers in drug discovery and development. From target identification through drug marketing J Clin Pharmacol 2003 43 329 341 12723454
- Morel NM Holland JM van der Greef J Introduction to systems biology – A new approach to understanding disease and treatment Mayo Clin Proc 2004 79 651 658 15132407
- Naylor S Biomarkers: current perspectives and future prospects Expert Rev Mol Diagn 2003 3 525 529 14510173
- Zolg JW Langen H How industry is approaching the search for new diagnostic markers and biomarkers Mol Cell Proteomics 2004 3 345 354 14749446
- Trull AK Biomarkers of Disease An Evidence-based Approach 1st ed Cambridge, UK Cambridge University Press 2002
- Nohaile M The biomarker is not the end Drug Discov Today 2011 16 878 883 21888986
- Lee JM Han JJ Altwerger G Kohn EC Proteomics and biomarkers in clinical trials for drug development J Proteomics 2011 74 2632 2641 21570499
- Naylor S Systems biology, information, disease and drug discovery Drug Discovery World 2005 6 23 33
- Samuel VT Shulman GI Mechanisms for insulin resistance: common threads and missing links Cell 2012 148 852 871 22385956
- Doria A Patti ME Kahn CR The emerging genetic architecture of type 2 diabetes Cell Metab 2008 8 186 200 18762020
- Staiger H Machicao F Fritsche A Häring HU Pathomechanisms of type 2 diabetes genes Endocr Rev 2009 30 557 585 19749172
- Conen D Rexrode KM Creager MA Ridker PM Pradhan AD Metabolic syndrome, inflammation, and risk of symptomatic peripheral artery disease in women: a prospective study Circulation 2009 120 1041 1047 19738135
- Ridker PM Buring JE Cook NR Rifai N C-reactive protein, the metabolic syndrome, and risk of incident cardiovascular events: an 8-year follow-up of 14 719 initially healthy American women Circulation 2003 107 391 397 12551861
- Holvoet P Kritchevsky SB Tracy RP The metabolic syndrome, circulating oxidized LDL, and risk of myocardial infarction in well-functioning elderly people in the Health, Aging, and Body Composition cohort Diabetes 2004 53 1068 1073 15047623
- Holvoet P Lee DH Steffens M Gross M Jacobs DRJr Association between circulating oxidized low-density lipoprotein and incidence of the metabolic syndrome JAMA 2008 299 2287 2293 18492970
- Duncan BB Schmidt MI Pankow JS Low-grade systemic inflammation and the development of type 2 diabetes: the atherosclerosis risk in communities study Diabetes 2003 52 1799 1805 12829649
- Pradhan AD Manson JE Rifai N Buring JE Ridker PM C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus JAMA 2001 286 327 334 11466099
- Lango H UK Type 2 Diabetes Genetics Consortium Palmer CN Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk Diabetes 2008 57 3129 3135 18591388
- van Hoek M Dehghan A Witteman JC Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study Diabetes 2008 57 3122 3128 18694974
- Buijsse B Simmons RK Griffin SJ Schulze MB Risk assessment tools for identifying individuals at risk of developing type 2 diabetes Epidemiol Rev 2011 33 46 62 21622851
- Heilbronn LK Cambell LV Adipose tissuemacrophages, low grade inflammation and insulin resistance in human obesity Curr Pharm Des 2008 14 1225 1230 18473870
- Reilly MP Lehrke M Wolfe ML Rohatgi A Lazar MA Rader DJ Resistin is an inflammatory marker of atherosclerosis in humans Circulation 2005 111 932 939 15710760
- De Luca C Olefsky JM Inflammation and insulin resistance FEBS Lett 2008 582 97 105 18053812
- Furukawa S Fujita T Shimabukuro M Increased oxidative stress in obesity and its impact on metabolic syndrome J Clin Invest 2004 114 1752 1761 15599400
- Heneghan HM Miller N Kerin MJ Role of microRNAs in obesity and the metabolic syndrome Obes Rev 2010 11 354 361 19793375
- Ling C Groop L Epigenetics: a molecular link between environmental factors and type 2 diabetes Diabetes 2009 58 2718 2725 19940235
- Bonauer A Boon RA Dimmeler S Vascular microRNAs Curr Drug Targets 2010 11 943 949 20415654
- Lyons TJ Basu A Biomarkers in diabetes: hemoglobin A1c, vascular and tissue markers Transl Res 2012 159 303 312 22424433
- Colagiuri S Optimal management of type 2 diabetes: the evidence Diabetes Obes Metab 2012 14 Suppl 1 3 8 22118704
- Lipska KJ Kosiborod M Hypoglycemia and adverse outcomes: marker or mediator Rec Cardiovasc Med 2011 12 132 135
- Rhee EP Gerszten RE Metabolomics and cardiovascular biomarker discovery Clin Chem 2012 58 139 147 22110018
- Herder C Karakas M Koenig W Biomarkers for the prediction of type 2 diabetes and cardiovascular disease Clin Pharmacol Ther 2011 90 52 66 21654741
- Schulze MB Weikert C Pischon T Use of multiple metabolic and genetic markers to improve the prediction of type 2 diabetes: the EPIC-Potsdam study Diab Care 2009 32 2116 2119
- Matthews PM Rabiner I Gunn R Non-invasive imaging in experimental medicine for drug development Curr Opin Pharmacol 2011 11 501 507 21570913
- Kolberg JA Jorgensen T Gerwien RW Development of a type 2 diabetes risk model from a panel of serum biomarkers from the Inter99 cohort Diab Care 2009 32 1207 1212
- Younus S Rodgers G Biomarkers associated with cardiometabolic risk in obesity Am Heart Hosp J 2011 9 E28 E32 21823073
- Cocucci E Racchetti G Meldolesi J Shedding microvesicles: artefacts no more Trends Cell Biol 2008 19 43 51 19144520
- Piccin A Murphy WG Smith OP Circulating microparticles: pathophysiology and clinical implications Blood Rev 2007 21 157 171 17118501
- Laulagnier K Motta C Hamdi S Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization Biochem J 2004 380 161 171 14965343
- Hogan MC Manganelli L Woolard JR Characterization of PKD protein-positive exosome-like vesicles J Am Soc Nephrol 2009 20 278 288 19158352
- Zhou R O’Hara SP Chen XM MicroRNA regulation of innate immune responses in epithelial cells Cell Mol Immunol 2011 8 371 379 21725335
- Aupeix K Hugel B Martin T The significance of shed membrane particles during programmed cell death in vitro and in vivo in HIV-1 infection J Clin Invest 1997 99 1546 1554 9119998
- Pan BT Johnstone RM Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor Cell 1983 33 967 978 6307529
- Masyuk AI Huang BQ Ward C Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia Am J Physiol Gastrointest Liver Physiol 2010 299 G990 G999 20634433
- Rupp AK Rupp C Keller S Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage Gynecol Oncol 2011 122 437 446 21601258
- Thery C Exosomes: secreted vesicles and intercellular communications F1000 Biol Rep 2011 3 15 21876726
- Rabinowits G Gercel-Taylor C Day JM Taylor DD Kloecker GH Exosomal microRNA: a diagnostic marker for lung cancer Clin Lung Cancer 2009 10 42 46 19289371
- Valadi H Ekström K Bossios A Sjöstrand M Lee JJ Lötvall JO Exosome- mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells Nat Cell Biol 2007 9 654 659 17486113
- Schorey JS Bhatnagar S Exosome function: from tumor immunology to pathogen biology Traffic 2008 9 871 881 18331451
- Gonzales PA Wang WY Mao YW Large-scale proteomics and phosphoproteomics of urinary exosomes J Am Soc Nephrol 2009 20 363 379 19056867
- Mathivanan S Fahner CJ Reid GE Simpson RJ ExoCarta 2012: database of exosomal proteins, RNA and lipids Nucleic Acids Res 2011 40 D1241 D1244 21989406
- Simons M Raposo G Exosomes-vesicular carriers for intercellular communication Curr Opin Cell Biol 2009 21 575 581 19442504
- Sudhof TC Rothman JE Membrane fusion: grappling with SNARE and SM proteins Science 2009 323 474 477 19164740
- Fukuda R McNew JA Weber T Functional architecture of an intracellular membrane t-SNARE Nature 2000 407 198 202 11001059
- Sutton RB Fasshauer D Jahn R Brunger AT Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution Nature 1998 395 347 353 9759724
- Ostrowski M Carmo NB Krumeich S Rab27a and Rab27b control different steps of the exosome secretion pathway Nat Cell Biol 2010 12 19 30 19966785
- Hsu C Morohashi Y Yoshimura S Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C J Cell Biol 2010 189 223 232 20404108
- Dykstra M Cherukuri A Sohn HW Tzeng SJ Pierce SK Location is everything: lipid rafts and immune cell signaling Annu Rev Immunol 2003 21 457 481 12615889
- Allende D Jumping to rafts: gatekeeper role of bilayer elasticity Trends Biochem Sci 2004 29 325 330 15276187
- Pike L Lipid rafts: heterogeneity on the high seas Biochem J 2004 378 281 292 14662007
- Rao R Logan B Forrest K Roszman TL Goebel J Lipid rafts in cytokine signaling Cytokine Growth Factor Rev 2004 15 103 110 15110794
- Hemler ME Tetraspanin functions and associated microdomains Nat Rev Mol Cell Biol 2005 6 801 811 16314869
- Tonoli H Barrett JC CD82 metastasis suppressor gene: a potential target for new therapeutics Trends Mol Med 2005 11 563 570 16271511
- Huang CL Liu D Masuva D MRP-1/CD9 gene transduction downregulates Wnt signal pathways Oncogene 2004 23 7475 7483 15334057
- Escola JM Kleijmeer MJ Stoorvogel W Griffith JM Yoshie O Geuze HJ Selective enrichment of tetraspanin proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes J Biol Chem 1998 273 20121 20127 9685355
- Chairoungdua A Smith DL Pochard P Hull M Caplan MJ Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling J Cell Biol 2010 190 1079 1091 20837771
- Fevrier B Vilette D Archer F Cells release prions in association with exosomes Proc Natl Acad Sci U S A 2004 101 9683 9688 15210972
- Vidal M Mangeat P Hoekstra D Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation J Cell Sci 1997 110 1867 1877 9296387
- Johnstone RM Adam M Hammond JR Orr L Turbide C Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem 1987 262 9412 9420 3597417
- Booth AM Fang Y Fallon JK Yang YM Hildreth JE Gould SJ Exosomes and HIV gag bud from endosome-like domains of the T cell plasma membrane J Cell Biol 2006 172 923 935 16533950
- Welsch S Keppler OT Habermann A Allespach I Krijnse-Locker J Krausslich HG HIV-1 buds predominantly at the plasma membrane of primary human macrophages PLoS pathogens 2007 3 e36 17381240
- Saksena S Sun J Chu T Emr SD ESCRTing proteins in the endocytic pathway Trends Biochem Sci 2007 32 561 573 17988873
- Shen B Wu N Yang JrM Gould SJ Protein targeting to exosomes/microvesicles by plasma membrane anchors J Biol Chem 2011 286 14383 14395 21300796
- Horstman LL Ahn YS Platelet microparticles: A wide-angle perspective Crit Rev Oncol Hematol 1999 30 111 142 10439058
- Lagos-Quintana M Rauhut R Lendeckel W Tuschl T Identification of novel genes coding form small expressed RNAs Science 2001 294 853 858 11679670
- Bartel DP MicroRNAs: genomics, biogenesis, mechanism, and function Cell 2004 116 281 297 14744438
- Hunter MP Ismail N Zhang X Detection of microRNA expression in human peripheral blood microvesicles PLoS One 2008 3 e3694 19002258
- Taylor DD Gercel-Taylor C MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer Gynecol Oncol 2008 110 13 21 18589210
- Mittelbrunn M Gutierrez-Vazquez C Villarroya-Beltri C Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells Nat Commun 2011 2 282 21505438
- Keller S Ridinger J Rupp AK Janssen JW Altevogt P Body fluid derived exosomes as a novel template for clinical diagnostics J Transl Med 2011 9 86 21651777
- Reid G Kirschner MB Van Zandwijk N Circulating microRNAs: Association with disease and potential use as biomarkers Crit Rev Oncol Hematol 2011 80 193 208 21145252
- Michael A Bajracharya SD Yuen PS Exosomes from human saliva as a source of microRNA biomarkers Oral Dis 2010 16 34 38 19627513
- Ciesla M Skrzypek K Kozakowska M Loboda A Jozkowicz A Dulak J MicroRNAs as biomarkers of disease onset Anal Bioanal Chem 2011 401 2051 2061 21544542
- Li Q Lin X Yang X Chang J NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression Am J Physiol Heart Circ Physiol 2010 298 H1340 H1347 20173049
- Kuwabara Y Ono K Horie T Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage Circ Cardiovasc Genet 2011 4 446 454 21642241
- Kooijmans SAA Vader P van Dommelen SM van Solinge WW Schiffelers RM Exosome mimetics: a novel class of drug delivery systems Int J Nanomed 2012 7 1525 1541
- Krützfeldt J Rajewski N Braich R Silencing of microRNAs in vivo with ‘antagomirs’ Nature 2005 438 685 689 16258535
- Czech MP MicroRNAs as therapeutic targets N Engl J Med 2006 354 1194 1195 16540623
- Ploner A Ploner C Lukasser M Niederegger H Huttenhofer A Methodological obstacles in knocking down small noncoding RNAs RNA 2009 15 1797 1804 19690100
- Orom UA Kauppinen S Lund AH LNA-modified oligonucleotides mediate specific inhibition of microRNA function Gene 2006 372 137 141 16503100
- Zampetaki A Mayr M MicroRNAs in vascular and metabolic disease Circ Res 2012 110 508 522 22302757
- Cortez MA Bueso-Ramos C Ferdin J Lopez-berestein G Soos AK Calin GA MicroRNAs in body fluids – the mix of hormones and biomarkers Nat Rev Clin Oncol 2011 8 467 477 21647195
- Ikezawa H Glycosylphosphatidylinositol (GPI)-anchored proteins Biol Pharm Bull 2002 25 409 417 11995915
- Ferguson MA Williams AF Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures Annu Rev Biochem 1988 57 285 320 3052274
- Cross GA Glycolipid anchoring of plasma membrane proteins Annu Rev Cell Biol 1990 6 1 39 2148872
- Low MG Glycosyl-phosphatidylinositol: a versatile anchor for cell surface proteins FASEB J 1989 3 1600 1608 2522071
- Diep DB Nelson KL Raja SM Pleshak EN Buckley JT Glycosylphosphatidylinositol anchors of membrane glycoproteins are binding determinants for the channel-forming aerolysin J Biol Chem 1998 273 2355 2360 9442081
- Gmachl M Sagan S Ketter S Kreil G The human sperm protein PH-20 has hyaluronidase activity FEBS Lett 1993 336 545 548 8282124
- Tozeren A Sung KL Dustin ML Chan PY Springer TA Chien S Micromanipulation of adhesion of a Jurkat cell to a planar bilayer membrane containing lymphocyte function-associated antigen 3 molecules J Cell Biol 1992 116 997 1006 1370839
- Müller G Bandlow W Lipolytic membrane release of two phosphatidylinositol-anchored cAMP receptor proteins in yeast alters their ligand-binding parameters Arch Biochem Biophys 1994 308 504 514 8109981
- Wang X Jansen G Fan J Variant GPI structure in relation to membrane-associated functions of a murine folate receptor Biochemistry 1996 35 16305 16312 8973205
- Kondoh G Tojo H Nakatani Y Angiotensin-converting enzyme is a GPI-anchored protein releasing factor crucial for fertilization Nat Med 2005 11 160 166 15665832
- Chan BL Lisanti MP Rodriguez-Boulan E Saltiel AR Insulin- stimulated release of lipoprotein lipase by metabolism of its phosphatidylinositol anchor Science 1988 241 1670 1672 2843987
- Romero G Luttrell L Rogol A Zeller K Hewlett E Larner J Phosphatidylinositol-glycan anchors of membrane proteins: potential precursors of insulin mediators Science 1988 240 509 511 3282305
- Musaka R Umeda M Endo T Kobata A Inoue K Characterization of glycosylphosphatidylinositol (GPI)-anchored NCAM on mouse skeletal muscle cell line C2C12: the structure of the GPI glycan and release during myogenesis Arch Biochem Biophys 1995 318 182 190 7726560
- Naghibalhossaini F Ebadi P Evidence for CEA release from human colon cancer cells by an endogenous GPI-PLD enzyme Cancer Lett 2006 234 158 167 15893415
- Bianco C Strizzi L Normanno N Khan N Salomon DS Cripto-1: an oncofetal gene with many faces Curr Top Dev Biol 2005 67 85 133 15949532
- Turner AJ Hooper NM The angiotensin-converting enzyme gene family: genomics and pharmacology Trends Pharmacol Sci 2002 23 177 183 11931993
- Kawagoe K Kitamura D Okabe M Glycosylphosphatidylinositol- anchor deficient mice: implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria Blood 1996 87 3600 3606 8611683
- Rosti V Tremml G Soares V Pandolfi PP Luzzatto L Bessler M Murine embryonic stem cells without Pig-a gene activity are competent for hematopoiesis with the PNH phenotype but not for clonal expansion J Clin Invest 1997 100 1028 1036 9276719
- Nozaki M Ohishi K Yamada N Kinoshita T Nagy A Takeda J Developmental abnormalities of glycosylphosphatidylinositol-anchor deficient embryos revealed by Cre/loxP system Lab Invest 1999 79 293 299 10092065
- Abdul-Aziz NM Turmaine M Greene ND Copp AJ Ephrin-EphA receptor interactions in mouse spinal neurulation: implications for neural fold fusion Int J Dev Biol 2009 53 559 568 19247962
- Watanabe K Bianco C Strizzi L Growth factor induction of Cripto-1 shedding by glycosylphosphatidylinositol-phospholase D and enhancement of endothelial cell migration J Biol Chem 2007 282 43 31643 31655 17720976
- Slein MW Logan GFJr Partial purification and properties of two phospholipases of Bacillus cereus J Bacteriol 1963 85 369 381 13989217
- Ikezawa H Yamanegi M Taguchi R Miyashita T Ohyabu T Studies on phosphatidylinositol phosphodiesterase (phospholipase C type) of Bacillus cereus: I. purification, properties and phosphatase-releasing activity Biochim Biophys Acta 1976 450 154 164 10986
- Futerman AH Low MG Ackermann KE Sherman WR Silman I Identification of covalently bound inositol in the hydrophobic membrane-anchoring domain of Torpedo acetylcholinesterase Biochem Biophys Res Commun 1985 129 312 317 4004881
- Robert WL Rosenberry TL Identification of covalently attached fatty acids in the hydrophobic membrane-binding domain of human erythrocyte acetylcholinesterase Biochem Biophys Res Commun 1985 133 621 627 4084290
- Tse AG Barclay AN Watts A Williams AF A glycophospholipid tail at the carboxyl terminus of the Thy-1 glycoprotein of neurons and thymocytes Science 1985 230 1003 1008 2865810
- Ferguson MA Haldar K Cross GA Trypanosoma brucei variant surface glycoprotein has a sn-1,2-dimyristyl glycerol membrane anchor at its COOH terminus J Biol Chem 1985 260 4963 4968 3988741
- Ferguson MA Low MG Cross GA Glycosyl-sn-1,2-dimyristylphos-phatidylinositol is covalently linked to Trypanosoma brucei variant surface glycoprotein J Biol Chem 1985 260 14547 14555 4055788
- Eisenhaber B Bork P Eisenhaber F Post-translational GPI lipid anchor modification of proteins in kingdoms of life: analysis of protein sequence data from complete genomes Protein Eng 2001 14 17 25 11287675
- Kinoshita T Inoue N Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis Curr Opin Struct Biol 2000 4 632 638
- Eisenhaber B Maurer-Stroh S Novatchkova M Schneider G Eisenhaber F Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins Bio Essays 2003 25 367 385
- Miyata T Takeda J Iida Y The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis Science 1993 259 1318 1320 7680492
- Takeda J Miyata T Kawagoe K Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria Cell 1993 73 703 711 8500164
- Bessler M Mason PJ Hillmen P Paroxysmal nocturnal haemoglobinuria (PNH) is caused by somatic mutations in the PIG-A gene EMBO J 1994 13 110 117 8306954
- Kinoshita T Inoue N Takeda J Defective glycosyl phosphatidylinositol anchor synthesis and paroxysmal nocturnal hemoglobinuria Adv Immunol 1995 60 57 103 8607375
- Rosse WF Ware RE The molecular basis of paroxysmal nocturnal hemoglobinuria Blood 1995 86 3277 3286 7579428
- Mortazavi Y Merk B McIntosh J Marsh JC Schrezenmeier H Rutherford TR The spectrum of PIG-A gene mutations in aplastic anemia/paroxysmal nocturnal hemoglobinuria (AA/PNH): a high incidence of multiple mutations and evidence of a mutational hot spot Blood 2003 101 2833 2841 12424196
- Luzzatto L Somatic mutations in paroxysmal nocturnal hemoglobinuria: a blessing in disguise? Cell 1997 88 1 4 9019395
- Barcellini W Increased resistance of PIG-A-bone marrow progenitors to tumor necrosis factor α and interferon γ: possible implications for the in vivo dominance of paroxysmal nocturnal hemoglobinuria clones Haematologica 2004 89 651 656 15194531
- Ait-Slimane T Galmes R Trugnan G Maurice M Basolateral internalization of GPI-anchored proteins occurs via clathrin-independent flotillin-dependent pathway in polarized hepatic cells Mol Biol Cell 2009 20 3792 3800 19605558
- Munro S Lipid rafts: elusive or illusive? Cell 2003 115 377 388 14622593
- Simons K Toomre D Lipid rafts and signal transduction Nat Rev Mol Cell Biol 2000 1 31 39 11413487
- Müller G Over S Wied S Frick W Association of (c)AMP-degrading glycosylphosphatidylinositol-anchored proteins with lipid droplets is induced by palmitate, H2O2 and the sulfonylurea drug, glimepiride, in rat adipocytes Biochemistry 2008 47 12774 12787
- Müller G Wied S Over S Frick W Inhibition of lipolysis by palmitate, H2O2 and the sulfonylurea drug, glimepiride, in rat adipocytes depends on cAMP degradation by lipid droplets Biochemistry 2008 47 1259 1273 18186616
- Müller G Wied S Walz N Jung C Translocation of glycosylphosphatidylinositol- anchored proteins from plasma membrane microdomains to lipid droplets in rat adipocytes is induced by palmitate, H2O2 and the sulfonylurea drug, glimepiride Mol Pharmacol 2008 73 1513 1529 18272749
- Müller G Jung C Straub J Wied S Induced release of membrane vesicles and exosomes from rat adipocytes containing lipid droplet, lipid raft and glycosylphosphatidylinositol-anchored proteins Cell Signal 2009 21 324 338 19010410
- Müller G Jung C Wied S Biemer-Daub G Induced translocation of glycosylphosphatidylinositol-anchored proteins from lipid droplets to adiposomes in rat adipocytes Br J Pharmacol 2009 158 749 770 19703169
- Brügger B Graham C Leibrecht I The membrane domains occupied by glycosylphosphatidylinositol-anchored prion protein and Thy-1 differ in lipid composition J Biol Chem 2004 279 7530 7536 14660659
- Szpurka H Schade AE Jankowska AM Maciejewski JP Altered lipid raft composition and defective cell death signal transduction in glycosylphosphatidylinositol anchor-deficient PIG-A mutant cells Br J Haematol 2008 142 413 422 18544084
- Lai EC Lipid rafts make for slippery platforms J Cell Biol 2003 162 365 370 12885764
- Varma R Mayor S GPI-anchored proteins are organized in submicron domains at the cell surface Nature 1998 394 798 801 9723621
- Friedrichson T Kurzchalia TV Microdomains of GPI-anchored proteins in living cells revealed by crosslinking Nature 1998 394 802 805 9723622
- Harder T Scheiffele P Verkade P Simons K Lipid domain structure of the plasma membrane revealed by patching of membrane components J Cell Biol 1998 141 929 942 9585412
- Pralle A Sphingolipidcholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells J Cell Biol 2000 148 997 1008 10704449
- Sharma P Varma R Sarasij RC Nanoscale organization of multiple GPI-anchored proteins in living cell membranes Cell 2004 116 4 577 589 14980224
- Ayuyan AG Cohen FS Raft composition at physiological temperature and pH in the absence of detergents Biophys J 2007 94 2654 2666 17993486
- Pinaud F Michalet X Gopal I Margeat E Moore H-P Weiss S Dynamic partitioning of a glycosyl-phosphatidylinositol-anchored protein in glycosphingolipid-rich microdomains imaged by single-quantum dot Tracking Traffic 2009 10 691 712
- Wang TY Leventis R Silvius JR Artificially lipid-anchored proteins can elicit clustering-induced intracellular signaling events in Jurkat thymphocytes independent lipid raft association J Biol Chem 2005 280 24 22839 22846 15817446
- Douglass AD Vale RD Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells Cell 2005 121 937 950 15960980
- Rooney IA Heuser JE Atkinson JP GPI-anchored complement regulatory proteins in seminal plasma. An analysis of their physical condition and the mechanisms of their binding to exogenous cells J Clin Invest 1996 97 7 1675 1686 8601633
- Zacharias DA Violon JD Newton AC Tsien RY Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells Science 2002 296 913 916 11988576
- Mayor S Riezman H Sorting of GPI-anchored proteins Nat Rev Mol Cell Biol 2004 5 110 120 15040444
- Leventhal I Grzybek M Simons K Greasing their way: lipid modifications determine protein association with membrane rafts Biochemistry 2010 49 6305 6316 20583817
- Bouma SR Drislane FW Huestis WH Selective extraction of membrane-bound proteins by phospholipid vesicles J Biol Chem 1977 252 19 6759 6763 893440
- Cook SL Bouma SR Huestis WH Cell to vesicle transfer of intrinsic membrane proteins: effect of membrane fluidity Biochemistry 1980 19 20 4601 4607 7426618
- Medof ME Nagarajan S Tykocinski ML Cell surface engineering with GPI-anchored proteins FASEB J 1996 10 5 574 586 8621057
- Kooyman DL Byrne GW McClelland S In vivo transfer of GPI-linked complement restriction factors from erythrocytes to the endothelium Science 1995 269 5220 89 92 7541557
- Rifkin MR Landsberger FR Trypanosome variant surface glycoprotein transfer to target membranes: a model for the pathogenesis of trypanosomiasis Proc Natl Acad Sci U S A 1990 87 801 805 2300563
- Sloand EM Maciejewski JP Dunn D Correction of the PNH defect by GPI-anchored protein transfer Blood 1998 92 11 4439 4445 9834251
- Sloand EM Mainwaring L Keyvanfar K Transfer of glycosylphosphatidylinositol- anchored proteins to deficient cells after erythrocyte transfusion in paroxysmal nocturnal hemoglobinuria Blood 2004 104 12 3782 3788 15304386
- Babiker AA Ronquist G Nilsson UR Nilsson B Transfer of prostasomal CD59 to CD59-deficient red blood cells results in protection against complement-mediated hemolysis Am J Reprod Immunol 2002 47 3 183 192 12069204
- Harding C Heuser J Stahl P Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes J Cell Biol 1983 97 2 329 339 6309857
- van Niel G Raposo G Candalh C Intestinal epithelial cells secrete exosome-like vesicles Gastroenterology 2001 121 2 337 349 11487543
- Wolfers J Lozier A Raposo G Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming Nat Med 2001 7 3 297 303 11231627
- Blanchard N Lankar D Faure F TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex J Immunol 2002 168 7 3235 3241 11907077
- Zitvogel L Regnault A Lozier A Eradication of established murine tumors using a novel cell-free vaccine: dendritic cells-derived exosomes Nat Med 1998 4 5 594 600 9585234
- Brasoveanu LI Fonsatti E Visintin A Melanoma cells constitutively release an anchor-positive soluble form of protectin (sCD59) that retains functional activities in homologous complement-mediated cytotoxicity J Clin Invest 1997 100 5 1248 1255 9276743
- Raposo G Nijman HW Stoorvogel W B lymphocytes secrete antigen-presenting vesicles J Exp Med 1996 183 3 1161 1172 8642258
- Ronquist G Brody I The prostasome: its secretion and function in man Biochim Biophys Acta 1985 822 203 218 2992593
- Rooney IA Atkinson JP Krul ES Physiologic relevance of the membrane attack complex inhibitory protein CD59 in human seminal plasma: CD59 is present on extracellular organelles (prostasomes), binds cell membranes, and inhibits complement-mediated lysis J Exp Med 1993 177 1409 1420 7683035
- Long KE Yomtovian R Kida M Knez JJ Medof ME Time-dependent loss of surface complement regulatory activity during storage of donor blood Transfusion 1993 33 4 294 300 7683151
- Bütikofer P Kuypers FA Xu CM Chiu DT Lubin B Enrichment of two glycosyl-phosphatidylinositol-anchored proteins, acetylcholinesterase and decay accelerating factor, in vesicles released from human red blood cells Blood 1989 74 5 1481 1485 2477079
- van den Berg CW Cinek T Hallett MB Horejsi V Morgan BP Exogenous glycosyl phosphatidylinositol-anchored CD59 associates with kinases in membrane clusters on U937 cells and becomes Ca(2+)-signaling competent J Cell Biol 1995 131 3 669 677 7593188
- Civenni G Test ST Brodbeck U Bütikofer P In vitro incorporation of GPI-anchored proteins into human erythrocytes and their fate in the membrane Blood 1998 91 5 1784 1792 9473247
- Premkumar DR Fukuoka Y Sevlever D Properties of exogenously added GPI-anchored proteins following their incorporation into cells J Cell Biochem 2001 82 2 234 245 11527149
- Zhang F Schmidt WG Hou Y Williams AF Jacobson K Spontaneous incorporation of the glycosyl-phosphatidylinositol-linked protein Thy-1 into cell membranes Proc Natl Acad Sci U S A 1992 89 12 5231 5235 1351678
- Bülow R Overath P Davoust J Rapid lateral diffusion of the variant surface glycoprotein in the coat of Trypanosoma brucei Biochemistry 1988 27 7 2384 2388 3382629
- Medof ME Kinoshita T Silber R Nussenzweig V Amelioration of lytic abnormalities of paroxysmal nocturnal hemoglobinuria with decay accelerating factor Proc Natl Acad Sci U S A 1985 82 9 2980 2984 2581259
- Dunn DE Yu J Nagarajan S Devetten M A knock-out model of paroxysmal nocturnal hemoglobinuria: Pig-a(-) hematopoiesis is reconstituted following intercellular transfer of GPI-anchored proteins Proc Natl Acad Sci U S A 1996 93 15 7938 7943 8755581
- De Broe ME Wieme RJ Logghe GN Spontaneous shedding of plasma membrane fragments by human cells in vivo and in vitro Clin Chim Acta 1977 81 237 245 923096
- Anderson SM Yu G Giattina M Miller JL Intercellular transfer of a glycosylphosphatidylinositol (GPI)-linked protein: release and uptake of CD4-GPI from recombinant adeno-associated virus-transduced HeLa cells Proc Natl Acad Sci U S A 1996 93 12 5894 5898 8650189
- Keller GA Siegel MW Caras IW Endocytosis of glycophospholipid-anchored and transmembrane forms of CD4 by different endocytic pathways EMBO J 1992 11 3 863 874 1532143
- Vakeva A Jauhiainen M Ehnholm C Lehto T Meri S High-density lipoproteins can act as carriers of glycophosphoinositol lipid-anchored CD59 in human plasma Immunology 1994 82 1 28 33 7519171
- Metzner C Mostegl MM Günzburg WH Salmons B Dangerfield JA Association of glycosylphosphatidylinositol (GPI)-linked protein with retroviral particles FASEB J 2008 22 8 2734 2739 18477763
- Chesebro B Trifilo M Race R Anchorless prion protein results in infectious amyloid disease without clinical scrapie Science 2005 308 5727 1435 1439 15933194
- McNally KL Ward AE Priola SA Cells expressing anchorless prion protein are resistant to scrapie infection J Virol 2009 83 9 4469 4475 19225008
- Gould SJ Booth AM Hildreth JE The Trojan exosome hypothesis Proc Natl Acad Sci U S A 2003 100 19 10592 10597 12947040
- Lauc G Heffer-Lauc M Shedding and uptake of gangliosides and glycosylphosphatidylinositol-anchored proteins Biochim Biophys Acta 2006 1760 584 602 16388904
- Camussi G Deregibus MC Bruno S Cantaluppi V Biancone L Exosomes/microvesicles as a mechanism of cell-to-cell communication Kidney Int 2010 78 838 848 20703216
- Lu P Sharom FJ Immunosuppression by YAC-1 lymphoma: role of shed gangliosides Cell Immunol 1996 173 22 32 8871598
- Deng W Li R Ladisch S Influence of cellular ganglioside depletion on tumor formation J Natl Cancer Inst 2000 92 11 912 917 10841826
- Simons M Friedrichson T Schulz JB Pitto M Masserini M Kurzchalia TV Exogenous administration of gangliosides displaces GPI-anchored proteins from lipid microdomains in living cells Mol Biol Cell 1999 10 10 3187 3196 10512859
- Crespo PM Zurita AR Daniotti JL Effect of gangliosides on the distribution of a glycosylphosphatidylinositol-anchored protein in plasma membrane from Chinese hamster ovary-K1 cells J Biol Chem 2002 277 47 44731 44739 12237294
- Ladisch S Gillard B Wong C Ulsh L Shedding and immunoregulatory activity of YAC-1 lymphoma cell gangliosides Cancer Res 1983 43 8 3808 3813 6861145
- Müller G Wied S Jung C Biemer-Daub G Frick W Transfer of glycosylphosphatidylinositol-anchored 5′-nucleotidase CD73 from adiposomes into rat adipocytes stimulates lipid synthesis Br J Pharmacol 2010 160 878 891 20590586
- Müller G Wied S Jung C Frick W Biemer-Daub G Inhibition of lipolysis by adiposomes containing glycosylphosphatidylinositol-anchored Gce1 protein in rat adipocytes Arch Physiol Biochem 2010 116 28 41 20053127
- Müller G Wied S Dearey E-A Biemer-Daub G Glycosylphosphatidylinositol- anchored proteins coordinate lipolysis inhibition between large and small adipocytes Metabolism 2011 60 1021 1037 21129759
- Müller G Schneider M Biemer-Daub G Wied S Upregulation of lipid synthesis in small rat adipocytes by microvesicle-associated CD73 from large adipocytes Obesity (Silver Spring) 2011 19 1531 1544 21372807
- Müller G Let’s shift lipid burden – from large to small adipocytes Eur J Pharmacol 2011 656 1 4 21295025
- Müller G Schneider M Gassenhuber J Wied S Release of exosomes and microvesicles harbouring specific RNAs and glycosylphosphatidylinositol- anchored proteins from rat and human adipocytes is controlled by histone methylation Am J Mol Biol 2012
- Müller G Personalised strategies for the diagnosis and therapy of type II diabetes and obesity Immunol Endocr Metabol Agents Med Chem 2012 12 80 109
- Müller G Novel target identification technologies for the personalised therapy of type II diabetes and obesity Immunol Endocr Metabol Agents Med Chem 2012 In press
- Skog J Würdinger T van Rijn S Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers Nat Cell Biol 2008 10 1470 1476 19011622
- Qazi KR Torregrosa Paredes P Dahlberg B Grunewald J Eklund A Gabrielsson S Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis Thorax 2010 65 1016 1024 20880880
- Al-Nedawi K Meehan B Kerbel RS Allison AC Rak J Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR Proc Natl Acad Sci U S A 2009 106 3794 3789 19234131
- Kogure T Lin WL Yan IK Braconi C Patel T Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth Hepatology 2011 54 1237 1248 21721029
- Castellana D Toti F Freyssinet JM Membrane microvesicles: Macromessengers in cancer disease and progression Thrombosis Res 2010 125 S84 S88
- Baj-Krzyworzeka M Szatanek R Weglarczyk K Baran J Zembala M Tumour-derived microvesicles modulate biological activity of human monocytes Immunol Lett 2007 113 76 82 17825925
- Yuan A Farber EL Rapoport AL Transfer of microRNAs by embryonic stem cell microvesicles PLoS One 2009 4 e4722 19266099
- Pegtel DM Cosmopoulos K Thorley-Lawson DA Functional delivery of viral miRNAs via exosomes Proc Natl Acad Sci U S A 2010 107 6328 6333 20304794
- Liu Y Xiang X Zhuang X Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells Am J Pathol 2010 176 2490 2499 20348242
- Lim PK Bliss SA Patel SA Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells Cancer Res 2011 71 1550 1560 21343399
- Yang M Chen J Su F Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells Mol Cancer 2011 10 117 21939504
- Deregibus MC Cantaluppi V Calogero R Endothelial progenitor of cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA Blood 2007 110 2440 2448 17536014
- Montecalvo A Larregina AT Shufesky WJ Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes Blood 2012 119 756 766 22031862
- Meckes DGJr Shair KH Marquitz AR Kung CP Edwards RH Raab-Traub N Human tumor virus utilizes exosomes for intercellular communication Proc Natl Acad Sci U S A 2010 107 20370 20375 21059916
- Pegtel DM Van De Garde MD Middeldorp JM Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion Biochim Biophys Acta 2011 1809 715 721 21855666
- Boulanger CM Scoazec A Ebrahimian T Circulating microparticles from patients with myocardial infarction cause endothelial dysfunction Circulation 2001 104 2649 2652 11723013
- Wilhelm OG Wilhelm S Escott GM Cellular glycosylphosphatidylinositol- specific phospholipase D regulates urokinase receptor shedding and cell surface expression J Cell Physiol 1999 180 2 225 235 10395292
- Omoto S Nomura S Shouzu A Nishikawa M Fukuhara S Iwasaka T 2002 Detection of monocyte-derived microparticles in patients with Type II diabetes mellitus Diabetologia 2011 45 550 555 12032632
- Schorey JS Bhatnagar S Exosome function: from tumor immunology to pathogen biology Traffic 2008 9 871 881 18331451
- Simpson RJ Lim JW Moritz RL Mathivanan S Exosomes: proteomic insights and diagnostic potential Expert Rev Proteomics 2009 6 3 267 283 19489699
- Timmers L Lim SK Arslan F Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium Stem Cell Res 2007 1 2 129 137 19383393
- Choo A Padmanabhan J Chin A Fong WJ Oh SK Immortalized feeders for the scale-up of human embryonic stem cells in feeder and feeder-free conditions J Biotechnol 2006 122 1 130 141 16233925
- Pisitkun T Shen RF Knepper MA Identification and proteomic profiling of exosomes in human urine Proc Natl Acad Sci U S A 2004 101 36 13368 13373 15326289
- Horstman LL Cell-derived microparticles and exosomes in neuroinflammatory disorders Int Rev Neurobiol 2007 79 227 268 17531844
- Ichim TE Zhong Z Kaushal S Exosomes as a tumor immune escape mechanism: possible therapeutic implications J Transl Med 2008 6 37 18644158
- Mitchell PJ Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med 2009 7 4 19138409
- Rabinowits G Gercel-Taylor C Day JM Taylor DD Kloecker GH Exosomal microRNA: a diagnostic marker for lung cancer Clin Lung Cancer 2009 10 42 46 19289371
- Simak J Gelderman MP Cell membrane microparticles in blood and blood products: Potentially pathogenic agents and diagnostic markers Transfusion Med Rev 2006 20 1 26
- Freyssinet JM Cellular microparticles: What are they bad or good for? J Thromb Haemost 2003 1 1655 1662 12871302
- Horstman LL Jy W Jimenez JJ Bidot C Ahn YS New horizons in the analysis of circulating cell-derived microparticles Keio J Med 2004 53 4 210 230 15647627
- Horstman LL Jy W Jimenez JJ Ahn YS Endothelial microparticles as markers of endothelial dysfunction Front Biosci 2004 9 1118 1135 14977533
- Morel O Toti F Hugel B Freyssinet JM Cellular microparticles: A disseminated storage pool of bioactive vascular effectors Curr Opin Hematol 2004 11 156 164 15257014
- Nomura S Fukuhara S Platelet microparticles Methods Mol Biol 2004 272 269 277 15226550
- Jimenez JJ Jy W Mauro LM Soderland C Horstmann LL Ahn YS Endothelial cells release phenotypically and quantitatively distinct microparticles in activation and apoptosis Thromb Res 2003 109 175 180 12757771
- Van der Pol E Van Gemmert MJ Sturk A Nieuwland R Van Leeuwen TG Single vs swarm detection of microparticles and exosomes by flow cytometry J Thromb Haemost 2012 10 919 930 22394434
- Leong HS Podor TJ Manocha B Lewis JD Validation of flow cytometric detection of platelet microparticles and liposomes by atomic force microscopy J Thromb Haemost 2011 9 2466 2476 21981726
- Blastos-Amador P Royo F Gonzalez E Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability J Proteom 2012 75 3574 3584
- Nomura S Function and clinical significance of platelet-derived microparticles Int J Hematol 2001 74 397 404 11794694
- Chargaff E West R The biological significance of the thromboplastic protein of blood J Biol Chem 1946 166 189 197 20273687
- Wolf P The nature and significance of platelet products in human plasma Br J Haematol 1967 13 269 288 6025241
- Bernal-Mizrachi L Jy W Jimenez JJ High levels of circulating endothelial microparticles in patients with acute coronary syndromes Am Heart J 2003 145 962 970 12796750
- Preston RA Jy W Jimenez JJ Effects of severe hypertension on endothelial and platelet microparticles Hypertension 2003 41 211 217 12574084
- Brodsky SV Zhang F Nasjietti A Gollgorsky MS Endotheliumderived microparticles impair endothelial function in vitro Am J Physiol Heart Circ Physiol 2004 286 H1910 H1915 15072974
- Wang Z Hill S Luther JM Hachey DL Schey KL Protemic analysis of urine exosomes by multidimensional protein identification technology (MudPIT) Proteomics 2012 12 329 338 22106071
- Zhang Y Li Y Qiu F Qiu Z Comprehensive analysis of low-abundance proteins in human urinary exosomes using peptide ligand library technology, peptide OFFGEL fractionation and nanoHPLC-chip-MS/MS Electrophoresis 2010 31 3797 3807 21082674
- Merchant ML Powell DW Wilkey DW Microfiltration isolation of human urinary exosomes for characterization by MS Proteomics Clin Appl 2010 4 84 96 21137018
- Verkman AS Dissecting the roles of aquaporins in renal pathophysiology using transgenic mice Semin Nephrol 2008 28 217 226 18519083
- Bolton K Segal D McMillan J Decorin is a secreted protein associated with obesity and type 2 diabetes Int I Obes (London) 2008 32 1113 1121
- Fisher LW Termine JD Young MF Deduced protein sequence of bone small proteoglycan I (biglycan) shows homology with proteoglycan II (decorin) and several nonconnective tissue proteins in a variety of species J Biol Chem 1989 264 4571 4576 2647739
- Rahmani M Wong BW Ang L Versican: signaling to transcriptional control pathway Can J Physiol Pharmacol 2006 84 77 92 16845893
- Kresse H Schonherr E Proteoglycans of the extracellular matrix and growth control J Cell Physiol 2001 189 266 274 11748584
- Iozzo RV Murdoch AD Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function Faseb J 1996 10 598 614 8621059
- King VL Hatch NW Chan HW de Beer MC de Beer FC Tannock LR A murine model of obesity with accelerated atherosclerosis Obesity (Silver Spring) 2010 18 35 41 19498343
- Zhang W Chuang YJ Jin T Antiangiogenic antithrombin induces global changes in the gene expression profile of endothelial cells Cancer Res 2006 66 5047 5055 16707426
- Bolton K Segal D Walder K The small leucine-rich proteoglycan, biglycan, is highly expressed in adipose tissue of Psammomys obesus and is associated with obesity and type 2 diabetes Biol Targ Therap 2012 6 67 72
- Esposito K Maiorino MI Di Palo C Effects of pioglitazone versus metformin on circulating endothelial microparticles and progenitor cells in patients with newly diagnosed type 2 diabetes – a randomized controlled trial Diabetes Obes Metab 2011 13 439 445 21255215
- Diamant M Nieuwland R Pablo RF Sturk A Smit JW Radder JK Elevated numbers of tissue-factor exposing microparticles correlate with components of the metabolic syndrome in uncomplicated type 2 diabetes mellitus Circulation 2002 106 2442 2447 12417540
- Diamant M Nieuwland R Pablo RF Sturk A Smit JW Radder JK Elevated numbers of tissue-factor exposing microparticles correlate with components of the metabolic syndrome in uncomplicated type 2 diabetes mellitus Circulation 2002 106 2442 2447 12417540
- Esposito K Ciotola M Giugliano D Pioglitazone reduces endothelial microparticles in the metabolic syndrome Arterioscler Thromb Vasc Biol 2006 26 1926 16857960
- Setzer F Oberle V Bläss M Platelet-derived microvesicles induce differential gene expression in monocytic cells: a DNA microarray study Platelets 2006 17 571 576 17127485
- Ferreira AC Peter AA Mendez AJ Postprandial hypertriglyceridemia increases circulating levels of endothelial cell microparticles Circulation 2004 110 3599 3603 15569844
- Freyssinet JM Toti F Formation of procoagulant microparticles and properties Thromb Res 2010 125 Suppl 1 S46 S48 20153515
- Agouni A Lagrue-Lak-Hal AH Ducluzeau PH Endothelial dysfunction caused by circulating microparticles from patients with metabolic syndrome Am J Pathol 2008 173 1210 1219 18772329
- Helal O Defoort C Robert S Increased levels of microparticles originating from endothelial cells, platelets and erythrocytes in subjects with metabolic syndrome: relationship with oxidative stress Nutr Metab Cardiovasc Dis 2010 21 665 671 20399083
- Cipoletta C Ryan KE Hanna EV Trimble ER Activation of peripheral blood CD14+ monocytes occurs in diabetes Diabetes 2005 54 2779 2786 16123369
- Sabatier F Darmon P Hugel B Type 1 and type 2 diabetic patients display different patterns of cellular microparticles Diabetes 2002 51 9 2840 2845 12196479
- Tan KT Tayebjee MH Lim HS Lip GY Clinically apparent atherosclerotic disease in diabetes is associated with an increase in platelet microparticle levels Diabet Med 2005 22 1657 1662 16401308
- Koga H Sugiyama S Kugiyama K Elevated levels of remnant lipoproteins are associated with plasma platelet microparticles in patients with type-2 diabetes mellitus without obstructive coronary artery disease Eur Heart J 2006 27 817 823 16434416
- Esposito K Ciotola M Schisano B Endothelial microparticles correlate with endothelial dysfunction in obese women J Clin Endocrinol Metab 2006 91 3676 3679 16822816
- Tramontano AF Lyubarova R Tsiakos J Palaia T Deleon JR Ragolia L Circulating endothelial microparticles in diabetes mellitus Mediators Inflamm 2010 2010 Article ID 250476
- Nomura S Suzuki M Katsura K Platelet-derived microparticles may influence the development of atherosclerosis in diabetes mellitus Atherosclerosis 1995 116 235 240 7575778
- Omoto S Nomura S Shouzu A Nishikawa M Fukuhara S Iwasaka T Detection of monocyte-derived microparticles in patients with type II diabetes mellitus Diabetologia 2002 45 550 555 12032632
- Morel O Hugel B Jesel L Sustained elevated amounts of circulating procoagulant membrane microparticles and soluble GPV after acute myocardial infarction in diabetes mellitus Thromb Haemost 2004 91 345 353 14961163
- Subbarayan S Kipnes M Sitagliptin: a review Expert Opin Pharmacother 2011 12 1613 1622 21651449
- Baetta R Corsini A Pharmacology of dipeptidyl peptidase-4 inhibitors: similarities and differences Drugs 2011 71 1441 1467 21812507
- Pisitkun T Johnstone R Knepper MA Discovery of urinary biomarkers Mol Cell Proteomics 2006 5 1760 1761 16837576
- Sun AL Deng JT Guan GJ Dipeptidyl peptidase-IV is a potential molecular biomarker in diabetic kidney disease Diabetes Vasc Dis Res 2012 In press
- Pisitkun T Shen RF Knepper MA Identification and proteomic profiling of microvesicles in human urine Proc Natl Acad Sci U S A 2004 101 13368 13373 15326289
- Pala L Mannucci E Pezzatini A Dipeptidyl peptidase-IV expression and activity in human glomerular endothelial cells Biochem Biophys Res Commun 2003 310 28 31 14511643
- Hu G Zhou R Liu J MixcroRNA-98 and let-7 confer cholangiocyte expression of cytokine–inducible Src homology 2-containing protein in response to microbial challenge J Immunol 2009 183 1617 1624 19592657
- Kota J Chivukula RR O’Donnell KA Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model Cell 2009 137 1005 1007 19524505
- Esau C Davis S Murray S miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting Cell Metabol 2006 3 87 98
- Trajkovski M Hausser J Soutschek J MicroRNAs 103 and 107 regulate insulin sensitivity Nature 2011 474 649 654 21654750
- Nathan DM Buse JB Davidsom MB Medical management of hyperglycemia in type 2 diabetes: A consensus algorithm for the initiation and adjustment of therapy: A consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes Diabetes Care 2009 32 193 203 18945920
- Leroyer AS Tedgui A Boulanger CM Microparticles and type 2 diabetes Diabetes Metab 2008 34 S27 31 18358424
- Müller G Wied S Dearey E-A Wetekam E-M Biemer-Daub G Lipid storage in large and small rat adipocytes by vesicle-associated glycosylphosphatidylinositol-anchored proteins results and problems in cell differentiation Richter W Beisiegel U Joost G Meyerhof J Sensory and Metabolic Control of Energy Balance Berlin Springer Press 2010 52 27 34
- De Ferranti S Mozaffarian D The perfect storm: obesity, adipocyte dysfunction, and metabolic consequences Clin Chem 2010 54 945 955 18436717
- Müller G Control of lipid storage and cell size between adipocytes by vesicle-associated glycosylphosphatidylinositol-anchored proteins Arch Physiol Biochem 2010 117 23 43 20883086
- Cui J Panse S Falkner B The role of adiponectin in metabolic and vascular disease: a review Clin Nephrol 2011 75 26 33 21176748
- Xie H Lim B Lodish HF MicroNAs induced during adipogenesis that accelerates fat cell development are downregulated in obesity Diabetes 2009 58 1050 1057 19188425
- Ortega FJ Moreno-Navarrete JM Pardo G MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation PLoS One 2010 5 e9022 20126310
- Aoki N Yokoyama R Asai N Adipocyte-derived microvesicles are associated with multiple angiogenic factors and induce angiogenesis in vivo and in vitro Endocrinology 2010 151 2567 2576 20382694
- Ogawa R Tanaka C Sato M Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation Biochem Biophys Res Commun 2010 398 723 729 20621060
- Hulsmans M De Keyzer D Holvoet P MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis FASEB J 2011 25 2515 2527 21507901
- Zhang Y Liu D Chen X Secreted monocytic miR-150 enhances targeted endothelial cell migration Mol Cell 2010 39 133 144 20603081
- Fichtlscherer S De RS Fox H Circulating microRNAs in patients with coronary artery disease Circ Res 2010 107 677 684 20595655
- Matkovich SJ Van Booven DJ Youker KA Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support Circulation 2009 119 1263 1271 19237659
- Ceolotto G Papparella I Bortoluzzi A Interplay between miR- 155, AT1R A1166C polymorphism, and AT1R expression in young untreated hypertensives Am J Hypertens 2010 24 241 246 20966899
- Wang X Zhang X Ren X MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion- induced cardiac injury Circulation 2010 122 1308 1318 20837890
- Heneghan HM Miller N Kerin MJ Role of microRNAs in obesity and the metabolic syndrome Obesity Rev 2009 11 354 361
- Lin Q Gao Z Alarcon RM Ye J Yun Z A role of miR-27 in the regulation of adipogenesis FEBS J 2009 276 2348 2358 19348006
- Esau C Kang X Peralta E MicroRNA-143 regulates adipocyte differentiation J Biol Chem 2004 279 52361 52365 15504739
- Xie H Lim B Lodish HF MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity Diabetes 2009 58 1050 1057 19188425
- Hugel B Martinez MC Kunzelmann C Freyssinet J-M Membrane microparticles: Two sides of the coin Physiology 2005 20 22 27 15653836
- Freyssinet JM Cellular microparticles: what are they bad or good for? J Thromb Haemost 2003 1 1655 1662 12871302
- Morel O Toti F Hugel B Freyssinet JM Cellular microparticles: a disseminated storage pool of bioactive vascular effectors Curr Opin Hematol 2004 11 156 164 15257014
- Thery C Zitvogel L Amigorena S Exosomes: composition, biogenesis and function Nat Rev Immunol 2002 2 569 579 12154376
- Aoki N Jin-no S Nakagawa Y Identification and characterization of microvesicles secreted by 3T3-L1 adipocytes: redox- and hormone-dependent induction of milk fat globule-epidermal growth factor 8-associated microvesicles Endocrinology 2007 148 3850 3862 17478559
- Record M Subra C Silvente-Poirot Poirot M Exosomes as intracellular signalosomes and pharmacological effectors Biochem Pharmacol 2011 81 1171 1182 21371441
- Prieur X Mok CY Velagapudi VR Differential lipid partitioning between adipocytes and tissue macrophages modulates macrophage lipotoxicity and M2/M1 polarization in obese mice Diabetes 2011 60 797 809 21266330
- Skurk T Alberti-Huber C Herder C Hauner H Relationship between adipocyte size and adipokine expression and secretion J Clin Endocrinol Metab 2007 92 1023 1033 17164304
- Luo N Liu J Chung BH Macrophage adiponectin expression improves insulin sensitivity and protects against inflammation and atherosclerosis Diabetes 2010 59 791 799 20350970
- Rosen ED Spiegelman BM Molecular regulation of adipogenesis Annu Rev Cell Dev Biol 2000 16 145 171 11031233
- Müller G Schneider M Biemer-Daub G Wied S Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis Cell Signal 2011 23 1207 1223 21435393
- Chen PY Meister G microRNA-guided posttranscriptional gene regulation Biol Chem 2005 386 36 38
- Müller G Wied S Jung C Over S Translocation of glycosylphosphatidylinositol- anchored proteins to lipid droplets and inhibition of lipolysis in rat adipocytes is mediated by reactive oxygen species Br J Pharmacol 2008 154 901 913 18454169
- Müller G Wied S Jung C Straub J Coordinated regulation of esterification and lipolysis by palmitate, H2O2 and the anti-diabetic sulfonylurea drug, glimepiride, in rat adipocytes Eur J Pharmacol 2008 597 6 18 18789917
- Cao J Li JL Li D Tobin JF Gimeno RE Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis Proc Natl Acad Sci U S A 2006 103 19695 19700 17170135
- Nishino N Tamori Y Tateya S Kawaguchi T Shibakusa T FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets J Clin Invest 2008 118 2808 2821 18654663
- Puri V Ranjit S Konda S Cidea is associated with lipid droplets and insulin sensitivity in humans Proc Natl Acad Sci U S A 2008 105 7833 7838 18509062
- Goichot B Grunebaum L Desprez D Circulating procoagulant microparticles in obesity Diabetes Metab 2006 32 82 85 16523191
- Wang JG Manly D Kirchhofer D Pawlinksi R Mackman N Levels of microparticle tissue factor activity correlate with coagulation activation in endotoxemic mice J Thromb Haemost 2009 7 1092 1098 19422446
- Heerwagen MJR Miller MR Barbour LA Friedman JE Maternal obesity and fetal metabolic programming: a fertile epigenetic soil Am J Physiol Regul Integr Comp Physiol 2010 299 R711 R722 20631295
- Stöger R Epigenetics and obesity Pharmacogenomics 2008 9 1851 1860 19072643
- Jenuwein T Allis CD Translating the histone code Science 2001 293 1074 1080 11498575
- Schreiber SL Bernstein BE Signaling network model of chromatin Cell 2002 111 771 778 12526804
- Martin C Zhang Y The diverse functions of histone lysine modification Nat Rev Mol Cell Biol 2005 6 838 849 16261189
- Jenuwein T The epigenetic magic of histone lysine methylation FEBS J 2006 273 3121 3135 16857008
- Cole PA Chemical probes for histone-modifying enzymes Nat Chem Biol 2008 4 590 597 18800048
- Spannhoff A Hauser AT Heinke R Sippl W Jung M The emerging therapeutic potential of histone methyltransferase and demethlyase inhibitors Chem Med Chem 2009 4 1568 1582 19739196
- Cho Y-W Hong SH Jin Q Histone methylation regulator PTIP is required for PPARγ and C/EBPα expression and adipogenesis Cell Metab 2009 10 27 39 19583951
- Ge K Epigenetic regulation of adipogenesis by histone methylation Biochim Biophys Acta 2012 1819 7 727 732 22240386
- Musri MM Carmona MC Hanzu FA Kaliman P Gomis R Parrizas M Histone demethylase LSD1 regulates adipogenesis J Biol Chem 2010 285 30034 30041 20656681
- Campion J Milagro FI Martinez JA Individuality and epigenetics in obesity Obesity Rev 2009 10 383 392
- Musri MM Gomis R Parrizas M A chromatin perspective of adipogenesis Organogenesis 2009 6 15 23 20592861
- Pereira-Lancha LO Campos-Ferraz PL Lancha AHJr Obesity: considerations about etiology, metabolism, and the use of experimental models Diab Metab Syn Obesity: Targets Ther 2012 5 75 87
- Puri V Virbasius JV Guilherme A Czech MP RNAi screens reveal novel metabolic regulators: RIP140, MAP4k4 and the lipid droplet associated fat specific protein (FSP) 27 Acta Physiol 2008 192 103 115
- Parra P Serra F Palou A Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice PLoS One 2010 5 e13005 20886002
- Toh SY Gong J Du G Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice PLoS One 2010 3 e2890 18682832
- Kuypers FA Larkin SK Emeis JJ Allison AC Interaction of an annexin V homodimer (Diannexin) with phosphatidylserine on cell surfaces and consequent anti-thrombotic activity Thromb Haemost 2007 97 478 486 17334517
- vanWjik MJ VanBavel E Sturk A Nieuwland R Microparticles in cardiovascular diseases Cardiovasc Res 2003 59 277 287 12909311
- Müller G The mode of action of glimepiride – beyond insulin secretion Curr Med Chem 2005 5 499 518
- van der Pol E Hoekstra AG Sturk A Otto C van Leeuwen TG Nieuwland R Optical and non-optical methods for detection and characterization of microparticles and exosomes J Thromb Haemost 2010 8 2596 2607 20880256
- Cheruvanky A Zhou H Pisitkun T Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator Am J Physiol Renal Physiol 2007 292 5 F1657 F1661 17229675
- Miranda KC Bond DT McKee M Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease Kidney Int 2010 78 191 199 20428099
- Gonzales PA Pisitkun T Hoffert JD Large-scale proteomics and phosphoprotemics of urinary exosomes J Am Soc Nephrol 2009 20 2 363 379 19056867
- Conde-Vancells J Rodriguez–Suarez E Gonzalez E Candidate biomarkers in exosome–like vesicles purified from rat and mouse urine samples Proteomics Clin Appl 2010 4 416 425 20535238
- Moon PG You S Lee JE Hwang D Baek MC Urinary exosomes and proteomics Mass Spectrom Rev 2011 30 1185 1202 21544848
- Orozco AF Lewis DE Flow cytometric analysis of circulating microparticles in plasma Cytometry 2010 77 502 514 20235276
- De M Rana S Akpinar H Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein Nat Chem 2009 1 461 465 20161380
- Anslyn EV Rotello VM Chemosensory models: approaches and applications of differential sensing Curr Opin Chem Biol 2010 14 683 684 20863741
- Miranda OR Creran B Rotello VM Array-based sensing with nanoparticles: ‘chemical noses’ for sensing biomolecules and cell surfaces Curr Opin Chem Biol 2010 14 728 736 20801707
- Müller G Glycosylphosphatidylinositol-anchored protein chips for patient–tailored multi–parameter proteomics J Biochip Tissue Chip 2011 S3 001 10.4172/2153-0777.S3-001
- Müller G (Glycosylphosphatidylinositol-based) protein chips and biosensors for biopharmaceutical process analytics J Bioprocess Biotec 2012 2 115 10.4172/2155-9821.1000115.
- Dragovic RA Gardiner C Brooks AS Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis Nanomedicine NBM 2011 7 780 788
- Hu G Drescher KM Chen X-M Exosomal miRNAs: biological properties and therapeutic potential Front Gene 2012 3 56
- Martinez MC Andriantsitohaina R Microparticles in angiogenesis: therapeutic potential Circ Res 2011 109 110 119 21700952
- György B Szabo TG Pasztoi M Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles Cell Mol Life Sci 2011 68 2667 2688 21560073
- Lee TH D’Asti E Magnus N Al-Nedawi K Meehan B Rak J Microvesicles as mediators of intracellular communication in cancer – the emerging science of cellular ‘debris’ Semin Immunopathol 2011 33 455 467 21318413
- Dowling P Clynes M Conditioned media from cell lines: a complementary model to clinical specimens for the discovery of disease-specific biomarkers Proteomics 2011 11 794 804 21229588
- Siljander PR Platelet-derived microparticles – an updated perspective Thromb Res 2011 127 S2 S30 S33 21193112
- Dignat-George F Boulanger CM The many faces of endothelial microparticles Arterioscler Thromb Vasc Biol 2011 31 1 27 33 21160065
- Keller S Sanderson MP Stoek A Altevogt P Immunol Lett 2006 107 102 108 17067686
- Meckes DGJr Raab-Traub N Microvesicles and viral infection J Virol 2011 85 12844 12854 21976651
- Pelchen-Matthews A Raposo G Marsh M Endosomes, exosomes and Trojan viruses Trends Microbiol 2004 12 310 316 15223058
- Stoorvogel W Kleijmeer MJ Geuze HJ Raposo G The biogenesis and functions of exosomes Traffic 2002 3 321 330 11967126
- Muralidharan–Chari V Clancy JW Sedgwick A D’Souza-Schorey C Microvesicles: mediators of extracellular communication during cancer progression J Cell Sci 2010 123 1603 1611 20445011
- Martinez MC Kunzelmann C Freyssinet JM Plasma membrane remodelling and cell stimulation Med Sci 2004 20 189 195
- Nosjean O Briolay A Roux B Mammalian GPI proteins: sorting, membrane residence and functions Biochim Biophys Acta 1997 1331 153 186 9325440
- Lakhan SE Sabharanjak S De A Endocytosis of glycosylphosphatidyl– anchored proteins J Biomed Sci 2009 16 93 109 19832981
- Ferguson MA The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosoma research J Cell Sci 1999 112 2799 2809 10444375
- Kinoshita T Fujita M Maeda Y Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress J Biochem 2008 144 287 294 18635593
- Paulick MG Bertozzi CR The glycosylphosphatidylinositol anchor: a complex membrane–anchoring structure for proteins Biochemistry 2008 47 6991 7000 18557633
- Müller G Jung C Wied S Welte S Jordan H Frick W Redistribution of glycolipid raft domain components induces insulin-mimetic signaling in rat adipocytes Mol Cell Biol 2001 21 4553 4567 11416134
- Müller G Dynamics of plasma membrane microdomains and crosstalk to the insulin signalling cascade FEBS Lett 2002 531 81 87 12401208
- Simons K Sampaio JL Membrane organisation and lipid rafts Cold Spring Harb Perspect Biol 2011 3 a004697 21628426
- Lajoie P Nabi IR Lipid rafts, caveolae, and their endocytosis Int Rev Mol Biol 2010 282 135 163
- Lingwood D Simons K Lipid rafts as a membrane-organizing principle Science 2010 327 46 50 20044567
- Lindner R Naim HY Domains in biological membranes Exp Cell Res 2009 315 2871 2878 19632223
- Foster LJ Lessons learned from lipid raft proteomics Expert Rev Proteomics 2008 5 541 543 18761463
- Epand RM Proteins and cholesterol-rich domains Biochim Biophys Acta 2008 1778 1576 1582 18423371
- Sun T Fu M Bookout AL Kliewer SA Mangelsdorf DJ MicroRNA let-7 regulates 3T3-L1 adipogenesis Mol Endocrinol 2009 23 925 931 19324969
- Li G Li Y Li X Ning X Li M Yang G MicroRNA identity and abundance in developing swine adipose tissue as determined by Solexa sequencing J Cell Biochem 2011 112 1318 1328 21312241
- Wang Q Li YC Wang J Kong J Qi Y Quigg RJ Li X miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130 Proc Natl Acad Sci U S A 2008 105 2889 2894 18287052
- Kim YJ Hwang SJ Bae YC Jung JS MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue Stem Cells 2009 27 3093 3102 19816956
- Lin Q Gao Z Alarcon RM Ye J Yun Z A role of miR-27 in the regulation of adipogenesis FEBS J 2009 276 2348 2358 19348006
- Lee EK Lee MJ Abdelmohsen K miR-130 suppresses adipogenesis by inhibiting PPARγ expression Mol Cell Biol 2010 31 626 638 21135128
- Kloting N Berthold S Kovacs P MicroRNA expression in human omental and subcutaneous adipose tissue PLoS One 2009 4 4699
- Estep M Armistead D Hossain N Differential expression of miRNAs in the visceral adipose tissue of patients with nonalcoholic fatty liver disease Aliment Pharmacol Ther 2010 32 487 497 20497147
- Chang CL Au LC Huang SW Kwok CF Ho LT Juan CC Insulin up-regulates heme oxygenase-1 expression in 3T3-L1 adipocytes via PI3-kinase- and PKC-dependent pathways and heme oxygenase-1-associated microRNA downregulation Endocrinology 2010 152 384 393 21147878
- Parra P Serra F Palou A Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice PLoS One 2010 5 e13005 20886002
- Jayachandran M Miller VM Heit JA Owen WG Methodology for isolation and characterization of microvesicles in peripheral blood J Immunol Meth 2012 375 207 214
- Mrvar-Brecko A Sustar V Jansa V Isolated microvesicles from peripheral blood and body fluids as observed by scanning electron microscope Blood Cells Mol Dis 2010 44 307 312 20199878
- Chen C Skog J Hsu C-H Microfluidic isolation and transcriptome analysis of serum microvesicles Lab Chip 2010 10 505 511 20126692
- Soo CY Song Y Zheng Y Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells Immunology 2012 136 192 197 22348503
- Simak J Gelderman MP Cell membrane microparticles in blood and blood products: potentially pathogenic agents and diagnostic markers Transfus Med Rev 2006 20 1 26 16373184
- Holme PA Solum NO Brosstad F Roger M Abdelnoor M Demonstration of platelet-derived microvesicles in blood from patients with activated coagulation and fibrinolysis using a filtration technique and western blotting Thromb Haemost 1994 72 666 671 7900071
- Grant R Ansa-Addo E Stratton D A filtration–based protocol to isolate human plasma membrane–derived vesicles and exosomes from plasma J Immunol Meth 2011 371 143 151
- Wang K Zhang S Weber J Baxter D Galas DJ Export of microRNAs and microRNA-protective protein by mammalian cells Nucleic Acids Res 2010 38 7248 7259 20615901
- Kosaka N Iguchi H Yoshioka Y Takeshita F Matsuki Y Ochiya T Secretory mechanisms and intercellular transfer of microRNAs in living cells J Biol Chem 2010 285 17442 17452 20353945