Abstract
Numerous biomarkers have been unveiled in the rapidly evolving biomarker discovery field, with an aim to improve the clinical management of disorders. In rare diseases, such as Duchenne muscular dystrophy, this endeavor has created a wealth of knowledge that, if effectively exploited, will benefit affected individuals, with respect to health care, therapy, improved quality of life and increased life expectancy. The most promising findings and molecular biomarkers are inspected in this review, with an aim to provide an overview of currently known biomarkers and the technological developments used. Biomarkers as cells, genetic variations, miRNAs, proteins, lipids and/or metabolites indicative of disease severity, progression and treatment response have the potential to improve development and approval of therapies, clinical management of DMD and patients’ life quality. We highlight the complexity of translating research results to clinical use, emphasizing the need for biomarkers, fit for purpose and describe the challenges associated with qualifying biomarkers for clinical applications.
Video abstract
Point your SmartPhone at the code above. If you have a QR code reader the video abstract will appear. Or use:
Background
Duchenne muscular dystrophy (DMD) is the most common form of muscular dystrophy with an incidence of 1 in 5000–9000 live born males per year.Citation1,Citation2 DMD is caused by mutations in the DMD gene located on the short arm of the X chromosome.Citation3 The disease is inherited in recessive X-linked manner, leading to rare cases of female patients mostly due to skewed inactivation of the chromosome carrying the non-mutated copy of the gene.Citation4 Several different mutations have been described, ranging from the most common out-of-frame deletions to duplication and point mutations.Citation5 Mutations lead to a DMD phenotype when the gene product dystrophin cannot be synthesized.Citation3 A milder form of the disease called Becker muscular dystrophy (BMD) is caused by mutations in the same gene causing shorter or partly functional dystrophin.Citation6 BMD patients can have very different clinical presentation with delayed muscular complaints leading to wheelchair dependency to almost asymptomatic cases with only elevated activity of creatine kinase (CK) in serum (a biomarker for muscle damage).Citation7 DMD patients experience a severe disease progression starting at young age with delayed motor development and proximal to distal weakness of skeletal muscles. Patients typically lose ambulation at about 9 years of age if untreated, while daily use of glucocorticoids (GC) prolongs the ambulatory phase with most of the affected individuals being able to walk up to 12 years of age and some patients up to age 15.Citation8 The life expectancy of DMD patients is improved thanks to GC treatment and better care, even though DMD patients die normally in their 30s due to cardiorespiratory insufficiency/complications.Citation9
In the last 20 years, research efforts converged in characterization of the disease mechanism and development of therapeutic strategies targeting the genetic defect (e.g., gene therapy,Citation10–Citation13 exon skipping,Citation14–Citation20 autologous genetically corrected stem cellsCitation21–Citation23 and stop codon read-throughCitation24–Citation26) or boosting compensating mechanisms (e.g., utrophin upregulation,Citation27 myostatin inhibitionCitation28,Citation29 and IGF-1 overexpressionCitation30). Less effort was dedicated to the development of outcome measures able to capture clinical benefit in clinical trials. This has recently changed with multiple investigators adapting and developing functional scales (e.g., the 6-minute walk test [6MWT]Citation31 and the performance of upper limbCitation32) and providing data enabling drug developers to better design and power interventional studies. The most used test in interventional studies, the 6MWT, has been found to be a good tool to monitor disease progression; however, the large variation between individuals, a strong and documented motivational component and the low potency of the drugs tested so far have not enabled to proceed to the full approval by regulatory agencies.Citation8,Citation31 Given this background, multiple groups are currently working on the identification of biomarkers, which could not only enrich the design of clinical trials, but also provide objective readouts to predict the likelihood of benefit due to the administration of experimental medicinal products. The availability of biomarkers would enable refined clinical trials design reducing the noise caused by patients with different characteristics and accelerate the evaluation and approval of medicinal products by detecting early signs of response to the drug and by anticipating clinical benefit. We will proceed to provide definitions to the known types of biomarkers to show what is currently available for DMD.
Types of biomarkers
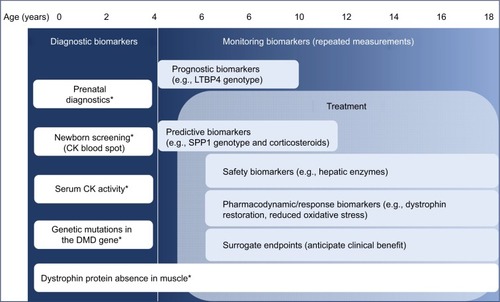
Biomarkers are measurable indicators of some biologic state or condition. The term “biomarker” has been often inappropriately used, leading to the recent release of the Biomarkers, EndpointS, and other Tools Resource documents by the US Food and Drug Administration (FDA)–National Institutes of Health Working Group aiming to clarify the differences between biomarker types.Citation33 The document discriminates between seven types of biomarkers, namely, 1) diagnostic, 2) monitoring, 3) pharmacodynamic/response, 4) predictive, 5) prognostic, 6) safety and 7) susceptibility/risk biomarkers.
Diagnostic biomarkers are used to detect or confirm the presence of a disease or condition of interest or to identify individuals with a subtype of the disease. Classical examples are blood pressure to diagnose hypertension and sweat chloride to confirm cystic fibrosis.
Monitoring biomarkers are typically serially measured in order to assess the status of a disease or evidence of exposure to (or effect of) a medical product (or an environmental agent). The concept of monitoring includes repetition of the test in time to detect changes in an individual or in the population. Examples are monitoring the drug concentration, drug toxicity and therapeutic response to a drug after treatment, as well as surveillance of serum cholesterol in patients with hypercholesterolemia. This definition includes multiple types of biomarkers such as pharmacodynamic, safety, prognostic and predictive biomarkers.
Pharmacodynamic/response biomarkers are used to show biologic response in individuals treated with a medicinal product or an environmental agent. This type of biomarker provides evidence that the drug has reached its target and produced the desired effect; however, it may not necessarily anticipate future clinical event of clinical benefit. Examples are quantification of the viral load in antiretroviral treatment or serum cholesterol levels after dosing patients with lipid-lowering drugs.Citation34 Pharmacodynamic biomarkers are often used in dose-finding studies to optimize the dose and show proof of principle of target engagement.
Predictive biomarkers are used to identify individuals who are more likely to receive benefit (or toxic effects) from the exposure to a drug compared to other subjects who do not have the biomarker. These types of biomarkers is not only useful in clinical trials, but also for patients’ care decision. Predictive biomarkers could be characteristics of the individuals (host characteristics), such as genetic variants, or characteristics of the disease process, such as mutations present in the tumor. An example is represented by mutation in the BRCA1 and BRCA2 genes that may be used to predict sensitivity to treatment with PARP inhibitors in women carrying platinum-sensitive ovarian cancer.Citation35 While designing randomized trials including only biomarker-positive patients could be a strategy to reduce the number of patients involved and observe larger effect sizes, it is still advisable to include patients who do not carry the biomarker to demonstrate the role of the biomarker. On the other hand, sole comparison of the experimental treatment between biomarker-positive and biomarker-negative patients may not clarify whether the biomarker is predictive, as biomarker-positive patients could perform better than biomarker-negative patients independently to the treatment, qualifying in this way the biomarker as a prognostic one instead of a predictive one.
Prognostic biomarkers are defined as biomarkers able to identify the likelihood of a clinical event in patients affected by a condition. These types of biomarkers is thus independent from the exposure to medicinal products; it may, however, be influenced by the stage of the disease, so the definition of the context is extremely important. Examples are BRCA1 and BRCA2 gene mutations as prognostic biomarkers to assess the odds of breast cancer in women who have already had breast cancer beforeCitation36 or C-reactive protein levels to assess the likelihood of recurrent coronary artery events in patients with unstable angina.Citation37 In certain cases, when, for example, the event rate is used as an endpoint, prognostic biomarkers can be used to include patients who are more likely to experience an event resulting in an enrichment of the clinical trial design.
Safety biomarkers are used to indicate the likelihood, presence or extent of toxicity after exposure to a medical product. An example is represented by the serum levels of hepatic enzymes for liver toxicity.Citation38,Citation39
Susceptibility/risk biomarkers are used to estimate the likelihood of a clinical event in otherwise healthy individuals. An example is represented by the factor V Leiden as a risk biomarker to develop thrombosis.Citation40
The use of biomarkers is especially important for rare diseases, not only to facilitate the diagnostic process of diseases rarely seen by general practitioners but also to enable better patient management in response to the available drugs and to support and accelerate the evaluation and marketization of medicinal products. In fact, mechanisms such as the accelerated approval by the FDA rely on the use of surrogate endpoints expected to anticipate clinical benefit, thus greatly reducing the time to commercialization. We present in an overview of the different types of biomarkers and the direct application to the DMD field.
Recent findings on DMD biomarkers
Biomarkers are able to describe both normal and pathologic conditions and provide information that characterizes patient health status at higher resolution than symptoms that are visible, palpable or physical tests. The increased resolution can describe alterations in organs, tissues or cells at a molecular level. The value of the biomarkers resides in more conclusive and precise measures that can aid clinical management of disorders, and they are less influenced by the patient’s ability to describe symptoms, their mood and ability and willingness to cooperate. Current research efforts have made major advances identifying cells, single nucleotide polymorphism (SNP), proteins, metabolites, lipids and miRNA as potential biomarkers. During the past 5 years, several scientific reports have testified existence of promising biomarkers, but only a few of them have been validated for use in a clinical setting. Validation of such markers implies not only a conclusive, clear link to a clinical endpoint or process, but also a specific, sensitive and robust measure as determined by an appropriate analytical method rendering reproducible results.
Tissue composition
Magnetic resonance spectroscopy (MRS) and magnetic resonance imaging (MRI) are nuclear magnetic resonance–based methods measuring re-emission of radiation from the nuclei of hydrogen atoms as a consequence of exposure to magnetic radiation. The emission is influenced by the molecular environment and, thus, can be used to distinguish between atoms in water molecules and atoms in lipid molecules. Exploiting these differences MRI can create images of tissues, enabling distinction between fat and muscle tissue. MRS, on the other hand, can detect signals from other hydrogen-containing molecules such as metabolites. Several protocols have been developed that after excitation with electromagnetic radiation measure the return of the tissue to the non-excited state either through spin–lattice (T1) or spin–spin (T2) process.Citation41–Citation44 The aggregated data collected from the analysis of protons in a tissue can be subsequently converted into two-dimensional images of the sample analyzed. Already in 2003, Tracy et al published the use of MRI to monitor tissue composition.Citation45 Today, quantitative MRI (qMRI) is able to estimate fat fraction based on chemical shift–based separation of water and fat signals. Among the different protocols and data analysis methods, the transverse relaxation time (MRI-T2), which represents the bulk of T2, has been reported to render higher signals in DMD patients in comparison to controls and to correlate with time-limited functional test, walking 30 ft, climbing four steps and rising from the floor.Citation44,Citation46,Citation47 The two- and three-point Dixon MRI and single-voxel 1H-MRS measure independent lipid infiltration which is found to be elevated in DMD boys.Citation43,Citation48,Citation49 qMRI measurement of fat in muscles from different organisms is consistent with the fat infiltration assessed by histologic analysis of biopsies.Citation50 Several longitudinal studies have reported that muscle fat and water quantification using MRI is a biomarker for DMD disease progression.Citation51,Citation52 Furthermore, the responsiveness of these biomarkers in respect to disease progression can be measured in different muscle groups in both lower and upper limbs, enabling assessment of disease progression not only in ambulant, but also non-ambulant patients incapable of performing physical tests.Citation51,Citation52 Decreased fat infiltration as a consequence of treatment with corticosteroids has been demonstrated in cross-sectional MRI/MRS analysis of lower limbs in DMD boys.Citation53 The effect of corticosteroids was measurable by MRI at 3 months after the initiation of treatment.Citation53 The correlations identified and the accuracy of the measurements make this biomarker suitable as a surrogate endpoint in clinical trials.Citation54,Citation55 Quantification of muscle fat and water estimated by qMRI offers the advantages of being less dependent on patient’s motivation, attention and coordination than functional tests, is reproducible across clinical centers and has a strong correlation with clinical parameters. However, patient experience of the MRI scan with respect to discomfort, duration they have to remain immobilized and repetition as well as the cost for MRI scans reaching up to £500 per hour are aspects that still need to be addressed.Citation56
Genomic markers
Genomic biomarkers, as biologic measurements, can be used to diagnose disease, qualify patients for specific therapies, as well as predict patient response to treatment and survival.Citation57 Within rare disorders, genome wide association studies on patient samples most likely underperform due to the limited population making identification of genomic biomarkers challenging.Citation58 The lack of statistical power in genome wide association studies for DMD has been partially overcome by combining and integrating information identified in animal models. Latent TGFβ binding protein 4 (LTBP4) has been identified as a genetic modifier that correlated with age of ambulation loss. In, particular, a haplotype constituted by four SNPs has been explored in depth in 254 non-ambulant patients from the United Dystrophinopathy Project cohort. The four SNPs composing this haplotype are missense variants. The first SNP can code for either valine (V) or isoleucine (I), the second and third variants for threonine (T) or alanine (A) and the last one for threonine (T) or methionine (M). Different haplotypes can arise from this configuration; however, the most frequent ones are VTTT and IAAM. Analysis of these variants in DMD patients showed that IAAM homozygous carriers were ambulant ~2 years longer than patients carrying other haplotypes.Citation59 The significance of LTBP4 as a biomarker has been confirmed by the analysis of 336 patients from five European clinical sitesCitation60 and more recently in a second cohort comprising 340 DMD subjects collected across three different continents.Citation61 This protective haplotype is associated with delayed loss of ambulation regardless of the steroid treatment. LTBP4 interacts directly with TGFβ which stimulates the expression of SPP1 in myoblasts, another genomic marker.Citation62 A polymorphism in the promoter region of the SPP1 gene, rs28357094, upstream the transcription start site affects the expression of SPP1. The GG/TG alleles were shown to be associated with rapid disease progression in comparison to the TT allele.Citation63,Citation64 This association, however, was not confirmed by more recent studies performed on European and American cohorts.Citation59,Citation60 Recently, Vianello et al clarified that SPP1 overexpression occurs in patients carrying the G allele as a consequence of GC treatment.Citation65 More recently, the minor allele of SNP rs1883832, lying in the 5′ untranslated region of the CD40 gene, was identified to be associated with early loss of ambulation.Citation58 Analysis of CD40 gene expression at the transcript and protein levels revealed diminished expression and altered B-cell and macrophage activation. Data obtained in animal models provide rational support for further genetic modifiers in DMD. A variant in the promoter region of the JAG1 locus was found to be associated with mild symptoms in the canine model Golden Retriever muscular dystrophy. This variant resulted in overexpression of JAG1 and consequently ameliorated the dystrophic phenotype in dogs and in the zebrafish model.Citation66 Jagged1 product was also found to be differentially represented in the serum of two independent DMD cohorts compared to healthy controls.Citation67 Another marker with effect on the DMD phenotype is the ACTN3 R577X null polymorphism. This genetic variant was associated with reduced muscle strength at baseline, but improved disease progression.Citation68 Annexin A6 (ANXA6) encodes a calcium-dependent membrane protein involved and is involved in sarcolemma repair.Citation69 Truncated isoforms of ANXA6 are synthesized as a result of two synonymous SNPs associated with alternative splicing events. Expression of the truncated isoform as a consequence of the SNPs hinders transport of the full-length annexin A6 to the sarcolemma and the repair process.Citation69 The biomarker value of ANXA6 might not reside in the predictive value of the genetic alteration, but the amount of protein isoforms expressed.Citation70
Proteins
The analysis of body fluids to identify protein biomarkers has been performed with many different technologies ranging from single immunoassaysCitation71 to small panels of targeted approaches,Citation72 to large-scale targeted approaches,Citation67,Citation73 to untargeted mass spectrometry (MS)–based approaches coupled to immunoassay-based validation.Citation74,Citation75 Many new potential candidates have been identified thanks to the new enabling technologies, and new evidence has been obtained for the already known biomarkers.Citation76
The most widely known and currently used biomarker is represented by the serum activity levels (units/L) of CK. CK activity levels (and also elevated protein levels of the muscle form of CK) indicate the muscle damage is ongoing. It is usually the first element leading to the diagnosis of Duchenne after confirming lack of dystrophin in muscle biopsies and a mutation in the DMD gene. CK activity in serum has been shown to correlate with other muscle-derived proteins (myofibrillar proteins as well as proteins involved in energy production) in the circulation, such as lactate dehydrogenase, carbonic anhydrase 3 or myosin light chain 3.Citation67,Citation73 Cross-sectional studies showed that muscle-related proteins are largely elevated in circulation at the time of diagnosis and decrease with age as the muscle mass is replaced by adipose tissue.Citation72 Due to seasonal variation, intraindividual variation, exercise-dependent fluctuation and progression toward healthy controls levels, CK is at the moment considered to be a good diagnostic biomarker, but a weak pharmacodynamic biomarker and surrogate endpoint.Citation72,Citation77,Citation78 Given the good performance of CK as a diagnostic biomarker, a test on dried blood spots has been recently developed to detect CK activity levels and the causative mutation.Citation79 While this assay will ensure early detection of DMD, it has also been shown to detect other forms of muscular dystrophy, such as limb girdle muscular dystrophy. As a biomarker, CK is a powerful and robust marker, but has not been widely adopted due to lack of approved early interventions and the perception that the test does not necessarily have consequences on improvement of motor function.Citation80–Citation82 A recent high-throughput cross-sectional study performed in serum samples of two independent cohorts has enabled the identification of 44 proteins discriminating DMD from healthy controls. Most of the identified proteins followed a CK-like profile; however, other proteins such as RET showed reduced levels compared to healthy controls, while some others such as GDF11 were characterized by a different progression with age.Citation67 It is still not clear whether these proteins could act as prognostic or predictive biomarkers or as surrogate endpoints. More studies are needed to understand the biologic meaning of the newly identified ones. The same researchers recently published a follow-up paper where they studied the response to treatment with GC in longitudinal settings and identified predictive biomarkers, mostly inflammatory ones such as CCL22, pharmacodynamic biomarkers, mostly related to the immune system such as FCER2, and safety biomarkers, related to GC side effects such as an increase in insulin and leptin.Citation83 The value of leptin as a safety parameter needs to be better evaluated, as an increase in serum leptin levels was reported in steroid-naïve patients.Citation84 Metalloproteinases (MMPs), especially MMP-9, have been found to be elevated in DMD patients and to increase with disease progression.Citation71,Citation85–Citation88 While the role of MMP-9 has been investigated in the context of satellite cells and muscle regeneration in murine models, it is still not clear how MMP-9 serum levels are related to disease progression.Citation89–Citation91 Analysis of other body fluids such as urine has also revealed an increased amount of myofibrillar proteins such as titin, which was confirmed in patient samples as well as in urine obtained from murine and canine models.Citation92,Citation93 A sandwich enzyme-linked immunosorbent assay was developed for quantification of the N-terminal titin fragment in urine, which detected 139.5 pmol/L.Citation94
Lipids and metabolites
While muscle/fat composition of the tissue is currently considered as one of the most promising biomarkers, its measurement by MRI relies on the lipid composition of the tissue. Besides fat/muscle fraction estimates described in the “Tissue composition” section, studies reveal specific lipids present in muscles, blood or urine also as potential interesting biomarker candidates.Citation95–Citation97 In comparison to healthy muscle, the dystrophic muscle is characterized by changed fatty acid composition, oxidative damage and elevated protein thiol oxidation.Citation98–Citation100 Furthermore, distribution of specific lipid compounds has been suggested to be useful for determining the metabolic state of muscle cells and fibers. Mdx muscle exhibits lower levels of carnosine, taurine, glycine, methionine and creatine in comparison to healthy muscle.Citation101,Citation102 In contrast, glutamate, glutamine, succinate, isoleucine, acetate, alanine and glycerol are increased in dystrophic muscle.Citation101,Citation103 Antioxidants such as vitamin E and the reduced form of coenzyme Q9 accumulate in tissues exposed to oxidative stress and can reveal regions with high oxidative activity in the muscle, whereas regions rich in triglycerides mark cells with high lipid metabolism such as adipose tissue.Citation104,Citation105 Cholesterol accumulation, likewise, can reveal regions undergoing muscle degeneration–regeneration, since it affects membrane deformability.Citation104 Trimethyl ammonium and creatine are two other metabolites altered in dystrophic muscle. The ratio of trimethyl ammonium to total creatine has a negative correlation to muscle function as demonstrated by analysis of 16 individuals (8 DMD and 8 controls).Citation106 The metabolic alterations recorded in dystrophic muscles are apparently detectable in body fluids such as serum, plasma or urine to some extent. Creatine levels that are lower in mdx muscle when compared to wild-type muscle are in contrast to serum levels that are higher in DMD patients compared to controls.Citation107 In a natural history study comprising 51 patients and 22 age-matched controls, several metabolites have been detected at altered levels. However, creatine to creatinine ratio increases with age in DMD patients, whereas it decreases in healthy individuals.Citation107 Besides serum, urine has also been analyzed within the context of DMD. Elimination of methyl-amino acids has been explored in urine. The levels of 3-methylhistidine urine decrease with age, whereas they tend to remain stable in healthy individuals.Citation108 Although metabolic biomarker candidates have been identified, it remains to not only confirm the findings and validate the most promising biomarkers, but also complement current knowledge with more targeted studies on the correlation of metabolite levels with clinical markers and disease symptoms.
MicroRNA
miRNAs or miRs have an intrinsic ability to function as gene silencers and modulate the synthesis of specific proteins. miRNAs are 21–25 nucleotides long RNA molecules that bind to the 3′ untranslated region of target mRNA through RNA-induced silencing complex and consequently alter the gene expression either through suppressed translation of targeted protein or decreased protein synthesis due to induced mRNA susceptibility to degradation. Advances in DNA sequencing technologies have contributed to identification of thousands of miRNAs in the human genome and have unveiled their impact on biologic processes.Citation109–Citation111 miRNAs’ control on protein synthesis, if deregulated as a consequence of altered expression, affects critical processes underlying cell development, proliferation, metabolism and differentiation.Citation110,Citation111 miRNAs represent ideal biomarkers since they are specifically expressed in different tissues and are released into the blood stream upon injuries.Citation112 In 2008, miRs were for the first time regarded as biomarkers and the first three tumor-associated miRs were identified.Citation113 The advantage of miR as biomarkers is their stability upon freezing and thawing, extreme pH conditions in both serum and plasma samples.Citation114
The intricate function of miRNAs makes them key players in several biologic processes such as regulation of cell cycle and apoptosis, development of tissue and organs including cardiac and skeletal muscle development, and biologic processes that are important and affected in DMD. Several miRNAs have been identified to be expressed 20-fold higher in muscle in comparison with the expression in other tissues and are considered to be muscle specific.Citation115 Altered expression of miRNAs in muscles as a consequence of diminished dystrophin expression in DMD has been suggested to contribute to leakage of miRNAs into the blood stream. The release of miRNAs from the skeletal muscle is likely to occur both through leakage and secretion, since many of them are associated with proteins and lipoproteins.Citation116
Several reports confirm the discovery of elevated miRNA blood levels in DMD patients in comparison to healthy individuals. miR-1, miR-30c, miR-31, miR-133, miR-181,Citation117 miR-206, miR-208a, miR-208b and miR-499Citation117–Citation120 are elevated in serum, whereas miR-95Citation121 is elevated in plasma. In contrast, miR-549 is less abundant in plasma from DMD patients compared to controls.Citation121 The majority of the miRNAs that are significantly elevated in DMD patients, miR-1, miR-133, miR-206, miR-208b and miR-499, can also differentiate between DMD and the milder BMD phenotype.Citation118,Citation122 Most of them are actively transcribed in myocytes and involved in proliferation and differentiation. miRNA abundance in muscle varies between muscle groups and does not necessarily correlate with the serum levels.Citation119 The expression of miR-16 in mdx muscle does not vary significantly in lower limb muscles and diaphragm.Citation119 In the same study, miR-1 and miR-133a showed decreased levels in soleus and tibialis anterior muscles and miR-206 was significantly increased in tibialis anterior and diaphragm. Fluctuation in miRNA expression in muscles is not entirely correlated with their release in blood. The decreased level of miRNAs in tissues suggests leakage or secretion of these markers into the blood stream rather than a change in expression levels. It is possible that the miRNA signature in blood is proportional to the amount of muscle regenerating fibers, which are particularly enriched in young dystrophic patients compared to older ones. This active secretion mechanism seems to be independent of the secretion of exosome/vesicles as most of the miRNAs in the circulation are extravesicular.Citation116 Some miRNAs have been found to be associated with clinical parameters indicative of disease progression. miR-30c was detected at higher levels in patients with better preserved motor function estimated by North Star Ambulatory Assessment (NSAA) and 6MWT, whereas miR181-a concentration was higher in patients with difficulties to climb and descend stairs, but it was not statistically significant.Citation117 Both observations were, however, not statistically significant. Similar results were obtained also from the analysis of miR-1, miR-206, miR-31, miR-133a and miR-133b in ambulant and non-ambulant patients, indicating lack of correlation with NSAA score.Citation123 Interestingly, serum levels of miR-133 showed positive correlation with fast-twitch muscle fiber composition and negative correlation with slow-twitch fiber composition, whereas miR-499 had negative correlation with fast-twitch muscle fiber composition.Citation118 These observations corroborate fluctuation in serum miRNA levels in association with muscle mass and fiber composition, which are affected during disease progression. Most importantly, levels of miR-1, miR-133a and miR-206 decrease over time in the serum of dystrophic animal models and DMD patients and they are potential markers for disease progression.Citation124 Further studies remain to be conducted for validation of miRNAs as biomarkers and explore their specificity and sensitivity.
Samples types and technologies
The plethora of biomarkers identified is the result of numerous efforts invested in samples collection harmonization, biobanks, collaboration of partners in multinational consortia and availability of technologies offering the possibility to go beyond the state of the art and enabling researchers to improve and refine their observations. In this section, we want to focus on the most used sample matrices and technologies.
Biologic samples
Muscle biopsies have been studied extensively through histology, immunostaining, proteomic and transcriptomic analyses. While the analysis of muscle biopsies has been key to identify candidate biomarkers such as SPP1, JAG1 and others,Citation63,Citation66,Citation125–Citation127 most of the effort is steered toward the analysis of biofluids that can be obtained less invasively, such as blood and urine. Blood and urine samples are retrieved relatively easily with little distress to the patient, offering more easily accessible biomarker measurements. Blood-derived sample matrices such as serum and plasma have been the most investigated ones. Here, differences exist between them as plasma samples can be obtained in different tube types depending on the downstream application. For example, plasma samples can be obtained in heparin–lithium tubes as well as EDTA tubes. While both tubes could potentially work for certain proteomic analysis (differences could still be present depending on the pipeline), the use of heparin tubes will seriously hamper polymerase chain reaction–based strategies to, for example, study miRNA, since heparin is a strong inhibitor of polymerases.Citation128 While certain pipelines have been optimized with a specific sample matrix, the obtained data should be still interpreted carefully as sample processing may affect the measurement independently from the technique used. As an example, certain MMPs are released during clotting, thus largely increasing the levels of MMPs in serum samples compared to plasma samples.Citation129 New sample matrices are being explored, such as PAX gene tubes in DMD, enabling analysis of RNA species in blood.Citation130,Citation131 The composition and complexity of these biologic samples influence both detection of the biomarkers and their stability.Citation132 Serum, plasma and urine carry residual molecules from different organs and tissues as potential biomarkers. Amount of biomarker molecules in body fluids constitutes merely a small fraction of the molecules that populate the samples as a consequence of dilution effects. Often, extra steps, such as globin depletion for PAX gene tubes derived RNA, are taken to reduce the dynamic range of the sample, especially when the downstream analysis is a high-throughput nontargeted analysis. Furthermore, while some biomarkers are affected by storage conditions, others are affected by parameters not necessarily related to the disorder, for example, age and gender of the individuals.Citation133
Technologies
Technological advances have empowered biomarker discovery and validation by offering more sensitive measurements, with improved detection limit and increased multiplexing capacity. With the advent of next-generation sequencing technology, whole genome and transcriptome sequencing has become achievable in only few hours. Massive parallel sequencing is characterized by generation of a large amount of data that, if inspected and interpreted, improve our knowledge regarding genomic markers and based on differential expression analysis render potential biomarkers and novel drug targets.Citation134 This technology has allowed development of personalized medicine, in particular for cancer.Citation135 Within DMD, however, the number of samples available is more restricted and new strategies for meta-analysis across platforms and more efficient exploration of the data generated are required through, for example, integration with other type of data on protein and metabolite levels.
MS is a widely used technology for analysis of biomolecules such as proteins, lipids and metabolites in complex samples, and biomarker discovery analysis has been performed on both tissue samples and body fluids using MS platforms such as MS/MS and matrix assisted laser desorption ionization-time of flight mass spectrometry or linear trap quadropole ion trap MS. The advantages offered by this technology are the ability to quantify biomolecules and distinguish between specific forms, for example, phosphorylated and nonphosphorylated forms of proteins. Sensitivity of the technology has been increased by improvement of ionization efficiency and spatial resolution combined with novel interrogate recorded data.Citation136 For protein biomarker discovery, targeted MS-based Multiple Reaction Monitoring allows simultaneous quantification of several biomarkers.Citation137 The increased analysis capacity of MS has been exploited to generate images of protein, lipid and metabolite distribution.Citation138
Beside MS, protein biomarkers have also been identified using affinity-based proteomics methods such as planar protein arrays and suspension bead array.Citation139 Massive efforts have been invested in generation of antibodies and affinity binders as aptamers, exponentially increasing the number of analytes that can be analyzed. Protein abundance measurements using these methods rely, however, on the quality of the affinity binders used. Current development of affinity binders addresses this issue by carefully designed validation strategies.Citation140
New directions
Although many biomarkers have been identified, they are not used in daily clinical practice and they are only occasionally included as exploratory biomarkers in clinical trials. The delay in the use of biomarkers becomes obvious when the number of FDA-approved biomarkes is compared with the number of biomarkers identified.Citation141 The lack of reliable biomarkers, within DMD, not only affects the immediate care of affected patients, but also hinders development of novel therapies and subsequent approval by regulatory authorities. One of the biggest challenges is the limited availability of samples for biomarker analysis. The majority of strategies used for identification and validation of biomarkers rely on inclusion of large number of patients, which is not achievable within rare disorders. Therefore, new strategies to interrogate samples from different perspectives and integrate molecular data at DNA, RNA, protein and metabolite levels are needed to extract the best performing biomarkers.
While genomic markers orchestrating gene expression are more useful for characterization of the phenotypes, stratification of patients and/or qualification for therapies, abundance of miRNA, protein and metabolite biomarkers can provide information about health status changes in time. Most of the biomarkers identified can discriminate between DMD and healthy individuals and even discriminate between the mild and severe DMD phenotypes, but not all of them clearly fit for monitoring disease progression. miR-18, miR-30c and miR-181a correlate with functional measures, whereas carbonic anhydrase 3, lactate dehydrogenase, the myosins and troponins correlate with disease milestones such as loss of ambulation. These findings raise the question whether functional tests are the most appropriate clinical parameter to be explored. Ideally, a combination of biomarkers and functional tests would have a better prognostic power than single readouts, enabling better modeling of individual disease trajectories.
What is still needed?
While the work done so far has made it possible to identify a number of candidate biomarkers, their daily applica tion in the clinics is still limited and major improvements are to be expected. New applications (such as analysis of dried blood spots) for the already known biomarkers (such as CK) are being developed with the aim to anticipate the diagnosis at birth; early diagnosis would enable better patient management and early treatment with GC, perhaps further postponing disease milestones.Citation79 The availability of qualified biomarkers would not only certainly benefit patient management, but also speed up the evaluation of medicinal products. Both FDA and European Medicines Agency have mechanisms in place for biomarker qualification, and even though the qualification process is not needed, the awareness of the qualification process forces researchers and clinicians to develop biomarkers for specific purposes and to evaluate them in a clear context of use.
Prognostic biomarkers able to forecast disease milestones such as loss of ambulation would enable improved patient selection for clinical trial design by either excluding patients who are more likely to lose ambulation (e.g., to ensure that muscle targeting drugs can have sufficient time and tissue target to improve muscle function) or by including patients who are more likely to lose ambulation (e.g., to increase the chance of showing efficacy in trials where the primary endpoint is the ratio between wheelchair-dependent patients and the total number of patients). Caution should be taken, however, as selection of patient groups may result in specific therapeutic indications, which could lead to drug availability only for a narrow subpopulation of DMD patients. Pharmacodynamic biomarkers could provide evidence that the drug is working (e.g., evidence of exon skipping and dystrophin restoration after treatment with antisense oligonucleotides) and they could be used in dose-finding studies to determine what is the best dose or regimen for the drug in use. Predictive biomarkers would largely accelerate the development of medicinal products for DMD patients and for patients affected by rare diseases in general. The availability of these types of biomarkers would improve the trial design by selecting those patients who are most likely to benefit from the treatment (e.g., SPP1 genotype for drugs targeting inflammatory pathways and/or compare GC treatment with other nonsteroidal anti-inflammatory drugs) and they will provide supporting evidence of response to the treatment. Lastly, biomarkers associated with patients’ performance that are strongly supportive of clinical benefit, such as surrogate endpoints, which show smaller interindividual variation would enable smaller and better controlled studies to be performed, thus accelerating the evaluation of medicinal products.
Disclosure
The authors report no conflicts of interest in this work.
References
- MercuriEMuntoniFMuscular dystrophiesLancet (London, England)20133819869845860
- MahJKKorngutLDykemanJDayLPringsheimTJetteNA systematic review and meta-analysis on the epidemiology of Duchenne and Becker muscular dystrophyNeuromuscul Disord201424648249124780148
- Tuffery-GiraudSBéroudCLeturcqFGenotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebaseHum Mutat200930693494519367636
- ViggianoEErgoliMPicilloEPolitanoLDetermining the role of skewed X-chromosome inactivation in developing muscle symptoms in carriers of Duchenne muscular dystrophyHum Genet2016135768569827098336
- Aartsma-RusAFokkemaIVerschuurenJTheoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutationsHum Mutat200930329329919156838
- FlaniganKMDunnDMvon NiederhausernAUnited Dys-trophinopathy Project ConsortiumNonsense mutation-associated Becker muscular dystrophy: interplay between exon definition and splicing regulatory elements within the DMD geneHum Mutat201132329930821972111
- van den BergenJCSchade van WestrumSMDekkerLClinical characterization of Becker muscular dystrophy patients predicts favorable outcome in exon-skipping therapyJ Neurol Neurosurg Psychiatry2014851929823667215
- GoemansNTuliniusMKroksmarkAKWilsonRvan den HauweMCampionGComparison of ambulatory capacity and disease progression of Duchenne muscular dystrophy subjects enrolled in the drisapersen DMD114673 study with a matched natural history cohort of subjects on daily corticosteroidsNeuromuscul Disord201727320321328169120
- BladenCLSalgadoDMongesSThe TREAT-NMD DMD global database: Analysis of more than 7,000 duchenne muscular dystrophy mutationsHum Mutat201536439540225604253
- FosterHSharpPSAthanasopoulosTCodon and mRNA sequence optimization of microdystrophin transgenes improves expression and physiological outcome in dystrophic mdx mice following AAV2/8 gene transferMol Ther200816111825183218766174
- SchinkelSBauerRBekeredjianRLong-term preservation of cardiac structure and function after adeno-associated virus serotype 9-mediated microdystrophin gene transfer in mdx miceHum Gene Ther201223656657522248393
- GregorevicPAllenJMMinamiErAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic miceNat Med200612778778916819550
- XiongFXuYZhengHMicrodystrophin delivery in dystrophin-deficient (mdx) mice by genetically-corrected syngeneic MSCs transplantationTranspl Proc201042727312739
- Aartsma-RusATherapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patientsHum Mol Genet200312890791412668614
- SpitaliPRimessiPFabrisMExon skipping-mediated dystrophin reading frame restoration for small mutationsHum Mutat200930111527153419760747
- WuBMoultonHMIversenPLEffective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomerProc Natl Acad Sci U S A200810539148141481918806224
- GoyenvalleABabbsAWrightJRescue of severely affected dystrophin/utrophin-deficient mice through scAAV-U7snRNA-mediated exon skippingHum Mol Genet201221112559257122388933
- MalerbaASharpPSGrahamIRChronic systemic therapy with low-dose morpholino oligomers ameliorates the pathology and normalizes locomotor behavior in mdx miceMol Ther201119234535421102560
- Tanganyika-de WinterCLHeemskerkHKarnaoukhTGLong-term exon skipping studies with 2′-O-Methyl phosphorothioate antisense oligonucleotides in Dystrophic mouse modelsMol Ther Nucleic Acids20121e4423344236
- WuBLiYMorcosP ADoranTJLuPLuQLOcta-guanidine morpholino restores dystrophin expression in cardiac and skeletal muscles and ameliorates pathology in dystrophic mdx miceMol Ther200917586487119277018
- TedescoFSHoshiyaHD’AntonaGStem cell–mediated transfer of a human artificial chromosome ameliorates muscular dystrophy201139696ra78
- TorrenteYBelicchiMSampaolesiMHuman circulating AC133(+) stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscleJ Clin Invest2004114218219515254585
- SampaolesiMBlotSD’AntonaGMesoangioblast stem cells ameliorate muscle function in dystrophic dogsNature2006444711957457917108972
- MalikVRodino-KlapacLRViolletLGentamicin-induced readthrough of stop codons in Duchenne muscular dystrophyAnn Neurol201067677178020517938
- FinkelRSRead-through strategies for suppression of nonsense mutations in Duchenne/Becker muscular dystrophy: aminoglycosides and ataluren (PTC124)J Child Neurol20102591158116420519671
- HaasMVlcekVBalabanovPEuropean medicines agency review of ataluren for the treatment of ambulant patients aged 5 years and older with Duchenne muscular dystrophy resulting from a nonsense mutation in the dystrophin geneNeuromuscul Disord201525151325497400
- GuiraudSSquireSEEdwardsBSecond-generation compound for the modulation of utrophin in the therapy of DMDHum Mol Genet201524154212422425935002
- MorineKJBishLTPendrakKSleeperMMBartonERSweeneyHLSystemic myostatin inhibition via liver-targeted gene transfer in normal and dystrophic micePLoS One201052e917620161803
- DumonceauxJMarieSBeleyCCombination of myostatin pathway interference and dystrophin rescue enhances tetanic and specific force in dystrophic mdx miceMol Ther201018588188720104211
- GregorevicPPlantDRLynchGSAdministration of insulin-like growth factor-I improves fatigue resistance of skeletal muscles from dystrophic mdx miceMuscle Nerve200430329530415318340
- GoemansNvan den HauweMWilsonRvan ImpeAKlingelsKBuyseGAmbulatory capacity and disease progression as measured by the 6-minute-walk-distance in Duchenne muscular dystrophy subjects on daily corticosteroidsNeuromuscul Disord201323861862323770101
- PaneMStacy MazzoneESivoSThe 6 minute walk test and performance of upper limb in ambulant duchenne muscular dystrophy boysPLoS Curr20146
- GroupF-NBWBEST (Biomarkers, EndpointS, and Other Tools) ResourceFood and Drug Administration (US)2016
- StoneNJRobinsonJGLichtensteinAH2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice GuidelinesCirculation201412925 Suppl 2S1S4524222016
- LedermannJHarterPGourleyCOlaparib maintenance therapy in platinum-sensitive relapsed ovarian cancerN Engl J Med2012366151382139222452356
- BasuNNInghamSHodsonJRisk of contralateral breast cancer in BRCA1 and BRCA2 mutation carriers: a 30-year semi-prospective analysisFam Cancer201514453153826239694
- HaverkateFThompsonSGPykeSDGallimoreJRPepysMBProduction of C-reactive protein and risk of coronary events in stable and unstable angina. European Concerted Action on Thrombosis and Disabilities Angina Pectoris Study GroupLancet (London, England)19973499050462466
- SeniorJREvolution of the Food and Drug Administration approach to liver safety assessment for new drugs: current status and challengesDrug Saf201437Suppl 1S9S1725352324
- McMillanHJGregasMDarrasBTKangPBSerum transaminase levels in boys with Duchenne and Becker muscular dystrophyPediatrics2011127e132e13621149430
- KujovichJLFactor V Leiden thrombophiliaGenet Med201113111621116184
- GarroodPHollingsworthKGEagleMMR imaging in Duchenne muscular dystrophy: quantification of T1-weighted signal, contrast uptake, and the effects of exerciseJ Magn Reson Imaging20093051130113819856446
- ForbesSCWalterGARooneyWDSkeletal muscles of ambulant children with Duchenne muscular dystrophy: validation of multicenter study of evaluation with MR imaging and MR spectroscopyRadiology2013269119820723696684
- TriplettWTBaligandCForbesSCChemical shift-based MRI to measure fat fractions in dystrophic skeletal muscleMagn Reson Med201472181924006208
- WrenTALBlumlSTseng-OngLGilsanzVThree-point technique of fat quantification of muscle tissue as a marker of disease progression in Duchenne muscular dystrophy: preliminary studyAJR Am J Roentgenol20081901W8W1218094282
- TracyBLIveyFMJeffrey MetterEFlegJLSiegelELHurleyBFA more efficient magnetic resonance imaging-based strategy for measuring quadriceps muscle volumeMed Sci Sports Exerc200335342543312618571
- ArpanIForbesSCLottDJT2 mapping provides multiple approaches for the characterization of muscle involvement in neuromuscular diseases: a cross-sectional study of lower leg muscles in 5–15-year-old boys with Duchenne muscular dystrophyNMR Biomed201326332032823044995
- KimHKLaorTHornPSRacadioJMWongBDardzinskiBJT2 mapping in Duchenne muscular dystrophy: distribution of disease activity and correlation with clinical assessmentsRadiology2010255389990820501727
- LottDJForbesSCMathurSAssessment of intramuscular lipid and metabolites of the lower leg using magnetic resonance spectroscopy in boys with Duchenne muscular dystrophyNeuromuscul Disord201424757458224798221
- TorrianiMTownsendEThomasBJBredellaMAGhomiRHTsengBSLower leg muscle involvement in Duchenne muscular dystrophy: an MR imaging and spectroscopy studySkeletal Radiol201241443744521800026
- SmithACParrishTBAbbottRMuscle-fat MRI: 1.5 Tesla and 3.0 Tesla versus histologyMuscle Nerve201450217017624677256
- RicottiVEvansMRSinclairCDUpper limb evaluation in duchenne muscular dystrophy: fat-water quantification by MRI, muscle force and function define endpoints for clinical trialsPLoS One2016119e016254227649492
- WokkeBHBosCReijnierseMComparison of dixon and T1-weighted MR methods to assess the degree of fat infiltration in duchenne muscular dystrophy patientsJ Magn Reson Imaging201338361962423292884
- ArpanIWillcocksRJForbesSCExamination of effects of corticosteroids on skeletal muscles of boys with DMD using MRI and MRSNeurology2014831197498025098537
- BonatiUHafnerPSchädelinSQuantitative muscle MRI: a powerful surrogate outcome measure in Duchenne muscular dystrophyNeuromuscul Disord201525967968526096788
- StraubVBalabanovPBushbyKStakeholder cooperation to overcome challenges in orphan medicine development: the example of Duchenne muscular dystrophyLancet Neurol201615888289027302365
- HollingsworthKGReducing acquisition time in clinical MRI by data undersampling and compressed sensing reconstructionPhys Med Biol20156021R297R32226448064
- BoutrosPCThe path to routine use of genomic biomarkers in the cancer clinicGenome Res201525101508151326430161
- BelloLFlaniganKMWeissRBAssociation study of exon variants in the NF-κB and TGFβ pathways identifies CD40 as a modifier of duchenne muscular dystrophyAm J Hum Genet20169951163117127745838
- FlaniganKMCecoELamarKMLTBP4 genotype predicts age of ambulatory loss in duchenne muscular dystrophyAnn Neurol201373448148823440719
- van den BergenJCHillerMBohringerSValidation of genetic modifiers for Duchenne muscular dystrophy: a multicentre study assessing SPP1 and LTBP4 variantsJ Neurol Neurosurg Psychiatry201586101060106525476005
- BarpABelloLPolitanoLGenetic modifiers of duchenne muscular dystrophy and dilated cardiomyopathyPLoS One20151010e014124026513582
- UaesoontrachoonKYooHJTudorEMPikeRNMackieEJPagelCNOsteopontin and skeletal muscle myoblasts: association with muscle regeneration and regulation of myoblast function in vitroInt J Biochem Cell Biol200840102303231418490187
- PegoraroEHoffmanEPPivaLSPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophyNeurology201176321922621178099
- BelloLPivaLBarpAImportance of SPP1 genotype as a covariate in clinical trials in Duchenne muscular dystrophyNeurology201279215916222744661
- VianelloSPanticBFustoASPP1 genotype and glucocorticoid treatment modify osteopontin expression in Duchenne muscular dystrophy cellsHum Mol Genet201726173342335128595270
- VieiraNMElversIAlexanderMSJagged 1 rescues the duchenne muscular dystrophy phenotypeCell201516351204121326582133
- HathoutYBrodyEClemensPRLarge-scale serum protein biomarker discovery in Duchenne muscular dystrophyProc Natl Acad Sci2015112237153715826039989
- HogarthMWHouwelingPJThomasKCEvidence for ACTN3 as a genetic modifier of Duchenne muscular dystrophyNat Commun201781414328139640
- Swaggart KAMcNallyEMModifiers of heart and muscle function: where genetics meets physiologyExp Physiol201499462162624213858
- VoAHMcNallyEMModifier genes and their effect on Duchenne muscular dystrophyCurr Opin Neurol201528552853426263473
- NadarajahVDvan PuttenMChaouchASerum matrix metalloproteinase-9 (MMP-9) as a biomarker for monitoring disease progression in Duchenne muscular dystrophy (DMD)Neuromuscul Disord201121856957821724396
- BurchPMPogoryelovaOGoldsteinRMuscle-derived proteins as serum biomarkers for monitoring disease progression in three forms of muscular dystrophyJ Neuromuscul Dis20152324125526870665
- AyogluBChaouchALochmüllerHAffinity proteomics within rare diseases: a BIO-NMD study for blood biomarkers of muscular dystrophiesEMBO Mol Med20146711924408963
- Cynthia MartinFHillerMSpitaliPFibronectin is a serum biomarker for Duchenne muscular dystrophyProteomics Clin Appl2014826927824458521
- OonkSSpitaliPHillerMComparative mass spectrometric and immunoassay-based proteome analysis in serum of Duchenne muscular dystrophy patientsProteomics Clin Appl201610329029926680509
- CarterNDHeathRJefferySJacksonMJNewhamDJEdwardsRHCarbonic anhydrase III in Duchenne muscular dystrophyClin Chim Acta19831332012086414742
- PercyMEAndrewsDFThompsonMWSerum creatine kinase in the detection of Duchenne muscular dystrophy carriers: effects of season and multiple testingMuscle Nerve19825158647057807
- NicholsonGAMorganGMeerkinMStraussEMcLeodJGThe creatine kinase reference interval. An assessment of intra- and inter-individual variationJ Neurol Sci1985712–32252314087024
- MendellJRShillingCLeslieNDEvidence-based path to newborn screening for duchenne muscular dystrophyAnn Neurol201271330431322451200
- MoatSJBradleyDMSalmonRClarkeAHartleyLNewborn bloodspot screening for Duchenne muscular dystrophy: 21 years experience in Wales (UK)Eur J Hum Genet201321101049105323340516
- PillersD-AMA new day for Duchenne’s?: The time has come for newborn screeningMol Genet Metab20141131–2111324974086
- KwonJMAbdel-HamidHZAl-ZaidySAClinical follow-up for duchenne muscular dystrophy newborn screening: a proposalMuscle Nerve201654218619127170260
- HathoutYConklinLSSeolHSerum pharmacodynamic biomarkers for chronic corticosteroid treatment of childrenSci Rep201663172727530235
- Rodríguez-CruzMCruz-GuzmánOREscobarRELópez-AlarcónMLeptin and metabolic syndrome in patients with Duchenne/Becker muscular dystrophyActa Neurol Scand2016133425326026133644
- HathoutYMarathiRLRayavarapuSDiscovery of serum protein biomarkers in the mdx mouse model and cross-species comparison to Duchenne muscular dystrophy patientsHum Mol Genet201423246458646925027324
- ZocevicARouillonJWongBServaisLVoitTSvinartchoukFEvaluation of the serum matrix metalloproteinase-9 as a biomarker for monitoring disease progression in Duchenne muscular dystrophyNeuromuscul Disord20159910
- SpitaliPAartsma-RusA’t HoenPACResponse to: evaluation of the serum matrix metalloproteinase-9 as a biomarker for monitoring disease progression in Duchenne muscular dystrophyNeuromuscul Disord201525544644725795139
- AndersonJSeolHGordish-DressmanHHathoutYSpurneyCFCINRG InvestigatorsInterleukin 1 Receptor-Like 1 Protein (ST2) is a Potential Biomarker for Cardiomyopathy in Duchenne Muscular DystrophyPediatr Cardiol20173881606161228821969
- LiHMittalAMakonchukDYBhatnagarSKumarAMatrix metalloproteinase-9 inhibition ameliorates pathogenesis and improves skeletal muscle regeneration in muscular dystrophyHum Mol Genet200918142584259819401296
- HindiSMShinJOguraYLiHKumarAMatrix metalloproteinase-9 inhibition improves proliferation and engraftment of myogenic cells in dystrophic muscle of mdx micePLoS One201388e7212123977226
- DahiyaSGivvimaniSBhatnagarSQipshidzeNTyagiSCKumarAOsteopontin-stimulated expression of matrix metalloproteinase-9 causes cardiomyopathy in the mdx model of duchenne muscular dystrophyJ Immunol201118752723273121810612
- RouillonJZocevicALegerTProteomics profiling of urine reveals specific titin fragments as biomarkers of Duchenne muscular dystrophyNeuromuscul Disord201424756357324813925
- RobertsonASMajchrzakMJSmithCMDramatic elevation in urinary amino terminal titin fragment excretion quantified by immunoassay in Duchenne muscular dystrophy patients and in dystrophin deficient rodentsNeuromuscul Disord201727763564528554556
- MaruyamaNAsaiTAbeCEstablishment of a highly sensitive sandwich ELISA for the N-terminal fragment of titin in urineSci Rep201663937527991570
- GriffinJLDes RosiersCApplications of metabolomics and proteomics to the mdx mouse model of Duchenne muscular dystrophy: lessons from downstream of the transcriptomeGenome Med2009133219341503
- McIntoshLMBakerREAndersonJEMagnetic resonance imaging of regenerating and dystrophic mouse muscleBiochem Cell Biol1998762–35325419923723
- El-ShafeyAFArmstrongAETerrillJRGroundsMDArthurPGScreening for increased protein thiol oxidation in oxidatively stressed muscle tissueFree Radic Res201145999199921696323
- TouboulDPiednoëlHVoisinVChanges of phospholipid composition within the dystrophic muscle by matrix-assisted laser desorption/ionization mass spectrometry and mass spectrometry imagingEur J Mass Spectrom (Chichester, Eng)2004105657664
- RodriguezMCTarnopolskyMAPatients with dystrophinopathy show evidence of increased oxidative stressFree Radic Biol Med20033491217122012706502
- TerrillJRRadley-CrabbHGGroundsMDArthurPGN-Acetylcysteine treatment of dystrophic mdx mice results in protein thiol modifications and inhibition of exercise induced myofibre necrosisNeuromuscul Disord201222542743422206641
- Martins-BachABBloiseACVainzofMRahnamaye RabbaniSMetabolic profile of dystrophic mdx mouse muscles analyzed with in vitro magnetic resonance spectroscopy (MRS)Magn Reson Imaging20123081167117622673895
- TerrillJRGroundsMDArthurPGTaurine deficiency, synthesis and transport in the mdx mouse model for Duchenne Muscular DystrophyInt J Biochem Cell Biol20156614114826239309
- GriffinJLWilliamsHJSangEClarkeKRaeCNicholsonJKMetabolic profiling of genetic disorders: a multitissue (1)H nuclear magnetic resonance spectroscopic and pattern recognition study into dystrophic tissueAnal Biochem20012931162111373073
- TouboulDBrunelleAHalgandFDe La PorteSLaprévoteOLipid imaging by gold cluster time-of-flight secondary ion mass spectrometry: application to Duchenne muscular dystrophyJ Lipid Res20054671388139515834124
- MurphyMEKehrerJPOxidative stress and muscular dystrophyChem Biol Interact1989692–31011732649259
- HsiehTJJawTSChuangHYJongYJLiuGCLiCWMuscle metabolism in Duchenne muscular dystrophy assessed by in vivo proton magnetic resonance spectroscopyJ Comput Assist Tomogr331150154
- BocaSMNishidaMHarrisMCorrection: discovery of metabolic biomarkers for duchenne muscular dystrophy within a natural history studyPLoS One2016117e015989527434074
- InoueRMiyakeMKanazawaASatoMKakimotoYDecrease of 3-methylhistidine and increase of NG,NG-dimethylarginine in the urine of patients with muscular dystrophyMetabolism1979288801804454517
- LondinELoherPTelonisAGAnalysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAsProc Natl Acad Sci U S A201511210E1106E111525713380
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- WangJSenSMicroRNA functional network in pancreatic cancer: from biology to biomarkers of diseaseJ Biosci201136348149121799259
- SchölerNLangerCDöhnerHBuskeCKuchenbauerFSerum microRNAs as a novel class of biomarkers: a comprehensive review of the literatureExp Hematol201038121126113020977925
- LawrieCHGalSDunlopHMDetection of elevated levels of tumor-associated microRNAs in serum of patients with diffuse large B-cell lymphomaBr J Haematol2008141567267518318758
- MitchellPSParkinRKKrohEMCirculating microRNAs as stable blood-based markers for cancer detectionProc Natl Acad Sci U S A200810530105131051818663219
- McCarthyJJMicroRNA-206: the skeletal muscle-specific myomiRBiochim Biophys Acta200817791168269118381085
- RobertsTCGodfreyCMcCloreyGExtracellular microRNAs are dynamic non-vesicular biomarkers of muscle turnoverNucleic Acids Res201341209500951323945935
- Llano-DiezMOrtezCIGayJADigital PCR quantification of miR-30c and miR-181a as serum biomarkers for Duchenne muscular dystrophyNeuromuscul Disord2017271152327979502
- LiXLiYZhaoLCirculating Muscle-specific miRNAs in Duchenne Muscular Dystrophy PatientsMol Ther Nucleic Acids20143e17725050825
- MizunoHNakamuraAAokiYIdentification of muscle-specific MicroRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophyPLoS One201163e1838821479190
- HuJKongMYeYHongSChengLJiangLSerum miR-206 and other muscle-specific microRNAs as non-invasive biomarkers for Duchenne muscular dystrophyJ Neurochem2014129587788324460924
- Jeanson-LehLLamethJKrimiSSerum profiling identifies novel muscle miRNA and cardiomyopathy-related miRNA biomarkers in golden retriever muscular dystrophy dogs and Duchenne muscular dystrophy patientsAm J Pathol2014184112885289825194663
- CacchiarelliDLegniniIMartoneJmiRNAs as serum biomarkers for Duchenne muscular dystrophyEMBO Mol Med20113525826521425469
- ZaharievaITCalissanoMScotoMDystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular DystrophyPLoS One2013811e8026324282529
- Coenen-StassAMLBettsCALeeYFSelective release of muscle-specific, extracellular microRNAs during myogenic differentiationHum Mol Genet201625183960397427466195
- DoranPWiltonSDFletcherSOhlendieckKProteomic profiling of antisense-induced exon skipping reveals reversal of pathobiochemical abnormalities in dystrophic mdx diaphragmProteomics2009967168519132684
- KotelnikovaEShkrob MAPyatnitskiyM AFerliniADaraseliaNNovel approach to meta-analysis of microarray datasets reveals muscle remodeling-related drug targets and biomarkers in Duchenne muscular dystrophyPLoS Comput Biol201282e100236522319435
- BakayMZhaoPChenJHoffmanEPA web-accessible complete transcriptome of normal human and DMD muscleNeuromuscul Disord200212Suppl 1S125S14112206807
- SefriouiDBeaussireLClatotFHeparinase enables reliable quantification of circulating tumor DNA from heparinized plasma samples by droplet digital PCRClin Chim Acta2017472757928729136
- MannelloFTanus-SantosJEMeschiariCATontiGADifferences in both matrix metalloproteinase 9 concentration and zymographic profile between plasma and serum with clot activators are due to the presence of amorphous silica or silicate salts in blood collection devicesAnal Biochem20083741566318082127
- WongBGilbertDLWalkerWLGene expression in blood of subjects with Duchenne muscular dystrophyNeurogenetics200910211712519067001
- LiuDZStamovaBHuSMicroRNA and mRNA expression changes in steroid naïve and steroid treated DMD patientsJ Neuromuscul Dis20152438739627858746
- ErcanMAkbulutEDAbuşoğluSStability of urine specimens stored with and without preservatives at room temperature and on ice prior to urinalysisClin Biochem20154813–1491992226025772
- IshikawaMMaekawaKSaitoKPlasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and agePLoS One201493e9180624632803
- AndersonMWSchrijverINext generation DNA sequencing and the future of genomic medicineGenes (Basel)201011386924710010
- UhlenMZhangCLeeSA pathology atlas of the human cancer transcriptomeScience20173576352
- CrutchfieldCAThomasSNSokollLJChanDWAdvances in mass spectrometry-based clinical biomarker discoveryClin Proteomics201613126751220
- MermelekasGVlahouAZoidakisJSRM/MRM targeted proteomics as a tool for biomarker validation and absolute quantification in human urineExpert Rev Mol Diagn201515111441145426472065
- BakerTCHanJBorchersCHRecent advancements in matrix-assisted laser desorption/ionization mass spectrometry imagingCurr Opin Biotechnol201743626927690313
- WingrenCAntibody-Based ProteomicsAdv Exp Med Biol201692616317927686812
- SchumacherSSeitzHQuality control of antibodies for assay developmentN Biotechnol2016335 Pt A54455026873787
- FüzéryAKLevinJChanMMChanDWTranslation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challengesClin Proteomics20131011324088261