Abstract
One of the main fundamental mechanisms of antibiotic resistance in Gram-negative bacteria comprises an effective change in the membrane permeability to antibiotics. The Gram-negative bacterial complex cell envelope comprises an outer membrane that delimits the periplasm from the exterior environment. The outer membrane contains numerous protein channels, termed as porins or nanopores, which are mainly involved in the influx of hydrophilic compounds, including antibiotics. Bacterial adaptation to reduce influx through these outer membrane proteins (Omps) is one of the crucial mechanisms behind antibiotic resistance. Thus to interpret the molecular basis of the outer membrane permeability is the current challenge. This review attempts to develop a state of knowledge pertinent to Omps and their effective role in antibiotic influx. Further, it aims to study the bacterial response to antibiotic membrane permeability and hopefully provoke a discussion toward understanding and further exploration of prospects to improve our knowledge on physicochemical parameters that direct the translocation of antibiotics through the bacterial membrane protein channels.
Introduction
Antibiotic resistance can be defined as the capability of any microbial organism to counterattack effects of antimicrobial drugs (antibiotics) () used against them.Citation1,Citation2 This phenomenon has become a global communal health threat due to an enormous increase in annual death rate.Citation2 The emergence of highly resistant organisms has led to the requirement of new antibacterial drugs.Citation1 Due to the slow progress of the current antibiotic research, there exists an enormous gap between bacterial evolution and the rate of development of novel antibiotic drugs.Citation1,Citation3,Citation4 Only about two new classes of antibiotics have been brought to the market in the last three decades. On the technical front, there is an urgent need for a greater understanding of how antibiotics work, how bacteria progress with resistance against these antibiotics, and what molecular machinery could be exploited to get around bacterial defense mechanisms.Citation1–Citation4 The current innovative way of improving the potential of antibiotics is to effectively introduce them into bacteria and further prevent them from degradation by bacterial enzymes before they reach their targets. There is an extreme necessity for counteracting the problem of multi-antibiotic resistance.Citation1,Citation4 The important mechanism () of resistance toward antibiotics known till date includes the enzymes-mediated deactivation of antibiotics for example, β-lactamase enzymes which hydrolyze and confer resistance against a diverse variety of antibiotics including penicillins, cephalosporins, carbapenems, and many more.Citation4–Citation7 The outer membrane vesicles (), these native vesicles released by Gram-negative bacteria, are mainly composed of periplasmic and outer membrane components including lipopolysaccharides, proteins, lipids, and other molecules.Citation8–Citation11 They help the producer cells while communicating with other cells concerning pathogenesis, secretion, nutrients acquisition, and self-defense.Citation5,Citation8–Citation10 These moieties protect bacteria from various environmental stress factors including antibiotics, for example, gentamicin, imipenem, ampicillin, melittin, colistin, and many more.Citation8–Citation14 Further, resistance mechanism is also mediated by reducing the entry of antibiotics into the target site of bacteria which is mainly effected by specific alteration of outer membrane permeability (). Efflux pumps effectively contribute towards resistance mechanism by antibiotic expulsion. In addition, antibiotic target proteins, for example, penicillin-binding proteins, are altered inside the bacterial cells, leading to antibiotic resistance.Citation2,Citation3,Citation5,Citation6,Citation15–Citation21
Figure 1 (A) Antibiotic resistance (an overview). (B) Various mechanisms of antibiotic resistance employed by Gram-negative bacteria (an overview). (C) Structural representation of outer membrane vesicles.
Abbreviation: Omps, outer membrane proteins.
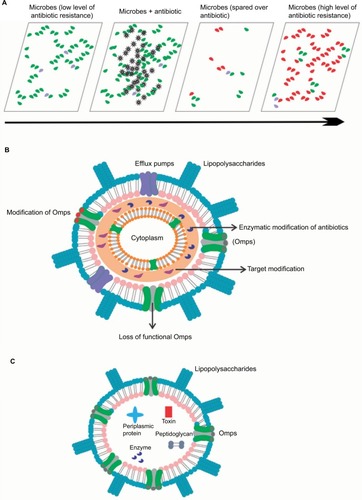
Figure 2 Antibiotic resistance mechanism associated with Omps modification. Antibiotic β-lactam molecules are represented by green stars, and Omps as trimers by gray cylinder. The width of the straight arrows imitating the level of β-lactam penetration via Omps. The curved arrows exemplify the uptake failure/reduce uptake occurring with the following: B: decrease in the level of wild-type Omps expression; C: expression of restricted-channel Omps; D: mutation or modification of the functional properties of a porin channel; and E: synthesis of modified Omps with significant constriction.
Abbreviation: Omps, outer membrane proteins.
In this review, we present a systemic overview of the role of different membrane protein transporters responsible for antibiotic transport, present in the outer membrane of Gram-negative bacteria.Citation4–Citation6,Citation22 We highlight the different achievements of the scientific community in understanding the uptake of different solutes including antibiotics.Citation7,Citation17,Citation22 This active knowledge of the role of outer membrane influx in antibiotic transport in Gram-negative bacteria can be useful for antibiotic drug development in the future, where the computed data can be employed toward understanding the detailed mechanism of bacterial membrane transport, and to further design novel antibiotics with an effective permeability profile.
Gram-negative bacteria
Gram-negative bacteria have a multifaceted cell envelope comprising an outer membrane that restricts the access to the periplasm by acting as a molecular filter, thus forming an efficient selective permeation barrier.Citation4–Citation6,Citation23,Citation24 This outer membrane, like other biological membranes, is fundamentally built up of a bilayer of lipids.Citation6,Citation18,Citation25,Citation26 As such, this lipid bilayer membrane is mostly impermeable to hydrophilic molecules including nutrients.Citation22,Citation25,Citation27 The effective intake of hydrophilic molecules is mainly controlled by specific water-filled open channels termed as outer membrane proteins (Omps) or porins.Citation22,Citation27–Citation29 These Omps are intensively characterized in Gram-negative bacteria and are further distinguished as nonspecific and specific Omps in accordance with their functional structure (monomeric or trimeric),Citation6,Citation7,Citation22,Citation24–Citation26,Citation28 substrate specificity, regulation, and expression.Citation15,Citation18,Citation29,Citation30 These membrane proteins do not show any hydrophobic stretches in their amino acid sequences and majorly form hollow β-barrel structures with a hydrophobic outer surface.Citation28,Citation31 The barrel structure encompasses the transmembranous pore-type structure with a crucial function of facilitating the passive flux of hydrophilic substancesCitation22,Citation28 and further acting as a functional diffusional barrier for nonpolar solutes.Citation6,Citation28 These proteins might show specific selectivity in general for either cations or anions.Citation5,Citation22,Citation28
Bacterial adaptation to reduce influx through these Omps is an increasing problem that contributes, together with efflux systems, to antibiotic resistance.Citation3–Citation5,Citation20,Citation23,Citation32–Citation34 An existing challenge for drug design is to interpret membrane permeability at molecular level to get a better insight into the role of membrane transport () in bacterial resistance mechanism.Citation4–Citation7,Citation20,Citation35 Like other hydrophilic molecules, polar antibiotics including β-lactam antibiotics and fluoroquinolones majorly sneak into Gram-negative bacteria using these Omps.Citation5,Citation31,Citation33 Any slight modification by the bacteria in the responsible Omps can significantly affect the antibiotic drug therapy.Citation33 Many clinically pertinent bacterial species including Enterobacter aerogenes, Escherichia coli, Enterobacter cloacae, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii have been sequenced for determining the effective key Omps () present in the outer membrane.Citation3–Citation6,Citation23,Citation28,Citation31–Citation33,Citation36,Citation37 Further, bacterial bugs including Pseudomonas aeruginosa, and Acinetobacter baumannii possess an innate low vulnerability toward β-lactams, through reduced outer membrane permeability.Citation5,Citation6,Citation20,Citation22,Citation38 For instance, reduced membrane permeability in Pseudomonas aeruginosa as compared to Enterobacteriaceae mainly occurs due to less number of Omps present in the outer membrane and their distinct physicochemical properties.Citation22,Citation38–Citation41 In other Gram-negative bugs, for example, Escherichia coli, Enterobacter and Klebsiella pneumoniae, susceptibility toward β-lactam molecules is closely related to the presence of nonspecific diffusion Omps, for example, OmpF and OmpC.Citation5,Citation6,Citation22
Table 1 Crucial Omps studied in different Gram-negative bacterial species
Previous works showing the effective role of different Omps () in molecular influx of different antibiotics are shown in . We discuss the achievements of the scientific community in this area by studying the role of different Omps in outer membrane permeability, using separate set of theoretical and experimental techniques including molecular simulation (MS), electrophysiology, minimum inhibitory concentration assay, liposome swelling assay, X-ray crystallography, and fluorescence resonance energy transfer.
Table 2 Conclusive investigations with different Omps studied in different Gram-negative bacterial species
Discussion
Computing influx
Typical antibiotic activity toward bacterial cell occurs in micromolar concentration range, thereby representing values that are approximately limited to a thousand molecules inflowing the cell in few minutes to hours.Citation7,Citation22 Such numbers are considerably beneath the detection limit of most of the techniques and thus require significant amplification of the signal.Citation4,Citation7,Citation22,Citation110 Measuring the flux of small molecules across the outer cell membrane can be possibly achieved by different approaches including whole-cell assays, which require computation of flux using genetically engineered bacterial cell.Citation7,Citation111,Citation112 These methods involve soaking bacteria in antibiotics for a fixed time followed by a separation process to remove the external media from the internalized antibiotics.Citation7 However, the quality of the separation method is crucial for improving permeability.Citation7,Citation111,Citation112 There are several published studies employing whole-cell assays to quantify the uptake, and their quality has been intensively compared.Citation7,Citation110–Citation116 Once the separation technique allows collecting sufficient amounts of internalized antibiotics, several biophysical methods can be used to quantify the intracellular antibiotics.Citation7,Citation113–Citation118 One of the promising tools for studying intracellular accumulation is mass spectrometry. The technique was successfully applied in measuring the uptake of antibiotics;Citation117,Citation118 for example, a work demonstrated cellular uptake of linezolid by E. coli using liquid chromatography–mass spectrometry.Citation118
The discussed methods allow quantifying the total turnover of a cell uptake which represents the relevant actual effective concentration seen by the bacteria. On the contrary, the comprehensive flux depends on a multitude of parameters and renders the molecular understanding difficult.Citation7,Citation22 To understand the molecular origin of the antibiotic uptake, we need information on the role of each individual involved component. For example, the so-called liposome swelling assay provides information on a model system.Citation35,Citation52,Citation55,Citation60,Citation80,Citation97,Citation105 The method involves reconstitution of batches of purified Omps into (multilamellar) liposomes.Citation7,Citation22 Under isosmotic addition, the diffusion of substrate inside the liposome results in alteration of the light-scattering pattern. The effective change in light-scattering signal is then correlated with the relative permeability of the molecules. The main disadvantage of this method is that it requires a large quantity of material and is only effective for uncharged molecules, whereas for charged molecules, the effect of counterion flow affects the quality of the measurement. Moreover, the assay can only determine average turnover numbers and often does not provide conclusive values.Citation7
Moreover, using conventional electrophysiology, computation of rate of flux of discrete small molecules across Omps present in bacterial outer cell membrane involves measurement of flux values at single molecular level.Citation7,Citation36,Citation45,Citation52,Citation56,Citation66,Citation67 Here, electrophysiological measurement using single Omps provides the best high-resolution () signal-to-noise ratio,Citation7,Citation18,Citation40,Citation73,Citation74,Citation83 thereby suggesting the higher efficacy of this method in sensing and understanding uptake at molecular level.Citation7,Citation15,Citation22 The method includes reconstitution of a single or multiple Omps into an artificial planar lipid bilayer and further uses transmembrane potential-driven ion current across the channel as a detection probe.Citation7,Citation67 Using ion current as a probe specifically demonstrates very well-characterized electrophysiological properties of the Omps,Citation15,Citation34,Citation45,Citation65,Citation66,Citation84,Citation106,Citation119–Citation121 including size,Citation122,Citation123 single-channel conductance, channel ion selectivity,Citation58,Citation75,Citation76,Citation90,Citation99–Citation101 channel gating dynamics, and more.Citation47,Citation95,Citation109 Likewise, the size of Omps is a key factor defining transport through the channel.Citation107,Citation108 This factor plays a key role in antibiotic susceptibility.Citation72–Citation74 Determination of the size of Omps using electrophysiology provides a crucial insight into the maximum size of molecule they can transport.Citation122,Citation123 This, further, helps in evaluating the inner structure including constriction site.Citation122–Citation125 Further, single-channel conductance of Omps, ion selectivity,Citation58,Citation75,Citation76,Citation84,Citation89 and gating dynamicsCitation35,Citation47,Citation94,Citation95,Citation109 give an insight into the channel–substrate binding and channel–substrate interactions.Citation35,Citation71,Citation83,Citation85,Citation97,Citation99,Citation101 An insight into the channel conductance can be obtained, specifically using staircase electrophysiology (), where real-time insertions of single channels at constant voltage can be attained.Citation59,Citation123 The conductance of any channel can be termed as its unique characteristic. This allows a better understanding of the open/close states of the channel and its gating dynamics which can then be employed in studying channel structure–activity relationship.Citation35,Citation71,Citation107,Citation108
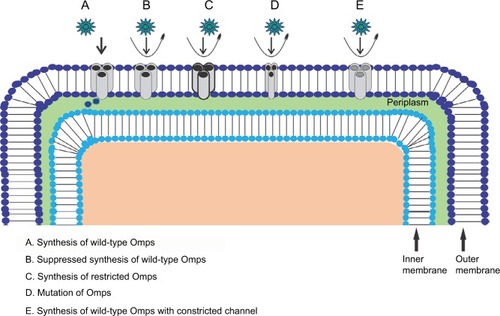
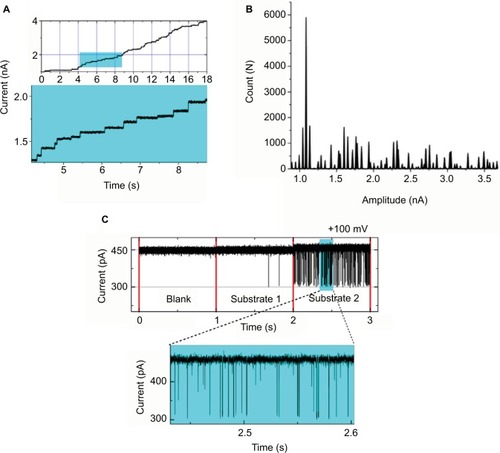
Figure 3 (A) Current recorded using staircase electrophysiology. A graphical representation depicting insertion of Omp over real time under applied potential. Recording time: 18 seconds. (B) Current histogram for the trace with each peak resembling a single Omp, showing, in total, approximately 45 Omps. (C) OmpF single channel–substrate interaction comparison: without substrate (blank), substrate 1 depicting no blockages, and substrate 2 inducing well-resolved channel blockage; a clear difference between the two substrates can be seen.
Abbreviation: Omp, outer membrane protein.
Using these functions, a proper insight into the channel interaction with different substrates can be obtained including substrate-induced partial or full blockage () of channelCitation52,Citation53,Citation67 and substrate-induced gating.Citation67 The function of these pores has been well documented on the basis of pore characteristics, chemical modification, and genetic mutations.Citation15 These parameters were further used to elaborate transport of the following antibiotics: meropenem,Citation52 imipenem,Citation52 cefotaxime,Citation48 cefpirome,Citation44 ceftriaxone,Citation44,Citation45 cefepime,Citation45 ceftazidime,Citation44,Citation45 ciprofloxacin,Citation45 norfloxacin,Citation45 and enrofloxacinCitation45 through OmpC; imipenem,Citation74 meropenem,Citation53 ceftazidime,Citation44,Citation45,Citation74 cefepime,Citation45,Citation55,Citation74 ceftriaxone,Citation44,Citation45 cefpirome,Citation44 ampicillin,Citation57,Citation60,Citation61,Citation67 benzylpenicillin,Citation60 amoxicillin,Citation57 carbenicillin,Citation57 azlocillin,Citation57 piperacillin,Citation57 ciprofloxacin,Citation45 norfloxacin,Citation45,Citation56,Citation126 enrofloxacin,Citation7,Citation45,Citation54,Citation66,Citation72 moxifloxacin,Citation65 different poly arginines,Citation64 polyamines,Citation106 and antimicrobial peptidesCitation51 through OmpF; ceftriaxoneCitation80 through Omp35; cefepimeCitation16 through Omp36; imipenemCitation91 and meropenemCitation91 through OccD3; imipenemCitation86 through OccD1; and meropenem,Citation104 glutamic acid,Citation104 arginine,Citation104 and imipenemCitation104 through CarO Omp ().
In contrast, single-channel recording provides the best signal-to-noise ratio and intrinsic data on Omp–substrate interaction.Citation40,Citation45,Citation65–Citation67 But the interpretation of molecule translocation cannot be made directly as the chances of molecule exit on the entry side are almost identical when compared to the transport of the molecule across the pore.Citation7 Whereas in the case of charged molecules, direct conclusion of translocation can be made as the increasing voltage will reduce the residence time of the molecules inside the Omp, which might provide some evidence of transport across the Omp. In addition, using channel selectivity, that is, channel inherent selection of either anion or cation, a quantitative flux assessment of the charged molecules can be made using electrophysiological reversal potential measurements.Citation36,Citation59 Using this approach, flux of β-lactamase inhibitors across OmpF and OmpC was estimated, showing the role of Omps in their transport across bacterial biobarrier.Citation36,Citation59 However, most of the molecules did not carry a net charge or show low intrinsic solubility which makes them trivial to measure and thus excludes them from screening via this method. Furthermore, the finite time resolution of electrophysiology also makes the method limited in screening of antibiotics uptake.Citation7,Citation45,Citation66,Citation67
Molecular simulation
In the current scenario, MS is well suited to obtain a particular information at an atomic scale.Citation121 Thus far, knowledge of the antibiotic translocation problem has pointed essentially toward three mechanisms including diffusion with molecule binding, a mechanism based on pore dehydration induced by the permeating molecule, and slow diffusion with molecule binding.Citation50,Citation61,Citation62,Citation70,Citation71,Citation97,Citation99,Citation121 Further, to discriminate among these mechanisms, and to attain a better description of the Omps behavior and their role in substrate transport, understanding the communication between pore and substrate is essential.Citation119–Citation121,Citation127,Citation128 Thanks to the high-resolution, molecular modeling simulations, detailed characterization is possible in terms of energetics ( from Ghai et al)Citation36 and bond formation including hydrogen bonds, hydrophobic contacts, and more.Citation50,Citation62,Citation71,Citation121
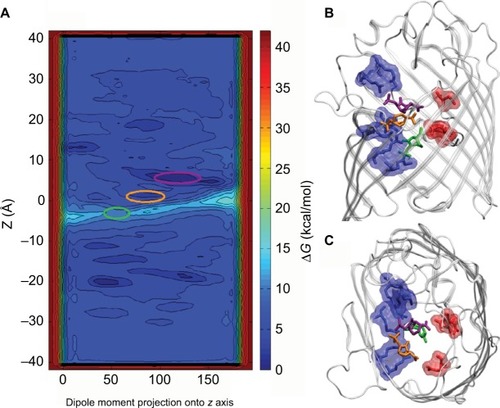
Figure 4 (A) Intrinsic depiction of the two-dimensional free energy of translocation of β-lactamase inhibitor (avibactam), reassembled from metadynamic simulations. (B) Lateral view and (C) topmost view of the avibactam inside OmpF pore in the two lowest minima near the constriction region and at the subsequent transition state. Reprinted with permission from Ghai I, Pira A, Scorciapino MA, et al. General method to determine the flux of charged molecules through nanopores applied to beta-lactamase inhibitors and OmpF. J Phys Chem Lett. 2017;8(6):1295–1301.Citation36 Copyright (2017) American Chemical Society.
The complete control over the characteristics of the system allows MS to explain the impact of pinpoint mutations and the effects that arise due to different domains of the same proteins.Citation95,Citation100,Citation101 Further, MS significantly allows understanding and interpreting available experimental data.Citation50,Citation61,Citation62,Citation70,Citation121 When combined with experimental approach, MS proves to be a complementary method. For instance, together with electrophysiology,Citation36,Citation48,Citation54,Citation55,Citation57,Citation58,Citation60,Citation61,Citation65–Citation67,Citation71–Citation73,Citation95,Citation97,Citation99–Citation101 MS was used for understanding the transport of β-lactamase inhibitors (), interaction of substrates with Omps (enrofloxacin, moxifloxacin, ampicillin, benzylpenicillin, carbenicillin, amoxicillin, azlocillin, piperacillin, ertapenem, imipenem, meropenem, cefepime, cefpirome, cefotaxime, ceftazidime, cefoxitin, and cefepime with OmpF;Citation7,Citation19,Citation21,Citation36,Citation50,Citation54,Citation55,Citation57,Citation58,Citation60–Citation63,Citation65–Citation67,Citation70–Citation73 cefotaxime, imipenem, and meropenem with OmpC;Citation19,Citation48 natural amino acids with OccD1),Citation87 ion transport including transport of phosphate potassium and chloride ion via OprPCitation90,Citation100–Citation102 and OprO,Citation99 and interaction of glycine ornithine, glucose, and imipenem with CarO isoforms.Citation105 Further, for liposome swellingCitation55,Citation60,Citation97,Citation105 and minimum inhibitory concentration assayCitation48,Citation60,Citation73 (not described), MS was helpful for understanding and interpreting the experimental results.
Rationalizing the process of permeation of antibiotics into Gram-negative bacteria via MS requires an accurate and exhaustive description of some key molecular properties of the antibiotic molecule.Citation121 MS is the best alternative tool to obtain homogenously derived physical–chemical descriptors for molecules with or without experimental approach.Citation121,Citation127,Citation128 MS based on all-atom empirical force fields with the resolution in microsecond time range and beyond could potentially provide a good level of description of the structural and dynamical properties of biological systems.Citation119,Citation121,Citation127,Citation128
Toward translational research
Translational research on understanding antimicrobial resistance has led to implausible development in recent yearsCitation4,Citation129 together with the expansion of novel techniques including proteomic analyses, high-sensitivity mass spectrometry, computational bioinformatics, and many more approaches.Citation4 For the most part, the discovery of novel technologies, the development of new infrastructures, along with the training of budding scientists have reinforced this evolution.Citation1,Citation4,Citation129,Citation130 But the transition is still not complete, and roadblocks still exist on the path to scientific progress, for example, combining different data into a shared database that can be intrinsically used to understand how Omps located in the outer membrane of Gram-negative bacteria are able to filter molecular influx.Citation24 The imperative need for new, effective Gram-negative antibacterial drugs comes at a time when techniques needed for innovative assays can provide significant crucial data over understanding the effective bottleneck.Citation4 Ideally, the overall penetration–efflux puzzleCitation4 will form part of a larger understanding of the Gram-negative cell envelope as well as direction on how to create small molecules that can easily penetrate across the outer membranes.Citation4 This information should move the antibacterial research community toward more rational approaches, which may enable the delivery of new agents to treat life-threatening infections.Citation1,Citation4,Citation129,Citation130
Conclusive remarks
This review summarizes the progressive scientific evidence explaining the role of Omps in membrane permeability of Gram-negative bacteria. The control of bacterial membrane permeability is a complex process that is strongly structured by an intricate network of arrangements that senses and retorts to pH, osmotic shock, temperature, and external chemical stress. Bacteria majorly make use of cultured regulated cascades that perceive and distinguish toxic compounds and respond through various resistance mechanisms including regulation of Omps.Citation6,Citation7,Citation15,Citation18,Citation22 The information on the role of effective Omps in substrate uptake and their structural relationship associated with their role in transport highlights the efforts of the scientific upfront in the direction of understanding the bacterial resistance.Citation6,Citation7,Citation15,Citation18,Citation22 Translocation across the Omps can be assumed as the first step in the journey of an antibiotic along the defined pathway toward its target. Consequently, interpretation of antibiotic translocation through porins at the molecular level is crucial for understanding the correlation between influx and antibiotic activities within bacteria. The function of the general diffusion pores has been well studied based on pore characteristics, chemical modification, and genetic mutations. Our understanding of the structure of the pore-forming complex has tremendously improved over the last decade with the emergence of MS, state-of-the-art X-ray data, mass spectrometry assay protocols, and novel high-resolution experimental approaches including electrophysiology. However, a better understanding of the transportation mechanism by outer membrane pores is required. The molecular basis of the antibiotic transport via specific porins is still completely open at present, and further rigorous studies are needed to give insight into the structure–activity relationship of pores associated with antibiotic transport. The data computed for these Omps can be further employed to elucidate the antibiotic uptake pathway through Omps at molecular level, which could possibly empower rational drug design to further enhance permeation and support novel strategies to dodge “impermeability”-mediated resistance mechanism.
Acknowledgments
The publication of this article was funded by the Open Access fund of Leibniz Universität Hannover. The authors would like to thank Prof. Dr Mathias Winterhalter and Prof. Dr Richard Wagner for their constructive comments.
Disclosure
The authors report no conflicts of interest in this work.
References
- KostyanevTBontenMJO’BrienSThe Innovative Medicines Initiative’s New Drugs for Bad Bugs programme: European public-private partnerships for the development of new strategies to tackle antibiotic resistanceJ Antimicrob Chemother201671229029526568581
- VentolaCLThe antibiotic resistance crisis: part 1: causes and threatsP T201540427728325859123
- GootzTDThe global problem of antibiotic resistanceCrit Rev Immunol2010301799320370622
- StavengerRAWinterhalterMTRANSLOCATION project: how to get good drugs into bad bugsSci Transl Med20146228228ed7
- NikaidoHRole of permeability barriers in resistance to beta-lactam antibioticsPharmacol Ther19852721972312412244
- NikaidoHMolecular basis of bacterial outer membrane permeability revisitedMicrobiol Mol Biol Rev200367459365614665678
- WinterhalterMCeccarelliMPhysical methods to quantify small antibiotic molecules uptake into Gram-negative bacteriaEur J Pharm Biopharm201595Pt A636726036449
- ChattopadhyayMKJaganandhamMVVesicles-mediated resistance to antibiotics in bacteriaFront Microbiol2015675826257725
- KulkarniHMJagannadhamMVBiogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteriaMicrobiology2014160102109212125069453
- ChattopadhyayMKJagannadhamMVCorrigendum: vesicles-mediated resistance to antibiotics in bacteriaFront Microbiol2015675826257725
- McBroomAJKuehnMJRelease of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress responseMol Microbiol200763254555817163978
- MarediaRDevineniNLentzPVesiculation from Pseudomonas aeruginosa under SOSScientific World J2012201218
- KulkarniHMSwamyChVJagannadhamMVMolecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditionsJ Proteome Res20141331345135824437924
- SchwechheimerCKuehnMJOuter-membrane vesicles from Gram-negative bacteria: biogenesis and functionsNat Rev Microbiol2015131060561926373371
- BenzRStructure and function of porins from gram-negative bacteriaAnnu Rev Microbiol1988423593932849372
- JamesCEMahendranKRMolitorAHow beta-lactam antibiotics enter bacteria: a dialogue with the porinsPLoS One200945e545319434239
- SchirmerTGeneral and specific porins from bacterial outer membranesJ Struct Biol199812121011099615433
- SchulzGEThe structure of bacterial outer membrane proteinsBiochim Biophys Acta20021565230831712409203
- ScorciapinoMAD’AgostinoTAcosta-GutierrezSMallociGBodrenkoICeccarelliMExploiting the porin pathway for polar compound delivery into Gram-negative bacteriaFuture Med Chem20168101047106227303954
- SinghHThangarajPChakrabartiAAcinetobacter baumannii: a brief account of mechanisms of multidrug resistance and current and future therapeutic managementJ Clin Diagn Res20137112602260524392418
- ZiervogelBKRouxBThe binding of antibiotics in OmpF porinStructure2013211768723201272
- PagesJMJamesCEWinterhalterMThe porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteriaNat Rev Microbiol200861289390318997824
- DupontHChoinierPRocheDStructural alteration of OmpR as a source of ertapenem resistance in a CTX-M-15-producing Escherichia coli O25b:H4-ST131 clinical isolateAntimicrob Agents Chemother2017615e00014e0001728264855
- ChakradharSBreaking through: How researchers are gaining entry into barricaded bacteriaNat Med201723890791028777786
- NakaeTOuter membrane of Salmonella. Isolation of protein complex that produces transmembrane channelsJ Biol Chem1976251721762178773934
- NikaidoHVaaraMMolecular basis of bacterial outer membrane permeabilityMicrobiol Rev19854911322580220
- Nguyen-DistecheMPollockJJGhuysenJMSensitivity to ampicillin and cephalothin of enzymes involved in wall peptide crosslinking in Escherichia coli K12, strain 44Eur J Biochem19744134574634593965
- SchulzGEPorins: general to specific, native to engineered passive poresCurr Opin Struct Biol1996644854908794162
- GuillierMGottesmanSStorzGModulating the outer membrane with small RNAsGenes Dev200620172338234816951250
- FahieMAYangBMullisMHoldenMAChenMSelective detection of protein homologues in serum using an OmpG nanoporeAnal Chem20158721111431114926451707
- LiHLuoYFWilliamsBJBlackwellTSXieCMStructure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapiesInt J Med Microbiol20123022636822226846
- DupontHGaillotOGoetgheluckASMolecular characterization of carbapenem-nonsusceptible enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinationsAntimicrob Agents Chemother201560121522126482307
- HancockREWoodruffWARoles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosaRev Infect Dis19881047707752460909
- IredellJBrownJTaggKAntibiotic resistance in Enterobacteriaceae: mechanisms and clinical implicationsBMJ2016352h642026858245
- ZahnMBhamidimarri SatyaPBasléAWinterhalterMvan den BergBStructural insights into outer membrane permeability of Acinetobacter baumanniiStructure201624222123126805524
- GhaiIPiraAScorciapinoMAGeneral method to determine the flux of charged molecules through nanopores applied to beta-lactamase inhibitors and OmpFJ Phys Chem Lett2017861295130128240914
- KretschmerNDamianakosHChinouIComparison of the cytotoxicity and antimicrobial activity of several isohexenylnaphthazarinsPlanta Med20117712M199
- HancockREBrinkmanFSFunction of pseudomonas porins in uptake and effluxAnnu Rev Microbiol200256173812142471
- Fito-BoncompteLChapalainABouffartiguesEFull virulence of Pseudomonas aeruginosa requires OprFInfect Immun20117931176118621189321
- NestorovichEMSugawaraENikaidoHBezrukovSMPseudomonas aeruginosa porin OprF: properties of the channelJ Biol Chem200628124162301623716617058
- SugawaraENestorovichEMBezrukovSMNikaidoHPseudomonas aeruginosa porin OprF exists in two different conformationsJ Biol Chem200628124162201622916595653
- PagesJMPeslierSKeatingTALavigneJPNicholsWWRole of the outer membrane and porins in susceptibility of beta-lactamase-producing Enterobacteriaceae to ceftazidime-avibactamAntimicrob Agents Chemother20156031349135926666933
- Acosta GutierrezSBodrenkoIScorciapinoMACeccarelliMMacroscopic electric field inside water-filled biological nanoporesPhys Chem Chem Phys201618138855886426931352
- LovelleMMachTMahendranKRWeingartHWinterhalterMGameiroPInteraction of cephalosporins with outer membrane channels of Escherichia coli. Revealing binding by fluorescence quenching and ion conductance fluctuationsPhys Chem Chem Phys20111341521153021152583
- MahendranKRKreirMWeingartHFertigNWinterhalterMPermeation of antibiotics through Escherichia coli OmpF and OmpC porins: screening for influx on a single-molecule levelJ Biomol Screen201015330230720086208
- PinetEFranceschiCDavin-RegliAZambardiGPagesJMRole of the culture medium in porin expression and piperacillin-tazobactam susceptibility in Escherichia coliJ Med Microbiol201564111305131426242994
- LiuNSamartzidouHLeeKWBriggsJMDelcourAHEffects of pore mutations and permeant ion concentration on the spontaneous gating activity of OmpC porinProtein Eng200013749150010906344
- LouHChenMBlackSSAltered antibiotic transport in OmpC mutants isolated from a series of clinical strains of multi-drug resistant E. coliPLoS One2011610e2582522053181
- SugawaraEKojimaSNikaidoHKlebsiella pneumoniae major porins OmpK35 and OmpK36 allow more efficient diffusion of beta-lactams than their Escherichia coli homologs OmpF and OmpCJ Bacteriol2016198233200320827645385
- Aguilella-ArzoMGarcia-CelmaJJCerveraJAlcarazAAguilellaVMElectrostatic properties and macroscopic electrodiffusion in OmpF porin and mutantsBioelectrochemistry200770232032716769257
- ApetreiAAsandeiAParkYHahmKSWinterhalterMLuchianTUnimolecular study of the interaction between the outer membrane protein OmpF from E. coli and an analogue of the HP(2–20) antimicrobial peptideJ Bioenerg Biomembr201042217318020180000
- BajajHScorciapinoMAMoynieLMolecular basis of filtering carbapenems by porins from beta-lactam-resistant clinical strains of Escherichia coliJ Biol Chem201629162837284726645688
- BodrenkoIBajajHRuggeronePWinterhalterMCeccarelliMAnalysis of fast channel blockage: revealing substrate binding in the microsecond rangeAnalyst2015140144820482725717496
- BrauserASchroederIGutsmannTModulation of enrofloxacin binding in OmpF by Mg2+ as revealed by the analysis of fast flickering single-porin currentJ Gen Physiol20121401698222689827
- BredinJSaintNMalleaMAlteration of pore properties of Escherichia coli OmpF induced by mutation of key residues in anti-loop 3 regionBiochem J2002363Pt 352152811964152
- CamaJBajajHPagliaraSQuantification of fluoroquinolone uptake through the outer membrane channel OmpF of Escherichia coliJ Am Chem Soc201513743138361384326478537
- DanelonCNestorovichEMWinterhalterMCeccarelliMBezrukovSMInteraction of zwitterionic penicillins with the OmpF channel facilitates their translocationBiophys J20069051617162716339889
- DanelonCSuenagaAWinterhalterMYamatoIMolecular origin of the cation selectivity in OmpF porin: single channel conductances vs. free energy calculationBiophys Chem2003104359160312914905
- GhaiIWinterhalterMWagnerRProbing transport of charged beta-lactamase inhibitors through OmpC, a membrane channel from E. coliBiochem Biophys Res Commun20174841515528109883
- HajjarEBessonovAMolitorAToward screening for antibiotics with enhanced permeation properties through bacterial porinsBiochemistry201049326928693520604536
- HajjarEMahendranKRKumarABridging timescales and length scales: from macroscopic flux to the molecular mechanism of antibiotic diffusion through porinsBiophys J201098456957520159153
- ImWRouxBIon permeation and selectivity of OmpF porin: a theoretical study based on molecular dynamics, Brownian dynamics, and continuum electrodiffusion theoryJ Mol Biol2002322485186912270719
- KumarAHajjarERuggeronePCeccarelliMMolecular simulations reveal the mechanism and the determinants for ampicillin translocation through OmpFJ Phys Chem B2010114299608961620590090
- LamichhaneUIslamTPrasadSWeingartHMahendranKRWinterhalterMPeptide translocation through the mesoscopic channel: binding kinetics at the single molecule levelEur Biophys J201342536336923271514
- MachTNevesPSpigaEFacilitated permeation of antibiotics across membrane channels—interaction of the quinolone moxifloxacin with the OmpF channelJ Am Chem Soc200813040133011330918788798
- MahendranKRHajjarEMachTMolecular basis of enrofloxacin translocation through OmpF, an outer membrane channel of Escherichia coli—when binding does not imply translocationJ Phys Chem B2010114155170517920349984
- NestorovichEMDanelonCWinterhalterMBezrukovSMDesigned to penetrate: time-resolved interaction of single antibiotic molecules with bacterial poresProc Natl Acad Sci U S A200299159789979412119404
- NevesPBerkaneEGameiroPWinterhalterMde CastroBInteraction between quinolones antibiotics and bacterial outer membrane porin OmpFBiophys Chem2005113212312815617818
- NevesPSousaIWinterhalterMGameiroPFluorescence quenching as a tool to investigate quinolone antibiotic interactions with bacterial protein OmpFJ Membr Biol2009227313314019148694
- PezeshkiSChimerelCBessonovANWinterhalterMKleinekathoferUUnderstanding ion conductance on a molecular level: an all-atom modeling of the bacterial porin OmpFBiophys J20099771898190619804720
- PhalePSPhilippsenAWidmerCPhaleVPRosenbuschJPSchirmerTRole of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulationBiochemistry200140216319632511371193
- SinghPRCeccarelliMLovelleMWinterhalterMMahendranKRAntibiotic permeation across the OmpF channel: modulation of the affinity site in the presence of magnesiumJ Phys Chem B2012116154433443822369436
- VidalSBredinJPagesJMBarbeJBeta-lactam screening by specific residues of the OmpF eyeletJ Med Chem20054851395140015743183
- WeichbrodtCBajajHBaakenGAntibiotic translocation through porins studied in planar lipid bilayers using parallel platformsAnalyst2015140144874488125834843
- BenzRDarveauRPHancockREOuter-membrane protein PhoE from Escherichia coli forms anion-selective pores in lipid-bilayer membranesEur J Biochem198414023193246325185
- DarveauRPHancockREBenzRChemical modification of the anion selectivity of the PhoE porin from the Escherichia coli outer membraneBiochim Biophys Acta1984774167746329296
- BornetCDavin-RegliABosiCPagesJMBolletCImipenem resistance of Enterobacter aerogenes mediated by outer membrane permeabilityJ Clin Microbiol20003831048105210698994
- LavigneJPSottoANicolas-ChanoineMHBouzigesNPagesJMDavin-RegliAAn adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolatesInt J Antimicrob Agents201341213013623280442
- ThiolasABornetCDavin-RegliAPagesJMBolletCResistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenesBiochem Biophys Res Commun2004317385185615081418
- BornetCSaintNFetnaciLOmp35, a new Enterobacter aerogenes porin involved in selective susceptibility to cephalosporinsAntimicrob Agents Chemother20044862153215815155215
- ArunmaneeWPathaniaMSolovyovaASGram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesisProc Natl Acad Sci U S A201611334E5034E504327493217
- CastanheiraMMendesRESaderHSLow frequency of ceftazidime-avibactam resistance among Enterobacteriaceae isolates carrying blaKPC collected in U.S. hospitals rom 2012 to 2015Antimicrob Agents Chemother2017613
- ErenEParkinJAdelanwaAToward understanding the outer membrane uptake of small molecules by Pseudomonas aeruginosaJ Biol Chem201328817120421205323467408
- LiuJWolfeAJErenECation selectivity is a conserved feature in the OccD subfamily of Pseudomonas aeruginosaBiochim Biophys Acta20121818112908291622824298
- BiswasSMohammadMMPatelDRMovileanuLvan den BergBStructural insight into OprD substrate specificityNat Struct Mol Biol200714111108110917952093
- HuangHHancockREThe role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosaJ Bacteriol199617811308530908655484
- SamantaSScorciapinoMACeccarelliMMolecular basis of substrate translocation through the outer membrane channel OprD of Pseudomonas aeruginosaPhys Chem Chem Phys20151737238672387626306922
- ErenEVijayaraghavanJLiuJSubstrate specificity within a family of outer membrane carboxylate channelsPLoS Biol2012101e100124222272184
- LiuJErenEVijayaraghavanJOccK channels from Pseudomonas aeruginosa exhibit diverse single-channel electrical signatures but conserved anion selectivityBiochemistry201251112319233022369314
- ModiNBenzRHancockREKleinekathoferUModeling the ion selectivity of the phosphate specific channel OprPJ Phys Chem Lett20123233639364526290999
- SoundararajanGBhamidimarriSPWinterhalterMUnderstanding carbapenem translocation through OccD3 (OpdP) of Pseudomonas aeruginosaACS Chem Biol20171261656166428440622
- ChalhoubHPletzerDWeingartHMechanisms of intrinsic resistance and acquired susceptibility of Pseudomonas aeruginosa isolated from cystic fibrosis patients to temocillin, a revived antibioticSci Rep201774020828091521
- ChenekeBRIndicMvan den BergBMovileanuLAn outer membrane protein undergoes enthalpy- and entropy-driven transitionsBiochemistry201251265348535822680931
- ChenekeBRvan den BergBMovileanuLAnalysis of gating transitions among the three major open states of the OpdK channelBiochemistry201150224987499721548584
- PothulaKRDhanasekarNNLamichhaneUSingle residue acts as gate in OccK channelsJ Phys Chem B2017121122614262128257208
- PothulaKRKleinekathoferUTheoretical analysis of ion conductance and gating transitions in the OpdK (OccK1) channelAnalyst2015140144855486425781224
- SamantaSD’AgostinoTGhaiIHow to get large drugs through small pores? Exploiting the porins pathway in Pseudomonas aeruginosaBiophys J20171123 Suppl 1416a27955889
- TamberSMaierEBenzRHancockRECharacterization of OpdH, a Pseudomonas aeruginosa porin involved in the uptake of tricarboxylatesJ Bacteriol2007189392993917114261
- ModiNGangulySBarcena-UribarriIStructure, dynamics, and substrate specificity of the OprO porin from Pseudomonas aeruginosaBiophys J201510971429143826445443
- ModiNBarcena-UribarriIBainsMBenzRHancockREKleinekathoferURole of the central arginine R133 toward the ion selectivity of the phosphate specific channel OprP: effects of charge and solvationBiochemistry201352335522553223875754
- ModiNBarcena-UribarriIBainsMBenzRHancockREKleinekathoferUTuning the affinity of anion binding sites in porin channels with negatively charged residues: molecular details for OprPACS Chem Biol201510244145125333751
- PongprayoonPBecksteinOWeeCLSansomMSSimulations of anion transport through OprP reveal the molecular basis for high affinity and selectivity for phosphateProc Natl Acad Sci U S A200910651216142161819966228
- Catel-FerreiraMNehmeRMolleVDeciphering the function of the outer membrane protein OprD homologue of Acinetobacter baumanniiAntimicrob Agents Chemother20125673826383222564848
- Catel-FerreiraMCoadouGMolleVStructure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumanniiJ Antimicrob Chemother20116692053205621705362
- ZahnMD’AgostinoTErenEBasleACeccarelliMvan den BergBSmall-molecule transport by CarO, an abundant eight-stranded beta-barrel outer membrane protein from Acinetobacter baumanniiJ Mol Biol2015427142329233925846137
- IyerRDelcourAHComplex inhibition of OmpF and OmpC bacterial porins by polyaminesJ Biol Chem19972723018595186019228026
- SaintNLouKLWidmerCLuckeyMSchirmerTRosenbuschJPStructural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterizationJ Biol Chem19962713420676206808702817
- LouKLSaintNPrilipovAStructural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysisJ Biol Chem19962713420669206758702816
- PhalePSSchirmerTPrilipovALouK-LHardmeyerARosenbuschJPVoltage gating of Escherichia coli porin channels: role of the constriction loopProc Natl Acad Sci U S A19979413674167459192635
- StockJBRauchBRosemanSPeriplasmic space in Salmonella typhimurium and Escherichia coliJ Biol Chem19772522178507861334768
- TestaCAJohnsonLJA whole-cell phenotypic screening platform for identifying methylerythritol phosphate pathway-selective inhibitors as novel antibacterial agentsAntimicrob Agents Chemother20125694906491322777049
- MortimerPGSPiddockLJVA comparison of methods used for measuring the accumulation of quinolones by enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureusJ Antimicrob Chemother19912856396531663928
- GoessensWHvan der BijAKvan BoxtelRAntibiotic trapping by plasmid-encoded CMY-2 beta-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coliAntimicrob Agents Chemother20135783941394923733461
- ShinJRLimKJKimDJChoJHKimSCDisplay of multimeric antimicrobial peptides on the Escherichia coli cell surface and its application as whole-cell antibioticsPLoS One201383e5899723516591
- CroxenMALawRJScholzRKeeneyKMWlodarskaMFinlayBBRecent advances in understanding enteric pathogenic Escherichia coliClin Microbiol Rev201326482288024092857
- FoxDTSchmidtENTianHSub-inhibitory fosmidomycin exposures elicits oxidative stress in Salmonella enterica serovar Typhimurium LT2PLoS One201494e9527124751777
- DavisTDGerryCJTanDSGeneral platform for systematic quantitative evaluation of small-molecule permeability in bacteriaACS Chem Biol20149112535254425198656
- ZhouYJoubranCMiller-VedamLThinking outside the “bug”: a unique assay to measure intracellular drug penetration in gram-negative bacteriaAnal Chem20158773579358425753586
- BezrukovSMBerezhkovskiiAMSzaboADiffusion model of solute dynamics in a membrane channel: mapping onto the two-site model and optimizing the fluxJ Chem Phys20071271111510117887882
- O’SheaRMoserHEPhysicochemical properties of antibacterial compounds: implications for drug discoveryJ Med Chem200851102871287818260614
- ScorciapinoMAAcosta-GutierrezSBenkerrouDRationalizing the permeation of polar antibiotics into Gram-negative bacteriaJ Phys Condens Matter2017291111300128155846
- KrasilnikovOVSabirovRZTernovskyVIMerzliakPGMuratkhodjaevJNA simple method for the determination of the pore radius of ion channels in planar lipid bilayer membranesFEMS Microbiol Immunol199251–3931001384601
- Barcena-UribarriITheinMMaierEBondeMBergstromSBenzRUse of nonelectrolytes reveals the channel size and oligomeric constitution of the Borrelia burgdorferi P66 porinPLoS One2013811e7827224223145
- KrasilnikovOVDa CruzJBYuldashevaLNVarandaWANogueiraRAA novel approach to study the geometry of the water lumen of ion channels: colicin Ia channels in planar lipid bilayersJ Membr Biol1998161183929430623
- KrasilnikovOVSizing channels with neutral polymersKasianowiczJJKellermayerMDeamerDWStructure and Dynamics of Confined PolymersDordrechtSpringer200297115
- BajajHAcosta GutierrezSBodrenkoIBacterial outer membrane porins as electrostatic nanosieves: exploring transport rules of small polar moleculesACS Nano20171165465547328485920
- SchwarzGDanelonCWinterhalterMOn translocation through a membrane channel via an internal binding site: kinetics and voltage dependenceBiophys J20038452990299812719230
- TranQTWilliamsSFaridRErdemliGPearlsteinRThe translocation kinetics of antibiotics through porin OmpC: insights from structure-based solvation mapping using WaterMapProteins201381229129923011778
- FontanaJMAlexanderESalvatoreMTranslational research in infectious disease: current paradigms and challenges aheadTransl Res2012159643045322633095
- GoldmanMThe innovative medicines initiative: a European response to the innovation challengeClin Pharmacol Ther201291341842522318619
