Abstract
Introduction
The emergence and spread of Klebsiella pneumoniae strains resistant to multiple antimicrobial agents are considered as a serious challenge for nosocomial infections.
Materials and methods
In this study, 175 nonrepetitive clinical isolates of K. pneumoniae were collected from hospitalized patients in Kerman, Iran. Extended-spectrum β-lactamases (ESBLs), AmpC, and carbapenemase-producing isolates were recognized by phenotypic methods. The resistance genes including efflux pumps oqxA/oqxB, 16S rRNA methylase, ESBL, AmpC, and carbapenemase were detected by PCR-sequencing method. Molecular typing was performed by enterobacterial repetitive intergenic consensus-PCR and multilocus sequence typing methods among blaNDM-positive isolates.
Results
Thirty-seven (21.14%) isolates along with sequence types (STs): ST43, ST268, ST340, ST392, ST147, and ST16 were harbored blaNDM. ST43 in 2015 and ST268 during 2016–2017 were the most frequent STs among New Delhi metallo-beta-lactamase (NDM)-positive isolates. We found the distribution of some isolates with blaNDM, blaCTX-M, blaSHV, blaOXA, blaTEM, blaCMY, rmtC, and oqxA/oqxB. Enterobacterial repetitive intergenic consensus-PCR represented seven clusters (A–G) plus four singletons among NDM-positive isolates. This study provides the first report of blaNDM-1-positve K. pneumoniae along with ST268 as well as the spread of nosocomial infections with six different STs harboring blaNDM-1 and other resistance genes in hospital settings especially neonatal intensive care unit.
Conclusion
The dissemination of various clones of NDM-producing K. pneumoniae can contribute to increase the rate of their spread in health care settings. Therefore, molecular typing and detection of resistance genes have an important role in preventing and controlling infection by limiting the dissemination of multidrug-resistant isolates.
Keywords:
Introduction
Infections caused by multidrug-resistant bacteria have declared a substantial threat to public health worldwide.Citation1 Carbapenems are the most important antibiotics used for the treatment of infections caused by extended-spectrum β-lactamases (ESBLs) and AmpC-producing Gram-negative bacteria.Citation2 Several mechanisms including the loss of outer membrane proteins and carbapenemase such as KPC, GES, VIM, IMP, GIM, New Delhi metallo-beta-lactamase (NDM), and OXA-types are involved in resistance to carbapenems in Enterobacteriaceae.Citation3 Carbapenemase-producing bacteria usually cause life-threatening infections and long-time hospitalization in health care settings.Citation1 For the first time, the NDM has been identified in carbapenem-resistant Klebsiella pneumoniae in Sweden and then has been reported in other Gram-negative bacteria.Citation4–Citation7 In Iran, the first NDM-producing K. pneumoniae was identified in March 2011 from Tehran.Citation1 NDM-producing K. pneumoniae are broadly considered as multidrug-resistant bacteria that have been commonly associated with additional resistance mechanisms such as AmpC, ESBLs, and methylation of 16S rRNA by armA, rmtA, rmtB, and rmtC.Citation8 Several typing methods have been introduced and developed for epidemiological investigation of K. pneumoniae including enterobacterial repetitive intergenic consensus amplification (ERIC-PCR) and multilocus sequence typing (MLST).Citation9,Citation10 MLST is one of the best molecular typing methods for long-term and global epidemiological investigations, and ERIC-PCR is usually used for local outbreaks over a short period of time.Citation10 In this study, we investigated the molecular epidemiology from NDM-1-producing clones among carbapenem-resistant K. pneumoniae isolates in Kerman hospitals, Iran, and we emphasized on the clonal relatedness of these isolates.
Materials and methods
Bacterial isolates
In this study, 175 nonduplicated isolates of K. pneumoniae were collected from hospitalized patients in four referral hospitals (Shafa, Afzalipoor, Bahonar, and Kashani) during February 2015 to November 2017 in Kerman, Iran. All the isolates were identified as K. pneumoniae by standard microbiological tests.Citation11
Antibiotic susceptibility testing
Antibacterial susceptibility test of isolates to cefepime (30 µg), cefotaxime (30 µg), cefoxitin (30 µg), ceftazidime (30 µg), ceftizoxime (30 µg), cefpodoxime (10 µg), imipenem (10 µg), meropenem (10 µg), ertapenem (10 µg), gentamicin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), and norfloxacin (10 µg) (Mast Group Ltd., Bootle, UK) was determined by disk diffusion method on Müller–Hinton agar media (Laboratorios CONDA, Madrid, Spain) according to the Clinical and Laboratory Standards Institute (CLSI).Citation12 Minimum inhibitory concentration (MIC) of isolates to cefotaxime, cefepime, and imipenem was determined by microbroth dilution method according to CLSI. To determine MIC of colistin and tigecycline by microbroth dilution method, we used the European Committee on Antimicrobial Susceptibility Testing recommendations (http://www.eucast. org/clinical-breakpoints). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as standard strains in antibacterial susceptibility testing.
Detection of ESBLs, AmpC, and carbapenemase-producing isolates
ESBLs and carbapenemase-producing isolates were determined according to CLSI recommendations by combination disk with clavulanate and Carba NP test, respectively.Citation12 AmpC disk test was used to detect AmpC β-lactamase-producing isolates.Citation13
Genomic DNA extraction
The genomic DNA was extracted using Exgene Clinic SV (GeneAll Biotechnology, Co., Ltd., Seoul, Republic of Korea; Kat: 106-152) according to the manufacturer’s guidelines.
Detection of resistance genes by PCR sequencing
Antibiotic resistance genes including ESBLs (blaTEM, blaSHV, blaCTX-M, blaOXA-1, and blaPER), caebapenemase (blaKPC, blaGES, blaOXA-48, blaIMP, blaVIM, blaNDM, blaSPM, blaSIM, blaGIM, and blaAIM), efflux pump (oxqA/B), 16S rRNA meth-ylase (rmtA, rmtB, rmtC, and armA), and mcr-1 (colistin resistance gene) were detected by PCR. The primers used for amplification of resistance genes are listed in . The AmpC β-lactamase genes including blaCMY, blaFOX, blaACC, blaACT, blaDHA, blaEBC, and blaCIT were detected by using multiplex PCR as previously described, and furthermore, PCR products were confirmed by sequencing (Bioneer Corporation, Daejeon, Republic of Korea).Citation29
Table 1 Sequence of primers used in this study for the detection of resistance genes in PCR method
MLST of NDM-producing isolates
MLST of isolates was performed using seven conserved housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) according to protocols available at the MLST Pasteur website (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html) for NDM-producing isolates. Products of the above genes in MLST were sequenced by Bioneer, Co. Sequences of each housekeeping gene in both directions were analyzed by Sequence Scanner Software v.2.0 (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, USA) and assembled by Lasergene 6 software (DNASTAR). The sequence types (STs) of each isolate were determined based on the seven studied loci described at http://bigsdb.pasteur. fr/klebsiella/klebsiella.html.
Molecular typing of blaNDM-positive isolates by ERIC-PCR
ERIC-PCR using ERIC2 primer (5′-AAGTAAGT-GACTGGGGTGAGC-3′) was used for molecular typing of NDM-positive isolates.Citation30 The results of ERIC-PCR were analyzed in http://insilico.ehu.eus/dice_upgma/using the Dice similarity coefficient. Clusters were defined as DNA patterns sharing ≥80% similarity.
Results
In this study, 175 nonduplicated isolates of K. pneumoniae were recovered from hospitalized patients in four referral hospitals in Kerman, Iran. The isolates were collected from different specimens including burning wounds 9 (5.1%), urine 126 (72%), blood 21 (12%), bronchoalveolar lavage 16 (9.1%), and cerebrospinal fluid 3 (1.7%).
Antimicrobial susceptibility testing
The rate of resistance to antibiotics was the following: cefpodoxime 83 (47.4%), cefotaxime 80 (45.7), ceftizoxime 78 (44.6%), ceftazidime 68 (38.9%), cefoxitin 64 (36.6%), cefepime 71 (40.5%), imipenem 45 (25.7%), meropenem 33 (18.9%), ertapenem 30 (17.1%), amikacin 68 (38.9%), gentamicin 59 (33.7%), norfloxacin 33 (18.9%), and ciprofloxacin 31 (17.7%). The ranges of MIC to imipenem, cefepime, and cefotaxime were 4–128 µg/mL, 16–2,048 µg/mL, and 8–2,048 µg/mL, respectively. MIC to colistin was increased in seven (4%) isolates with range 2–16 µg/mL and among other isolates were ≤0.5 µg/mL. All isolates were sensitive to tigecycline with MIC ≤0.5 µg/mL. The MIC results of the clinical isolates are shown in .
Table 2 The MIC of clinical isolates of Klebsiella pneumoniae resistance to imipenem, cefotaxime, cefepime, and colistin
Phenotypic confirmatory tests
Among the 175 K. pneumoniae isolates, 72 (41.1%) strains produced ESBLs, 12 (6.8%) isolates produced AmpC, and 8 (4.5%) isolates produced both ESBLs and AmpC β-lactamase. Out of 175 K. pneumoniae isolates, 37 (21.1%) isolates were considered as positive carbapenemases with Carba NP test.
PCR amplification of antibiotic resistance genes
Based on the PCR assays, the prevalence of ESBL genes was as follows: blaCTX-M 46.28% (n=81), blaSHV 41.1% (n=72), blaTEM 38.9% (n=68), and blaOXA-1 21.7% (n=38). The only carbapenemase gene found in isolates was blaNDM-1 21.14% (n=37). The major AmpC β-lactamase genes found were blaCMY 2.85% (n=5), followed by blaFOX 1.1% (n=2) and blaACC, blaACT 0.6% (n=1). The efflux pump genes including oqxA/oqxB were detected in 36.6% (n=64) and 19.4% (n=34) of isolates. Aminoglycoside-resistant genes (16S rRNA methylase) including rmtC and armA were observed in 5.7% (n=10) and 1.1% (n=2) of isolates, respectively. The rest of the antibiotic resistance genes (blaEBC, blaCIT, blaVIM, blaIMP, blaGIM, blaAIM, blaSPM, blaSIM, blaGES, blaKPC, blaOXA-48, blaPER, blaDHA, rmtA, rmtB, and mcr-1) were negative.
Some sequences of the antibiotic resistance genes including blaNDM, blaTEM, blaCTX-M, blaOXA-1, blaSHV, armA, and rmtC were submitted to the GenBank under accession numbers MG515599, MG515594, MG515597, MG515600, MG515593, MG515596, and MG515592, respectively.
Molecular typing of NDM-producing isolates
In this study, we described the first NDM-producing K. pneumoniae isolates belonging to the ST268 (n=14), which was the major ST. The other STs were as follows: ST43 (n=9), ST340 (n=7), ST392 (n=5), ST147 (n=1), and ST16 (n=1).
According to the eBURST results, ST268 is triple-locus variants of ST16 reporting NDM-producing K. pneumoniae previously. In this study, in comparison with other STs, most isolates of K. pneumoniae ST268 carrying rmtC gene were associated with neonatal intensive care unit (NICU), whereas one of K. pneumoniae ST43 isolate coproducing armA and blaNDM genes was associated with surgical unit ().
Table 3 Distribution and genetic characterization of 37 NDM-producing Klebsiella pneumoniae strains isolated from hospitalized patients
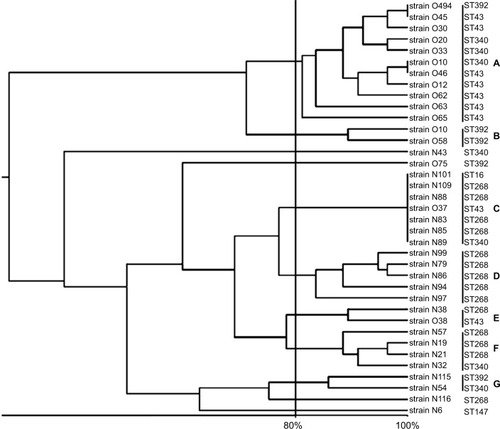
showed distribution and genetic characterization of 37 NDM-producing K. pneumoniae strains. ERIC-PCR findings showed that the 37 NDM-producing strains were divided into 7 clusters A to G (11 strains in clusters A, 2 strains in clusters B, E, G, 7 strains in cluster C, 5 strains in cluster D, 4 strains in cluster F, and 3 strains were selected to represent sporadic strains) (). ST43 was divided into three clusters (A, C, E), ST268 divided into four clusters (C, D, E, F), ST340 divided into four clusters (A, C, F, G), ST392 divided into three clusters (A, B, G), ST16 was subdivided into one cluster, and ST147 showed as one singleton ( and ).
Figure 1 Corresponding dendrogram generated with UPGMA clustering method in clinical blaNDM-positive isolates of Klebsiella pneumoniae.
Note: ERIC-PCR represented seven clusters (A–G) plus four singletons among NDM-positive isolates.
Abbreviations: ERIC-PCR, enterobacterial repetitive intergenic consensus amplification; NDM, New Delhi metallo-beta-lactamase.
Discussion
During the past decades, carbapenem resistance among K. pneumoniae is typically caused by the emergence of transmissible carbapenemases, such as blaKPC and blaNDM.Citation5 NDM specially comprises of the most rapidly growing group of metallo-beta-lactamases.Citation5 They have been increasingly detected in different countries, suggesting a worldwide dissemination.Citation1 Here, we reported the distribution of nosocomial infections caused by NDM-producing K. pneumoniae, especially in NICU from four referral hospitals in Kerman, Iran.
According to the previous studies, our findings showed that the most effective antibiotics against isolates were colistin and tigecycline.Citation31,Citation32 However, most isolates exhibited a high resistance level to other antimicrobial agents including extended spectrum cephalosporins, carbapenems, quinolones, and aminoglycosides. Similar to our findings, in the study in Egypt, all carbapenem-resistant K. pneumoniae isolates were sensitive to colistin and tigecycline.Citation32
In this study, eleven (17.46%) isolates indicated positive results for AmpC disk test. In our study, non-AmpC-producing isolates might be associated with other resistance mechanisms. Shi et al reported that cefoxitin resistance could be related to the change of cellular permeability to antibiotics, resulting from the loss or deficiency of outer membrane proteins.Citation33,Citation34
In our hospital settings, we found the emergence and establishment of NDM-producing K. pneumoniae along with rmtC and armA. Sporadic dissemination of NDM-1 in Iran was first described in 2013, which was resistant to the majority of antibiotics except for colistin.Citation1 In the current findings, we detected four clinical isolates being resistant to colistin, although one of them only harbored blaNDM-1 gene. This study also focused on epidemiological investigation of MLST and ERIC-PCR in the NDM-positive K. pneumoniae. To the best of our knowledge, the obvious report of the ST prevalence has not been yet accounted for NDM-producing K. pneumoniae in Iran. However, we reported the prevalence of different STs of NDM-producing K. pneumoniae among hospitalized patients during 3 years from March 2015 to November 2017 in Kerman.
According to this study, NDM-producing STs including ST16, ST147, and ST340 were found in India and Korea.Citation35,Citation36 On the contrary, ST147 was recently observed in NDM-positive K. pneumoniae in Iraq.Citation37 Our data showed a dissemination of a novel ST namely 268, which has not been reported in NDM-1-producing K. pneumoniae, during February 2016 to November 2017. The ST268 has been established as a major threat to NICUs from two referral hospitals after detecting the following STs including ST43, ST340, ST392, and ST14.
In general, the epidemiological trend of NDM-producing isolates in our hospital might be divided into three stages. From March 2015 to December 2015, the following STs ST43, ST340, ST392, and ST147 were found. From the beginning of February 2016 up till November 2017, the ST268 has been mainly investigated. Based on these findings, we supposed the ST268 was a successful clone to have recently been established in our hospital settings in 2017. This ST has been found in hypermucoviscous K. pneumonia, which was associated with invasive liver abscess syndrome in eastern Asia.Citation38 On the contrary, ST268 was recognized in capsule serotype K20 K. pneumoniae isolates, relating to primary meningitis in Taiwan.Citation39 Furthermore, these isolates were significantly associated with the virulence factors such as rmpA, rmpA2, and aerobactin.Citation39 Importantly, nine of ten rmtC-positive isolates in our current study belonged to K. pneumoniae, representing ST268, which often exhibited the most predominant ST of carbapenem-resistant K. pneumoniae isolates in NICU. Coproduction of 16S rRNA methylase resistance genes (rmtC, armA) among carbapenem-resistant K. pneumoniae with ST14 and ST340 was reported by Poirel et al.Citation40
In our findings, we observed another major ST, namely ST43, identified during 2015. This ST was able to carry virulence factors causing bacteremia.Citation41 In this study, most isolates belonging to ST43 have been detected from blood samples (). Recently, ST147 has been associated with blaCMY-4 gene and blaOXA-48 described in Tunisia.Citation42 In our study, ST147 was associated with blaNDM, blaCTX-M, and blaSHV, although this ST has been recognized as a serious threat to public health worldwide.Citation36 ST16 was the other type represented by one isolate from Afzalipoor Hospital. In this study, ST16 has been associated with blaNDM, blaCTX-M, and blaSHV. This ST has been recently reported in Italy, coproducing NDM-1 and OXA-232, recovering from blood and urine samples of a hospitalized patient with urosepsis.Citation43 However, Lester et alCitation34 and Hammerum et alCitation44 in New Zealand and Denmark showed that K. pneumoniae ST16 was recognized at several occasions, disseminating ESBLs and NDM-5 carbapenemase genes.
Our findings showed that ST340 has different ERIC-PCR patterns, that is a single-locus variant of ST11, which was found in Sweden and the UK.Citation45,Citation46 Additionally, NDM-producing isolates from ST340 detected in March 2015 were compared with isolates collected with the same ST in November 2017 to check for ERIC-PCR profile variations within NDM-positive K. pneumoniae. As shown in , ERIC-PCR pattern from ST340 displayed identical ERIC-PCR profiles among NDM-producing isolates with the other STs (43, 268, 392, and 16) in different clusters. Similar to this study, Richter et al in Italy showed that no ERIC-PCR profile variation was found between carbapenemase-producing K. pneumoniae strains from STs 258 and 37.Citation47
Lascols et al in India showed a diverse range of clones harboring carbapenemase-producing K. pneumoniae strains representing STs 147 and 340.Citation36,Citation40,Citation48,Citation49 The prevalence of blaNDM-1 is frequently associated with promiscuous plasmids related to a broad host range of clinical variants harboring blaNDM-1, hence our findings hypothesized that nosocomial acquisition of blaNDM by both outward sources including patients who have traveled to Iran specially neighboring countries and also have acquired a broad spectrum of different resistance genes.Citation48 We detected K. pneumoniae ST392, which has been previously associated with the dissemination of blaNDM-1, blaKPC, and blaOXA-48 genes.Citation50,Citation51 However, in this study, ST392 was detected among blaNDM- and blaOXA-positive isolates, recovering from one hospital with different ERIC-PCR patterns. In this study, some STs were observed to have different ERIC-PCR pattern types; therefore, our molecular typing results revealed that ERIC-PCR and MLST provided measures of genetic diversity, while they were not similar methods. These findings showed that ERIC-PCR displayed pattern discriminations for same STs; however, ERIC-PCR has provided a potential molecular typing method to distinguish greater ranges of genetic changes among NDM-positive K. pneumoniae in our hospital settings.Citation52 In this study, up to November 2017, 37 patients with at least one NDM-positive K. pneumoniae isolates were identified. Most isolates were recovered at Afzalipoor Hospital from NICU (). Interestingly, the most NDM-positive K. pneumoniae isolates belonged to the two STs; 268 and 340 displayed various ERIC-PCR patterns within different clusters. On the contrary, in different hospitals, K. pneumoniae isolates with similar STs revealed diverse ERIC-PCR patterns during 2015–2017. It was suggested that these isolates might be affected by coacquisition of some antibiotic resistance plasmids; therefore, it might affect the results of the ERIC-PCR patterns. Moreover, the molecular typing techniques such as ERIC-PCR and pulsed-field gel electrophoresis were used to identify alterations in short-term, while no obvious difference was observed in STs during 3 years, since MSLT is considered to evaluate the alternations in the most conserved genes, showing long-term variations.Citation52
Conclusion
In this study, the molecular characterization and epidemiological investigations revealed the dissemination of different clones of NDM-producing K. pneumoniae in our hospital settings. Due to the highly resistant nature of bacterial strains carrying the blaNDM, there are very limited antibiotics to combat these bacteria. In this study, MLST differentiated the 37 representative NDM-positive K. pneumoniae strains into 7 STs. The STs included ST268 (n=14), ST43 (n=9), ST340 (n=7), ST392 (n=5), and single isolates representing STs 147 and 16. To the best of our knowledge, the clinical isolates of K. pneumoniae representing ST268 have not been reported among NDM-producing K. pneumoniae. Distribution of blaNDM is obviously related to the promiscuity of many plasmids resulting in a wide range of Gram-negative bacteria containing diverse blaNDM harboring plasmids. However, our study suggests that dominant clones (STs 43 and 268) have had a potential role to monitor and continue a long-term survival of blaNDM-1 dis semination. In addition, our data highlight that the potential for local and neighboring countries such as Pakistan, India, and Iraq has been reported as the endemic dissemination of blaNDM-producing K. pneumoniae clones and plasmid-mediated resistance. Therefore, dissemination of different clones with blaNDM among carbapenem-resistant K. pneumoniae might have resulted in more frequent opportunities for the emergence of blaNDM-positive among other Gram-negative bacteria and it highlighted the need to ongoing epidemiological surveillance and comprehensive infection control guidelines. We also suggested the intrinsic genetic factors, causing a spread and establishment of ST268, as a new NDM-producing K. pneumoniae clone identified to increase our knowledge about it.
Ethical statement
The K. pneumoniae strains were originally taken as part of routine hospital procedure, and then specifically recovered for this work. This study was approved by ethical numbers: IR:KMU.REC.1395.436 and IR.KMU.REC.1395.806 in ethical committee of Kerman University of Medical Sciences.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Acknowledgments
We thank Student Research Committee Kerman University of Medical Sciences for supporting in financial aid for MLST. This research was supported by the research council and student research committee of Kerman University of Medical Sciences, Kerman, Iran, grant numbers: 95000056 and 95000522.
Disclosure
The authors report no conflicts of interest in this work.
References
- ShahcheraghiFNobariSRahmati GhezelgehFFirst report of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in IranMicrob Drug Resist2013191303622984942
- YanJPuSJiaXMultidrug resistance mechanisms of carbapenem resistant Klebsiella pneumoniae strains isolated in chongqing, ChinaAnn Lab Med201737539840728643488
- NordmannPNaasTPoirelLGlobal spread of carbapenemase-producing enterobacteriaceaeEmerg Infect Dis201117101791179822000347
- DortetLPoirelLNordmannPWorldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteriaBiomed Res Int2014201424985624790993
- YongDTolemanMAGiskeCGCharacterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneu-moniae sequence type 14 from IndiaAntimicrob Agents Chemother200953125046505419770275
- ZhengRZhangQGuoYOutbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan, ChinaAnn Clin Microbiol Antimicrob2016151026896089
- NordmannPCarbapenemase-producing Enterobacteriaceae: overview of a major public health challengeMed Mal Infect2014442515624360201
- KocsisEGužvinecMButićIblaNDM-1 Carriage on IncR plasmid in enterobacteriaceae strainsMicrob Drug Resist201622212312826484384
- BrisseSVerhoefJPhylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotypingInt J Syst Evol Microbiol200151Pt 391592411411715
- DiancourtLPassetVVerhoefJGrimontPABrisseSMultilocus sequence typing of Klebsiella pneumoniae nosocomial isolates multilocus sequence typing of Klebsiella pneumoniae nosocomial isolatesJ Clin Microbiol2005434178418216081970
- MahonCRLehmanDCManuselisGText Book of Diagnostic Microbiology3rd edSaundersPhiladelphia, PA2007
- CLSIPerformance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M10027th edWayne, PAClinical and Laboratory Standards Institute2017
- BlackJAMolandESThomsonKSAmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamasesJ Clin Microbiol20054373110311316000421
- MonsteinHJÖstholm-BalkhedANilssonMVMultiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in EnterobacteriaceaeAPMIS2007115121400140818184411
- FeizabadiMMDelfaniSRajiNDistribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, IranMicrob Drug Resist2010161495319961397
- PoirelLBonninRANordmannPAnalysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencingAntimicrob Agents Chemother201155911AAC-4229
- KhorvashFYazdaniMRSoudiAAShabaniSTavahenNPrevalence of acquired carbapenemase genes in Klebsiella Pneumoniae by multiplex PCR in IsfahanAdv Biomed Res201764128503496
- PoirelLWalshTRCuvillierVNordmannPMultiplex PCR for detection of acquired carbapenemase genesDiagn Microbiol Infect Dis201170111912321398074
- MirsalehianAFeizabadiMNakhjavaniFADetection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patientsBurns2010361707419524369
- GiskeCGFrödingIHasanCMDiverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United KingdomAntimicrob Agents Chemother20125652735273822354295
- LabuschagneCJWeldhagenGFEhlersMMDoveMGEmergence of class 1 integron-associated GES-5 and GES-5-like extended-spectrum beta-lactamases in clinical isolates of Pseudomonas aeruginosa in South AfricaInt J Antimicrob Agents200831652753018436436
- Rodríguez-MartínezJMDíaz de AlbaPBrialesAContribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniaeJ Antimicrob Chemother2013681687323011289
- RojasLJWrightMSde La CadenaEInitial assessment of the molecular epidemiology of bla NDM-1 in ColombiaAntimicrob Agents Chemother20166074346435027067339
- FritscheTRCastanheiraMMillerGHJonesRNArmstrongESDetection of methyltransferases conferring high-level resistance to ami-noglycosides in enterobacteriaceae from Europe, North America, and Latin AmericaAntimicrob Agents Chemother20085251843184518347105
- GuoYZhouHQinLFrequency, antimicrobial resistance and genetic diversity of Klebsiella pneumoniae in food samplesPLoS One2016114e015356127078494
- HidalgoLHopkinsKLGutierrezBAssociation of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UKJ Antimicrob Che-mother201368715431550
- KimJYParkYJKwonHJHanKKangMWWooGJOccurrence and mechanisms of amikacin resistance and its association with beta-lactamases in Pseudomonas aeruginosa: a Korean nationwide studyJ Antimicrob Chemother200862347948318606785
- LiuYYWangYWalshTREmergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological studyLancet Infect Dis201616216116826603172
- Pérez-PérezFJHansonNDDetection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCRJ Clin Microbiol20024062153216212037080
- MeachamKJZhangLFoxmanBBauerRJMarrsCFEvaluation of genotyping large numbers of Escherichia coli isolates by entero-bacterial repetitive intergenic consensus-PCRJ Clin Microbiol200341115224522614605168
- SamonisGMarakiSKarageorgopoulosDEVouloumanouEKFalagasMESynergy of fosfomycin with carbapenems, colistin, netilmicin, and tigecycline against multidrug-resistant Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa clinical isolatesEur J Clin Microbiol Infect Dis201231569570121805292
- GamalDFernández-MartínezMSalemDCarbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtFInt J Infect Dis201643172026686939
- ShiWLiKJiYCarbapenem and cefoxitin resistance of Kleb-siella pneumoniae strains associated with porin OmpK36 loss and DHA-1 β-lactamase productionBraz J Microbiol201344243544224294234
- LesterCHOlsenSSJakobsenLEmergence of extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae in Danish hospitals; this is in part explained by spread of two CTX-M-15 clones with multilocus sequence types 15 and 16 in ZealandInt J Antimicrob Agents201138218018221612893
- KimSYRheeJYShinSYKoKSCharacteristics of community-onset NDM-1-producing Klebsiella pneumoniae isolatesJ Med Microbiol201463Pt 1868924173426
- LeeC-RLeeJHParkKSKimYBJeongBCLeeSHGlobal dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methodsFront Microbiol20167DM0113026834723
- Gharout-SaitAAlsharapySABrasmeLEnterobacteriaceae isolates carrying the New Delhi metallo-β-lactamase gene in YemenJ Med Microbiol201463Pt 101316132325009193
- HiraiYAsahata-TagoSAinodaYFirst case of liver abscess with endogenous endophthalmitis due to non-K1/K2 serotype hypermuco-viscous Klebsiella pneumoniae clone ST268J Gastrointest Dig Syst201661383
- KuYHChuangYCChenCCKlebsiella pneumoniae isolates from meningitis: epidemiology, virulence and antibiotic resistanceSci Rep201771663428747788
- PoirelLDortetLBernabeuSNordmannPGenetic features of blaNDM-1-positive EnterobacteriaceaeAntimicrob Agents Chemother201155115403540721859933
- HoltKEWertheimHZadoksRNGenomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsi-ella pneumoniae, an urgent threat to public healthProc Natl Acad Sci U S A201511227E3574E358126100894
- GramiRMansourWBen Haj KhalifaAEmergence of ST147 Klebsiella pneumoniae-producing OXA-204 carbapenemase in a University Hospital, TunisiaMicrob Drug Resist201622213714026447939
- AvolioMVignaroliCCrapisMCamporeseACo-production of NDM-1 and OXA-232 by ST16 Klebsiella pneumoniae, Italy, 2016Future Microbiol2017121119112228876082
- HammerumAMHansenFOlesenBInvestigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencingJ Glob Antimicrob Resist20153321922127873714
- WoodfordNTurtonJFLivermoreDMMultiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistanceFEMS Microbiol Rev201135573675521303394
- GiskeCGFrödingIHasanCMDiverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United KingdomAntimicrob Agents Chemother20125652735273822354295
- RichterSNFrassonIFranchinEKPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care unitsGut Pathog201241722800501
- SartorALRazaMWAbbasiSAMolecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from PakistanAntimicrob Agents Chemother20145895589559324982081
- LascolsCHackelMMarshallSHIncreasing prevalence and dissemination of NDM-1 metallo-β-lactamase in India: data from the SMART study (2009)J Antimicrob Chemother20116691992199721676902
- RojasLJWrightMSde La CadenaEInitial assessment of the molecular epidemiology of blaNDM-1 in ColombiaAntimicrob Agents Chemother20166074346435027067339
- Bocanegra-IbariasPGarza-GonzálezEMorfín-OteroRMolecular and microbiological report of a hospital outbreak of NDM-1-carrying Enterobacteriaceae in MexicoPLoS One2017126e017965128636666
- KiddTJGrimwoodKRamsayKARaineyPBBellSCComparison of three molecular techniques for typing Pseudomonas aeruginosa isolates in sputum samples from patients with cystic fibrosisJ Clin Microbiol201149126326821084517
