Abstract
Background
IncR, IncFII, IncpA1763-KPC, and IncN1 plasmids have been increasingly found among Enterobacteriaceae species, but plasmids with hybrid structures derived from the above-mentioned incompatibility groups have not yet been described.
Methods
Plasmids p721005-KPC, p504051-KPC, and pA3295-KPC were fully sequenced and compared with previously sequenced related plasmids pHN84KPC (IncR), pKPHS2 (IncFIIK), pKOX_NDM1 (IncFIIY), pHN7A8 (IncFIIpHN7A8), and R46 (IncN1).
Results
The backbone of p721005-KPC/p504051-KPC was a hybrid of the entire 10-kb IncR-type backbone from pHN84KPC, the entire 64.3-kb IncFIIK-type maintenance, and conjugal transfer regions from pKPHS2, a 15.5-kb IncFIIY-type maintenance region from pKOX_NDM1 and a 5.6-kb IncpA1763-KPC-type backbone region from pA1763-KPC, and it contained a primary IncR replicon and two auxiliary IncpA1763-KPC and IncN1 replicons. The backbone of pA3295-KPC was a hybrid of a 7.2-kb IncFIIpHN7A8-type backbone region from pHN7A8, the almost entire 33.3-kb IncN1-type maintenance and conjugal transfer regions highly similar to R46, a 26.2-kb IncFIIK-type maintenance regions from pKPHS2, the above 15.5-kb IncFIIY-type maintenance region, and the above 5.6-kb IncpA1763-KPC-type backbone region, and it contained a primary Inc-FIIpHN7A8 replicon and two auxiliary IncpA1763-KPC and IncN1 replicons. Each of p721005-KPC, p504051-KPC, and pA3295-KPC acquired a wealth of accessory modules, carrying a range of intact and residue mobile elements (such as insertion sequences, unit transposons, and integrons) and resistance markers (such as blaKPC, tetA, dfrA, and qnr).
Conclusion
In each of p721005-KPC, p504051-KPC, and pA3295-KPC, multiple replicons in coordination with maintenance and conjugation regions of various origins would maintain a broad host range and a stable replication at a steady-state plasmid copy number.
Introduction
An IncR replicon alone is able to promote plasmid replication but often coexists with additional replicons such as IncC, IncFII, and IncH.Citation1 pEFER (GenBank accession number CU928144) is the first sequenced IncR single-replicon plasmid, but pHN84KPC (GenBank accession number KY296104) is more appropriate as the reference of IncR single-replicon plasmids because it contains the most complete IncR backbone, which is composed of repB (replication initiation) and parAB, umuCD, vagCD, resD, and retA (maintenance).Citation1 IncR single-replicon plasmids lack conjugal transfer genes, making it not self-transmissible.Citation2
IncFII plasmids are usually low copy number plasmids with a narrow host range and circulated mainly among Enterobacteriaceae species.Citation3 Due to significant variations at nucleotide and amino acid levels of backbone sequences, Inc-FII plasmids can be divided into multiple subgroups, namely IncFIIY, IncFIIK, IncFIIpHN7A8, and IncFIIp0716-KPC, represented by pKOX_NDM1 (GenBank accession number JQ314407),Citation4 pKPHS2 (GenBank accession number CP003224),Citation5 pHN7A8 (GenBank accession number JN232517), and p0716-KPC (GenBank accession number KY270849),Citation6,Citation7 respectively. A single replicon IncFIIpHN7A8, three replicons IncFIIK, IncFIB, and IncR, and a single replicon IncFIIY can be found in pHN7A8, pKPHS2, and pKOX_NDM1, respectively. p0716-KPC has a complex chimera backbone, which is composed of a primary replicon IncFIIp0716-KPC, the entire 64.3-kb IncFIIK-type maintenance, and conjugal transfer regions found in pKPHS2, a 15.5-kb IncFIIY-type maintenance region found in pKOX_NDM1, a 5.6-kb backbone region from pA1763-KPC (GenBank accession number MH909340), and an unknown 3.2-kb conjugal transfer region. The pA1763-KPC backbone can be divided into two parts: 1) the above 5.6-kb IncpA1763-KPC-type backbone region composed of a novel replicon IncpA1763-KPC and several maintenance genes including parA, ccdBA and resA; and 2) the 64.3-kb IncFIIK-type maintenance and conjugal transfer regions found in pKPHS2.
IncN plasmids can be further divided into three subgroups IncN1, IncN2, and IncN3, in which backbones have conserved gene organization but with limited nucleotide sequence homology.Citation8 The backbone of IncN1 reference plasmid R46 (GenBank accession number AY046276) includes regions of replication (repAIncN1), maintenance (mucAB, ardBR, ccgAE, and stbABC), and conjugal transfer (nuc, tivB, eex, dtr23, and rlx).Citation9
This study dealt with sequencing and genomic dissection of two IncR:IncpA1763-KPC:IncN1 multi-replicon plasmids p721005-KPC and p504051-KPC as well as an IncFIIpHN7A8:IncpA1763-KPC:IncN1 plasmid pA3295-KPC. The backbone of each plasmid displayed a very complex chimera structure, with integration of several accessory modules composed of mobile elements and associated resistance markers especially including blaKPC-2.
Materials and methods
Bacterial strains
Klebsiella pneumoniae 721005 was isolated in 2013 from urine specimens of a 60-year-old male with paraplegia in a public hospital in Ningbo city of China. K. pneumoniae 504051 was isolated in 2013 from a blood specimen of a 68-year-old male with pancreatic neoplasm in the hospital mentioned previously. K. pneumoniae A3295 was recovered in 2016 from a sputum specimen of a pneumonia patient in a public hospital in Beijing city.
Conjugal transfer
Conjugal transfer experiments were carried out with rifampin-resistant Escherichia coli EC600 used as a recipient and each of the blaKPC-positive 721005, 504051, and A3295 isolates as a donor. Three milliliters of overnight cultures of each of donor and recipient bacteria were mixed together, harvested, and resuspended in 80 µL of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 µm pore size (EMD Millipore) which was placed on BHI agar (BD Biosciences) plate and then incubated for mating at 37°C for 12–18 hours. Bacteria were washed from filter membrane and spotted on Muller-Hinton (MH) agar (BD Biosciences) plates containing 1,000 µg/mL rifampin together with 2 µg/mL meropenem, for selecting a blaKPC-carrying E. coli transconjugant.
Electroporation
Electroporation experiments were carried out for the 721005 and 504051 isolates. To prepare competent cells for electroporation, 200 mL of overnight culture of E. coli TOP10 in Super Optimal Broth (SOB) at an optical density (OD600) of 0.4–0.6 was washed three times with electroporation buffer (0.5 M mannitol and 10% glycerol) and concentrated into a final volume of 2 mL. One microgram of genomic DNA from the 721005 or 504051 isolate was mixed with 100 µL of competent cells for electroporation at 25 µF, 200 Ω, and 2.5 Kv. The resulting cells were suspended in 500 µL of SOB and an appropriate aliquot was spotted on SOB agar plates containing 1 µg/mL meropenem, for selecting a blaKPC-carrying E. coli electroporant.
Sequencing and sequence assembly
Genomic DNA was isolated from each of the 721005, 504051, and A3295 isolates using a Blood & Cell Culture DNA Maxi Kit (Qiagen, Hilden, Germany). Genomic DNA of strain 504051 or A3295 was sequenced from a mate-pair library with average insert size of 5 kb (ranged from 2 kb to 10 kb) using a MiSeq sequencer (Illumina, San Diego, CA, USA). Quality control, removing adapters and low quality reads, were performed using Trimmomatic 0.36.Citation10 The filtered clean reads were then assembled using Newbler 2.6,Citation11 followed by extraction of the consensus sequence with CLC Genomics Workbench 3.0 (Qiagen Bioinformatics). Gapfiller V1.11 was used for gap closure.Citation12
For the 721005 isolate, genome sequencing was performed with a sheared DNA library with average size of 15 kb (ranged from 10 kb to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, Menlo Park, CA, USA), as well as a paired-end library with an average insert size of 400 bp (ranged from 150 kb to 600 kb) on a HiSeq sequencer (Illumina). The paired-end short Illumina reads were used to correct long PacBio reads utilizing proovread,Citation13 and then the corrected PacBio reads were assembled denovo utilizing SMARTdenovo (available from: https://github.com/ruanjue/ smartdenovo).
Sequence annotation and comparison
Open reading frames and pseudogenes were predicted using RAST 2.0 with default parameters, combined with BLASTP/ BLASTN searches against the UniProtKB/Swiss-Prot database and the RefSeq database.Citation14–Citation17 Annotation of resistance genes, mobile elements, and other features was carried out using the online databases including CARD,Citation18 ResFinder,Citation19 ISfinder,Citation20 INTEGRALL,Citation21 and the Tn Number Registry.Citation22 Gene organization diagrams were drawn in Inkscape 0.48.1 (https://inkscape.org/en/).
Phenotypic assays
Activity of Ambler class A/B/D carbapenemases in bacterial cell extracts was determined by a modified CarbaNP test.Citation23 Bacterial antimicrobial susceptibility was tested by BioMérieux Vitek 2 and interpreted according to the Clinical and Laboratory Standards Institute guidelines.
Nucleotide sequence accession numbers
The complete sequences of plasmids p721005-KPC, p504051-KPC, and pA3295-KPC were submitted to Gen-Bank under accession numbers MG764550, MH477636, and MG764553, respectively.
Ethics statement
Ethics approval and informed consent were not required. All the bacterial isolates involved in this study were part of the routine hospital laboratory procedure.
Results and discussion
Overview of sequenced plasmids
High-throughput sequencing with genomic DNA of the 721005, 504051, and A3295 isolates generated the circular sequences of plasmids p721005-KPC, p504051-KPC, and pA3295-KPC, which were 64,198, 163, 588, and 153,274 bp in length, with average G+C contents of 53.92%, 53.82%, and 54.52%, and contained 74, 59, and 60 predicted open reading frames in total, respectively ( and Figure S1). Each plasmid was composed of the backbone regions, and the accessory modules that were recognized as acquired DNA regions were associated with adjacent mobile elements and inserted at different sites of the backbone (Figure S1). pA3295-KPC, but not p721005-KPC and p504051-KPC, could be transferred from the A3295 isolate into E. coli EC600 through conjugation, generating the transconjugant A3295-KPC-EC600. p721005-KPC/p504051-KPC could be transferred from the 72005 or 504051 isolate into E. coli TOP10 through electroporation, yielding the electroporant 721005-KPC-TOP10 or 504051-KPC-TOP10, respectively. All these strains had class A carbapenemase activity (data not shown), and were resistant to all the cephalosporin and carbapenem drugs tested (), which resulted from presence of blaKPC-2 in all these plasmids.
Table 1 Major features of plasmids analyzed
Table 2 Antimicrobial drug susceptibility profiles
Multi-replicon chimera structure
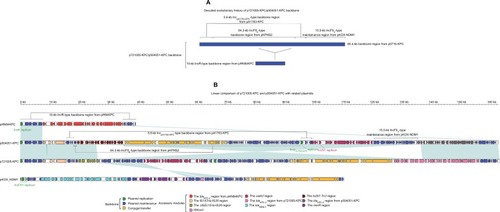
Each of p721005-KPC, p504051-KPC, and pA3295-KPC displayed a complex chimera structure. The sequences of p721005-KPC and p504051-KPC were almost identical (99% BLAST coverage and 99% nucleotide identity) to each other. The p721005-KPC/p504051-KPC backbone () was a hybrid of the entire 10-kb IncR-type back-bone found in pHN84KPC, and an 85.4-kb p0716-KPC-derived backbone region that was composed of the almost whole p0716-KPC backbone except for the IncFIIp0716-KPC replicon and the 3.2-kb conjugal transfer region. p721005-KPC/p504051-KPC acquired four separate accessory modules, namely the cryptic IS1X3–to–IS26 region, the cryptic ΔISEc15–to–IS26 region in p721005-KPC or the In207–ΔTn2 region in p504051-KPC, the catA2 region, and the blaKPC-2 region (). A 3.6-kb IncN1-type backbone region, including the IncN1 replicon, was found in the catA2 region. Taken together, p721005-KPC/ p504051-KPC contained a primary IncR replicon, as well as two auxiliary replicons IncpA1763-KPC and IncN1.
Figure 1 Modular structures of p7210005-KPC and p504051-KPC.
Notes: Shown are deduced evolutionary history of plasmid backbone (A) and linear comparison of sequenced plasmids (B). Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
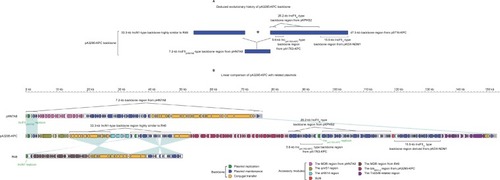
The backbone () of pA3295-KPC was a hybrid of a 7.2-kb IncFIIpHN7A8-type backbone region found in pHN7A8, a 33.3-kb IncN1-type backbone region highly similar to R46, and a 47.3-kb backbone region found in p0716-KPC.Citation7 The 7.2-kb region included an IncFIIpHN7A8 replicon, a partial maintenance locus pemIK, and a partial conjugal transfer remnant region (rlx, tivF16, and finO). The 33.3-kb region contained almost the entire IncN1-type maintenance and conjugal transfer regions. The 47.3-kb region could be further divided into a 26.2-kb IncFIIK-type maintenance region from pKPHS2, the 15.5-kb IncFIIY-type maintenance region from pKOX_NDM1, and the 5.6-kb backbone region from pA1763-KPC. These two 26.2-kb and 15.5-kb regions were also found in p721005-KPC/ p504051-KPC. A total of four accessory modules, namely the qnrS1 region, the dfrA14 region, the Tn6346-related region, and the blaKPC-2 region, were identified in pA3295-KPC. Remarkably, pA3295-KPC carried a primary Inc-FIIpHN7A8 replicon, together with two auxiliary replicons IncpA1763-KPC and IncN1.
Figure 2 Modular structure of pA3295-KPC.
Notes: Shown are deduced evolutionary history of plasmid backbone (A) and linear comparison of sequenced plasmids (B). Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
Abbreviation: MDR, multidrug resistance.
Accessory resistance regions
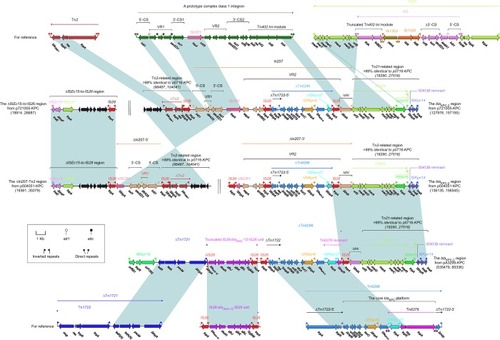
All of p721005-KPC, p504051-KPC, and pA3295-KPC contained the blaKPC-2 regions (). The blaKPC-2 region from p721005-KPC could be divided into a blaTEM-1-containing Tn2-related region and a mer-harboring Tn21-related region as observed in p0716-KPC,Citation7 and a complex class 1 inte-gron In207. In207 harbored two resistance regions, namely variable region 1 (VR1, containing a single gene cassette dfrA25) and VR2 (harboring qnrB52 and a blaKPC-2-carrying ΔTn6296).Citation7 The blaKPC-2 region from p504051-KPC differed from p721005-KPC but by two major modular changes: 1) insertion of IS26 into ISCR1 of In207, truncating In207 into VR1-carrying In207-5′ and VR2-carrying In207-3′; and 2) translocation of In207-5′ from the blaKPC-2 region to connect with the separate ΔISEc15-to-IS26 region (also found as a cryptic accessory module in p721005-KPC), constituting the ΔIn207–Tn2 region. The blaKPC-2 region of pA3295-KPC was organized sequentially as ISKPn19, tetA(A)-containing ΔTn1721,Citation24 a truncated IS26–blaSHV-12 –IS26 unit,Citation25 a blaKPC-2-carrying ΔTn6296 slightly differing from that from p721005-KPC/p504051-KPC, IS26, a tni remnant, and the Tn21-related region.
Figure 3 The blaKPC-2 regions and related regions from p7210005-KPC, p504051-KPC, and pA3295-KPC.
Notes: Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate nucleotide positions within corresponding plasmids.
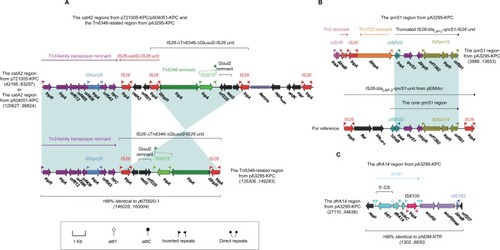
The Tn6346-related region (, also found in p675920-1) of pA3295-KPC was composed of a Tn3-family transposon remnant and the IS26–ΔTn6346–ΔGIsul2–IS26 unit.Citation26 The catA2 region () of p721005-KPC/ p504051-KPC differed from this Tn6346-related region by three major modular changes: 1) insertion of the IS26–catA2–IS26 unit; 2) inversion of the cryptic IS26– ΔTn6346–ΔGIsul2–IS26 unit; and 3) integration of an IncN1-type backbone region containing repAIncN1 and its iterons, ardK and mpr. The qnrs1 region (, also found in p675920-2) from pA3295-KPC was composed of a truncated IS26–blalap-2–qnrS1–IS26 unit and several unit transposon remnants and intact or residue ISs.Citation26 The dfrA14 region (, as observed in pNDM-BTR) from pA3295-KPC harbored In191 containing a single gene cassette dfrA14, ecoRII–ecoRIImet (antirestriction system), and ΔIS1X2.Citation27
Figure 4 Other accessory resistance regions from p7210005-KPC, p504051-KPC, and pA3295-KPC.
Notes: Shown are the catA2 regions from p721005-KPC/p504051-KPC and the Tn6346-related region from pA3295-KPC (A), the qnrS1 region from pA3295-KPC (B), and the dfrA14 region from pA3295-KPC (C). Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate nucleotide positions within corresponding plasmids.
Conclusion
Multi-replicon plasmids are increasingly recognized, and existence of multiple replicons is one means by which plasmids with a narrow host range can be restructured to achieve broad host range.Citation3,Citation28 In this study, a detailed comparative genomics analysis was subjected to three blaKPC-2-carrying chimera plasmids p721005-KPC, p504051-KPC, and pA3295-KPC, disclosing that their sequences were derived from different plasmids belonging to various incompatible groups. p721005-KPC/p504051-KPC might evolve from recombination of IncR plasmid pHN84KPC and IncFII plasmid p0716-KPC, displaying a IncR:IncpA1763-KPC:IncN1 multi-replicon structure. pA3295-KPC was a hybrid of IncFIIpHN7A8 plasmid pHN7A8, IncN1 plasmid R46, and IncFIIp0716-KPC plasmid p0716-KPC, carrying three replicons IncFIIpHN7A8, IncpA1763-KPC, and IncN1. IncFIIpHN7A8 replicon manifests as an antisense RNA-regulated replicon, for which an unstable antisense RNA prevents Rep translation at high concentrations by RNA interference, while IncR, IncN1, and IncpA1763-KPC replicons belong to iteron-regulated replicons, for which iterons (directly repeated sequences) are specifically bound by Rep monomers.Citation28 In each of these plasmids, replicons in coordination with maintenance and conjugation regions would maintain their stable replication at a steady-state plasmid copy number. Each of these plasmids integrated a wealth of accessory modules (), which were inserted at different sites across the backbone and carried a range of mobile elements (such as IS elements, unit transposons, and integrons) and associated resistance markers (such as blaKPC, tet, dfrA, and qnr; ), making host K. pneumoniae strains to be multidrug resistant.
Table 3 Drug resistance genes in sequenced plasmids
Author contributions
DZ and JH conceived the study and designed experimental procedures. DQ, YS, LH, XJ, and ZY performed the experiments. All the authors contributed to reagents and materials and data mining. DZ, DQ, YS, and JH wrote this manuscript. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFC1200100) and the National Science and Technology Major Project of China (2018ZX10733402).
Supplementary material
Figure S1 Schematic maps of sequenced plasmids.
Notes: Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and grey, respectively. The innermost circle presents GC-skew [(G–C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.
![Figure S1 Schematic maps of sequenced plasmids.Notes: Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and grey, respectively. The innermost circle presents GC-skew [(G–C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.](/cms/asset/91d6cc5d-3b99-4ec2-9126-fe987bdd0b43/didr_a_189168_sf0001_c.jpg)
Disclosure
The authors report no conflicts of interest in this work.
References
- CompainFFrangeulLDrieuxLComplete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniaeAntimicrob Agents Chemother20145874207421024752259
- AlvaradoAGarcillán-BarciaMPde La CruzFA degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settingsPLoS One201277e4043822792321
- VillaLGarcía-FernándezAFortiniDCarattoliAReplicon sequence typing of IncF plasmids carrying virulence and resistance determinantsJ Antimicrob Chemother201065122518252920935300
- HuangTWWangJTLauderdaleTLComplete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from TaiwanAntimicrob Agents Chemother20135784072407623752513
- LiuPLiPJiangXComplete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputumJ Bacteriol201219471841184222408243
- HeLPartridgeSRYangXComplete nucleotide sequence of pHN7A8, an F33:A-:B-type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from ChinaJ Antimicrob Chemother2013681465022984205
- FengJYinZZhaoQGenomic characterization of novel IncFII-type multidrug resistant plasmids p0716-KPC and p12181-KPC from Klebsiella pneumoniaeSci Rep201771583028725038
- JiangXYinZYinXSequencing of blaIMP-carrying IncN2 plasmids, and comparative genomics of IncN2 plasmids harboring class 1 integronsFront Cell Infect Microbiol2017710228424761
- DelverEPBelogurovAAOrganization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elementsJ Mol Biol1997271113309300052
- BolgerAMLohseMUsadelBTrimmomatic: a flexible trimmer for Illumina sequence dataBioinformatics201430152114212024695404
- NederbragtAJOn the middle ground between open source and commercial software – the case of the Newbler programGenome Biol2014154113225180324
- BoetzerMPirovanoWToward almost closed genomes with GapFillerGenome Biology2012136R5622731987
- EdgarRCMUSCLE: multiple sequence alignment with high accuracy and high throughputNucleic Acids Res20043251792179715034147
- BrettinTDavisJJDiszTRASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomesSci Rep201551836525666585
- BoratynGMCamachoCCooperPSBLAST: a more efficient report with usability improvementsNucleic Acids Res201341Web Server issueW29W3323609542
- BoutetELieberherrDTognolliMUniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry viewMethods Mol Biol20161374235426519399
- O’LearyNAWrightMWBristerJRReference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotationNucleic Acids Res201644D1D733D74526553804
- JiaBRaphenyaARAlcockBCARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance databaseNucleic Acids Res201745D1D566D57327789705
- ZankariEHasmanHCosentinoSIdentification of acquired antimicrobial resistance genesJ Antimicrob Chemother201267112640264422782487
- SiguierPPerochonJLestradeLMahillonJChandlerMISfinder: the reference centre for bacterial insertion sequencesNucleic Acids Research20063490001D32D3616381877
- MouraASoaresMPereiraCLeitaoNHenriquesICorreiaAINTEGRALL: a database and search engine for integrons, integrases and gene cassettesBioinformatics20092581096109819228805
- RobertsAPChandlerMCourvalinPRevised nomenclature for transposable genetic elementsPlasmid200860316717318778731
- FengWZhouDWangQDissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospitalSci Rep2016613341927641711
- PartridgeSRAnalysis of antibiotic resistance regions in Gram-negative bacteriaFEMS Microbiol Rev201135582085521564142
- FordPJAvisonMBEvolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosomeJ Antimicrob Chemother2004541697515163647
- FengJYinZZhanZStructure genomics of two chimera plasmids p675920-1 and p675920-2 coexisting in a multi-drug resistant Klebsiella pneumoniae isolateOncotarget2018
- ZhaoYWangLZhangZStructural genomics of pNDM-BTR harboring In191 and Tn6360, and other bla NDM-carrying IncN1 plasmidsFuture Microbiol201712141271128129027482
- PillaGTangCMGoing around in circles: virulence plasmids in enteric pathogensNat Rev Microbiol201816848449529855597
