Abstract
Purpose
New Delhi metallo-β-lactamase 5 (NDM-5) shows stronger resistance to carbapenems and broad-spectrum cephalosporins than NDM-1 because NDM-5 differs from NDM-1 by two amino acid substitutions. In this study, our aim was to characterize a NDM-5-producing Escherichia coli isolate KY1497 from a patient with urinary tract infection in Japan, who had no recent history of overseas travel.
Patients and Methods
NDM-5-producing E. coli isolate KY1497 was detected in the urine sample of a patient hospitalized in a tertiary hospital in Japan. The complete genome sequence of isolate KY1497 was determined by short- and long-read sequencing with hybrid assembly, followed by multilocus sequence typing (MLST), core-genome phylogeny analysis, plasmid analysis, and transconjugation experiments.
Results
KY1497 was classified as ST405 by MLST, and core-genome phylogeny exhibited the closest lineage to the clinical isolates in Nepal (IOMTU605) and Canada (FDAARGOS_448). KY1497 harbors blaNDM-5 in the IncFII-IncFIB(pB171) replicon plasmid (pKY1497_1, 123,767 base pairs). Plasmid analysis suggested that the cognate plasmids of pKY1497_1 have a minor plasmid background, rather than the globally disseminated IncX3 plasmid carrying blaNDM-5. Transconjugation analysis revealed that pKY1497_1 is transmissible to the recipient E. coli J53 strain.
Conclusion
We characterized a novel Inc replicon plasmid (IncFII-IncFIB[pB171]) carrying blaNDM-5 and its host E. coli strain. NDMs are associated with a high risk of infection worldwide because of their antibiotic resistance and untreatable and hard-to-treat infections. Other patients in the hospital showed negative results for carbapenem-resistant Enterobacteriaceae. As NDM-producing strains are only sporadically detected in Japan, attention should be provided to the community prevalence of NDM-producing E. coli strains to prevent nosocomial infections.
Introduction
Bacterial resistance due to β-lactamase is increasingly associated with carbapenemases encoded by various plasmids. Among these newly emerging carbapenemases, New Delhi metallo-β-lactamase 1 (NDM-1) was first reported in 2009.Citation1 NDMs can hydrolyze all β-lactams, but not monobactams, and are associated with a high risk of causing a global health crisis.
NDM-5 is a variant that differs from other NDM enzymes because it contains two substitutions (Val88Leu and Met154Leu) and shows increased resistance to carbapenems and broad-spectrum cephalosporins.Citation2 In 2011, NDM-5 was first identified in the UK in a strain of Escherichia coli isolated from a patient with a recent history of hospitalization in India.Citation2 Escherichia coli strains possessing NDM-5 were subsequently reported to be prevalent in Denmark, France, and Algeria.Citation3 In Japan, detection of an NDM-5-producing clinical isolate of E. coli was first reported in 2014; this isolate belonged to sequence type (ST) 540, and the patient had traveled to Bangladesh.Citation4 Herein, we report the first detection of an NDM-5-producing E. coli strain belonging to ST405 in Japan.
Materials and Methods
Bacterial Isolates
In October 2015, a 79-year-old man was admitted to Kitasato University Hospital with cervical spinal cord injury causing respiratory muscle paralysis and upper and lower limb weakness. The patient developed pneumonia on day 5 of hospitalization; therefore, empirical antimicrobial treatment with vancomycin (VCM) (1 g twice daily) and tazobactam/piperacillin (TAZ/PIPC) (3.5 g three times daily) was initiated. Notable pathogens including gram-positive pathogenic bacteria such as methicillin-resistant Staphylococcus aureus were not cultured from the blood, sputum, and pleural effusion specimens. Therefore, VCM treatment was discontinued after 6 days, while TAZ/PIPC treatment was continued to manage the patient’s condition for 29 days. On hospital day 52, the patient developed a urinary tract infection, and a strain of carbapenem-resistant E. coli (KY1497 strain) was isolated from a urine specimen, although the other patients in the hospital showed negative results for carbapenem-resistant Enterobacteriaceae. The carbapenem-resistant E. coli was continuously isolated from the patient’s stool and urine specimens as colonies. The cardiopulmonary function of the patient gradually weakened, respiratory failure progressed, and the patient died on day 149. He had been admitted directly to our hospital without a history of international travel.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of the isolate was determined by microdilution according to the Clinical and Laboratory Standards Institute (CLSI) reference methods,Citation5 except that European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpointsCitation6 were used to evaluate tigecycline and polymyxin B. Two disks of ceftazidime and sodium mercaptoacetic acid (Eiken Chemical Co., Ltd., Tokyo, Japan) were used as indicators of metallo-β-lactamase production.
Antimicrobial Resistance Gene Screening and Molecular Typing
Polymerase chain reaction (PCR) was performed to detect the blaIMP, blaVIM, blaNDM, blaOXA-48, and CTX-M-1 group genes in the isolate.Citation7–Citation9 The bacterial PFGE plug was digested with S1 nuclease, followed by PFGE using a previously reported method with some modifications,Citation10 and visible DNA bands, which were possible plasmids, were excised to extract DNA.
Whole-Genome Sequencing
DNA libraries were constructed using the Nextera XT sample prep kit according to the manufacturer’s instructions (Illumina, Inc., San Diego, CA, USA), followed by next-generation sequencing (Miseq, Illumina, Inc.).Citation11 We performed long-read sequencing with PacBio RSII and obtained the resulting unitigs with HGAP v. 4.0 de novo assembler, followed by error-correction and complete genome sequence determination by Illumina short-read sequencing. Genome annotation was carried out using DFAST.Citation12 Strain genotyping was determined in silico by MLST (http://cge.cbs.dtu.dk/services/MLST/).
Plasmid Conjugation
Plasmid conjugation using the broth methodCitation13 was carried out between the blaNDM5-positive isolate KY1497 and sodium azide-resistant E. coli J53 as the recipient strain. Transconjugants were selected on selection plates supplemented with a combination of ceftriaxone (8 mg/L) and sodium azide (100 mg/L), and the presence of the NDM-5 gene was confirmed by PCR.
Ethics Statement
This study was approved by the research ethics committee of Kitasato University Hospital (approval no. B17-123) and complied with the Declaration of Helsinki. Written informed consent was obtained from the patient for publication of this case report.
Results and Discussion
Comparative Analysis of the IncFII-IncFIB Plasmids Harboring Carbapenemases
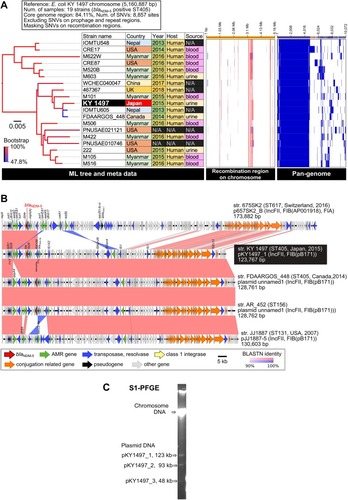
The complete genome sequence of KY1497 suggested that it belongs to ST405 and O102: H6 () and carries three plasmids. blaNDM-5 is located on the 123.7-kb plasmid pKY1497_1, which is an IncFII-IncFIB(pB171) replicon type (). S1-PFGE revealed three plasmid bands with lengths corresponding to those of the complete genome sequences (). Core-genome phylogeny of blaNDM-5-positive E. coli ST405 (19 strains in total) suggested that KY1497 had the most similar lineage to clinical isolate IOMTU605 in Nepal and FDAARGOS_448 in Canada, with 38 and 39 single nucleotide variants (SNVs), respectively (). These two strains carry the homologous blaNDM-5-positive plasmid (). Pair-wise alignment of pKY1467_1 displayed homologous regions with other IncFII-IncFIB(pB171) plasmids except for the qepA4 quinolone-resistance gene (). Regarding the IncFII-IncFIB(pB171) background, pKY1497_1 shares most of its plasmid backbone, whereas pJJ1887-5 in E. coli JJ1887 (ST131) carries other antimicrobial resistance genes rather than blaNDM-5, indicating that pJJ1887-5 is one of the most common ancestral plasmids for blaNDM-5 acquisition.Citation14 We detected blaNDM variants on plasmids >100-kb in size, with IncF, IncA/C, and untypeable replicons. Previous reports have indicated that the IncX3-type plasmid plays a major role in the global dissemination of NDM-producing Enterobacteriaceae.Citation11,Citation15,Citation16 In this study, molecular characterization of KY1497 revealed that it carried blaNDM-5 in a 123.7-kb plasmid harboring IncFII-IncFIB(pB171), suggesting that IncF has a similar role of dissemination as IncX3.
Figure 1 Comparative genomic analysis among 19 strains of blaNDM-5-positive E. coli ST405. (A) Core-genome SNV analysis and pan-genome analysis. Detected SNVs in the repeat and prophage regions were excluded. Recombination regions of the chromosome were predicted using Gubbins v. 2.3.4, followed by masking SNVs in the recombination regions. A maximum likelihood phylogenetic tree was constructed from 8,857 SNV sites in the core genome region. Pan-genomic analysis was performed using Roary v. 3.12.0. The recombination region and pan-genomic data were visualized using Phandango. The red and light blue bars indicate recombination regions of the ancestral type and single isolate, respectively. The blue bars in pan-genomic data indicate the presence of a gene cluster. (B) Comparative representation of complete blaNDM-5-positive plasmid sequences among five E. coli strains. The plasmid sequences were aligned using BLASTN, followed by visualization using Easyfig. Similarity of homologous and inversion blocks is indicated in red and blue, respectively. The backbone of the IncFII-IncFIB(pB171) plasmids is highly conserved among the different ST types. (C) S1-PFGE analysis of E. coli KY1497.
Strain Features
In the isolate, blaNDM was the only carbapenem-resistance gene detected. The MLST analysis classified E. coli (KY1497 strain) as ST405, suggesting that E. coli ST405 strains have the potential to become a reservoir for the blaNDM-5 gene. NDM-5-producing E. coli belonging to ST405 was previously detected in Spain and Italy.Citation17 Escherichia coli ST405 was found to carry blaCTX−M, blaNDM, and a repertoire of virulence genes comparable to those in O25b: H4ST131.Citation18 According to a previous study, among NDM-producing E. coli, ST405 was the fourth most common reported ST and the most abundant ST in Nepal and Europe, showing the highest distribution in the UK and Italy.Citation19 Sporadic occurrence of NDM-5 producers in Japan has been reported,Citation3,Citation4 however, NDM-5-producing E. coli belonging to ST405 has not been detected.
Transferability of blaNDM-5
KY1497 was resistant to fluoroquinolones and all β-lactams, including broad-spectrum cephalosporins and carbapenems, whereas it remained susceptible to tigecycline. The antimicrobial susceptibilities of transconjugants derived from E. coli J53 were similar to those of the donor KY1497 strain, particularly for penicillin, cephalosporin, and tigecycline (). The KY1497 strain successfully transferred the resistance plasmid at a frequency of 8.3 × 10–6, creating E. coli J53 KY1497T, suggesting the horizontal transfer of blaNDM-5 in the IncFII-IncFIB(pB171) plasmid. The IncF type plasmids were conjugatable, which may explain the rapid spread of the NDM-carrying isolates. Therefore, effective and feasible measures must be taken immediately to control the dissemination of these resistant plasmids.
Table 1 Antimicrobial Susceptibility Profile of E. coli Isolate with the blaNDM-5 Gene and Its Transconjugant
How Does blaNDM-5 Spread?
Travelers contribute significantly to the global spread of microbes and resistance genes. KY1497 was isolated from a non-traveler, suggesting that it is caused by an autochthonous strain or transmission by undetected carriers. In this case, TAZ/PIPC was prescribed for 29 days before detecting KY1497. Long-term broad-spectrum antibiotics may enable the detection of carbapenem-resistant strains.Citation20,Citation21 As the patient stayed in a private room, environmental investigation was performed after his death. Swab samples from a shelf close to the bed, hand-wash sink, drain ditch, inside of bedpan, and toilet-cleaning apparatus were tested by cultivation as possible sources of NDM-5 transmission; however, NDM-5-producing bacteria were not isolated. There have been previous reports of community-acquired NDM-producing isolates, indicating the existence of an undetected reservoir and potential transmission among colonized carriers in hospitals.Citation22,Citation23
One limitation of this study was that the undetected reservoirs formed by NDM-5-producing isolates were not investigated. Our results strongly emphasize that while strains producing NDM enzymes are rarely reported among hospitalized patients in Japan, attention should be paid to the community prevalence of such strains to monitor future trends and prevent further horizontal spread.
Conclusion
In this study, a self-transmissible IncFII-IncFIB plasmid carrying blaNDM-5 was detected in ST405 E. coli, which is a novel Inc replicon plasmid. We highlighted the dissemination potential of the IncFII-IncFIB plasmids harboring blaNDM-5. Effective infection control steps should be taken to prevent nosocomial infections.
Abbreviations
MLST, multilocus sequence typing; NDM-5, New Delhi metallo-β-lactamase 5; PCR, polymerase chain reaction; ST, sequence type; TAZ/PIPC, tazobactam/piperacillin; VCM, vancomycin.
Data Sharing Statement
The complete, annotated genome sequence of E. coli KY1497 has been deposited in a public database (chromosome, AP019803; pKY1497_1, AP019804; pKY1497_2, AP019805; and pKY1497_3: AP019806). The short- and long-read sequences for DNA-Seq have been deposited in the DNA Data Bank of Japan (DRA accession DRA008639; BioProject PRJDB8512; BioSample SAMD00178070; DRR accession DRR184076-DRR184080).
Acknowledgments
This work was supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development, AMED (grant numbers JP19fk0108048 and JP19fk0108103).
Disclosure
The authors report no conflicts of interest in this work.
References
- Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054.19770275
- Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55:5952–5954. doi:10.1128/AAC.05108-1121930874
- Uchida H, Tada T, Sugahara Y, et al. A clinical isolate of Escherichia coli co-harbouring mcr-1 and blaNDM-5 in Japan. J Med Microbiol. 2018;67:1047–1049. doi:10.1099/jmm.0.00079329972350
- Nakano R, Nakano A, Hikosaka K, et al. First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agents Chemother. 2014;58:7611–7612. doi:10.1128/AAC.04265-1425246390
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 25th Informational Supplement. M 100-S25. Wayne, PA: CLSI; 2015.
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters Version 9.0, valid from 2019-01-01. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf. Accessed 11st, 2019.
- Takayama Y, Adachi Y, Nihonyanagi S, et al. Modified Hodge test using Mueller-Hinton agar supplemented with cloxacillin improves screening for carbapenemase-producing clinical isolates of Enterobacteriaceae. J Med Microbiol. 2015;64:774–777. doi:10.1099/jmm.0.00006825934552
- Shibata N, Kurokawa H, Doi Y, et al. PCR classification of CTX-M-type beta-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrob Agents Chemother. 2006;50:791–795. doi:10.1128/AAC.50.2.791-795.200616436748
- Poirel L, Héritier C, Tolün V, et al. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi:10.1128/AAC.48.1.15-22.200414693513
- Bender JB, Hedberg CW, Besser JM, et al. Surveillance for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N Engl J Med. 1997;337:388–394. doi:10.1056/NEJM1997080733706049241128
- Zhang F, Xie L, Wang X, et al. Further spread of blaNDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front Microbiol. 2016;30:e424.
- Tanizawa Y, Fujisawa T, Kaminuma E, et al. DFAST and DAGA: web-based integrated genome annotation tools and resources. Biosci Microbiota Food Health. 2016;35:173–184. doi:10.12938/bmfh.16-00327867804
- Nakano R, Okamoto R, Nakano Y, et al. CFE-1, a novel plasmid-encoded AmpC-lactamase with an ampR gene originating from Citrobacter freundii. Antimicrob Agents Chemother. 2004;48::1151–1158. doi:10.1128/AAC.48.4.1151-1158.200415047515
- Rahman M, Shukla SK, Prasad KN, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44:30–37. doi:10.1016/j.ijantimicag.2014.03.00324831713
- Zhang Q, Lv L, Huang X, et al. Rapid increase in carbapenemase-producing Enterobacteriaceae in retail meat driven by the spread of the blaNDM-5-carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob Agents Chemother. 2019;63:e00573–19. doi:10.1128/AAC.00573-1931182541
- Li X, Fu Y, Shen M, et al. Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob Resist Infect Control. 2018;7:59. doi:10.1186/s13756-018-0349-629713466
- Barrado L, Pérez-Vázquez M, Del Pozo JL, et al. Clonal transmission of NDM-5-producing Escherichia coli belonging to high-risk sequence type ST405. Int J Antimicrob Agents. 2018;52:123–124. doi:10.1016/j.ijantimicag.2018.05.01829864499
- Roy Chowdhury P, McKinnon J, Liu M, Djordjevic SP. Multidrug resistant uropathogenic Escherichia coli ST405 with a novel, composite IS26 transposon in a unique chromosomal location. Front Microbiol. 2019;9:3212. doi:10.3389/fmicb.2018.0321230671039
- Dadashi M, Yaslianifard S, Hajikhani B, et al. Frequency distribution, genotypes and the most prevalent sequence types of New Delhi metallo-beta-lactamase-producing Escherichia coli among clinical isolates around the world: a review. J Glob Antimicrob Resist. 2019;19:284–293. doi:10.1016/j.jgar.2019.06.00831212107
- Cendejas E, Gómez-Gil R, Gómez-Sánchez P, Mingorance J. Detection and characterization of Enterobacteriaceae producing metallo-beta-lactamases in a tertiary-care hospital in Spain. Clin Microbiol Infect. 2010;16:181–183. doi:10.1111/j.1469-0691.2009.02888.x19624502
- Chusri S, Silpapojakul K, McNeil E, Singkhamanan K, Chongsuvivatwong V. Impact of antibiotic exposure on occurrence of nosocomial carbapenem-resistant Acinetobacter baumannii infection: a case control study. J Infect Chemother. 2015;21:90–95. doi:10.1016/j.jiac.2014.10.00225454216
- Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62:499–513. doi:10.1099/jmm.0.052555-023329317
- Poirel L, Hervé V, Hombrouck-Alet C, Nordmann P. Long-term carriage of NDM-1-producing Escherichia coli. J Antimicrob Chemother. 2011;66:2185–2186. doi:10.1093/jac/dkr23621653599
