Abstract
Introduction
Antimicrobial resistance (AMR), associated with antimicrobial use (AMU), is a major public concern. Surveillance and monitoring systems are essential to assess and control the trends in AMU and AMR. However, differences in the surveillance and monitoring systems between countries and sectors make comparisons challenging. The purpose of this article is to describe all surveillance and monitoring systems for AMU and AMR in the human and livestock sectors, as well as national surveillance and monitoring systems for AMR in food, in six European countries (Spain, Germany, France, the Netherlands, the United Kingdom and Norway) as a baseline for developing suggestions to overcome current limitations in comparing AMU and AMR data.
Methods
A literature search in 2018 was performed to identify relevant peer-reviewed articles and national and European grey reports as well as AMU/AMR databases.
Results
Comparison of AMU and AMR systems across the six countries showed a lack of standardization and harmonization with different AMU data sources (prescription vs sales data) and units of AMU and AMR being used. The AMR data varied by sample type (clinical/non-clinical), laboratory method (disk diffusion, microdilution, and VITEK, among others), data type, ie quantitative (minimum inhibition concentration (MIC) in mg/L/inhibition zone (IZ) in mm) vs qualitative data (susceptible-intermediate-resistant (SIR)), the standards used (EUCAST/CLSI among others), and/or the evaluation criteria adopted (epidemiological or clinical).
Discussion
A One Health approach for AMU and AMR requires harmonization in various aspects between human, animal and food systems at national and international levels. Additionally, some overlap between systems of AMU and AMR has been encountered. Efforts should be made to improve standardization and harmonization and allow more meaningful analyses of AMR and AMU surveillance data under a One Health approach.
Introduction
Antimicrobial use (AMU) in the last few decades is the main trigger for antimicrobial resistance (AMR) in humans and animals. For example, broad use of fluoroquinolones, effective antimicrobials against gram-positive and gram-negative bacteria, in humans and some animal populations has caused high resistance rates.Citation1 Antibiotics like colistin, that have issues with side effects but still have low resistance rates, have been reconsidered as a last-line drug due to a lack of alternative antimicrobials for multidrug-resistant Gram-negative bacteria.Citation2 This global threat includes both pathogenic and commensal bacteria. In order to tackle the AMR crisis, several global strategies have been developed such as the Global Action Plan (GAP) of the World Health Organization (WHO),Citation3 the new European One Health Action Plan against AMRCitation4 and the Central Asian and Eastern European Surveillance of Antimicrobial Resistance network (CAESAR).Citation5
SurveillanceCitation6 and monitoringCitation6 systems of AMU and AMR in humans and animals are essential to assess and subsequently control the global trends in the use of antimicrobials and antimicrobial susceptibility patterns of bacteria in different populations.Citation7 Using a One Health approach, zoonotic and indicator bacteria are of particular relevance.
Surveillance and monitoring systems are one of the five strategies of the GAP. However, even when the proper implementation of these systems enables the collection of reliable and good quality data, not all countries worldwide have surveillance and monitoring systems in place so it is not possible to perform a global comparison.
Several projects address the systems' evaluation of AMU and AMR in human, livestock and food sectors in Europe and also across European countries. As an illustration, the Ecology from Farm to Fork Of microbial drug Resistance and Transmission (EFFORT) projectCitation8 is a relevant work collecting AMU and AMR data from broilers, pigs, turkeys, veal calves, rainbow trout and companion animals at farm level across different EU countries. Additionally, the Antibiotic Resistance Dynamics: the influence of geographic origin and management systems on resistance gene flows within humans, animals and the environment (ARDIG)Citation9 project gathers AMU and AMR data from the human and animal sectors together with AMR data collection from food at European level. Likewise, another crucial work carried out at global level and in the animal sector is the Network on quantification of veterinary Antimicrobial usage at herd level and Analysis, CommunicaTion and benchmarkING to improve responsible usage (AACTING).Citation10 The latter initiative has generated a review of existing systems that collect AMU data at farm level.
This report follows on the work carried out in the ARDIG project and provides a review of AMU and AMR surveillance and monitoring systems, adopting a One Health approach, currently available in six European countries that perform routine surveillance, as well as systems at a European level.
It will make recommendations regarding the harmonization of surveillance and monitoring systems across Europe with a view to help overcome current limitations in comparing AMU and AMR data captured by these systems from different sectors and countries within Europe.
Materials and Methods
In this manuscript, we gathered key features of surveillance and monitoring systems on AMU and AMR in livestock and humans as well as AMR systems in food from Spain, Germany, France, the Netherlands, Norway, the United Kingdom (UK) together with its regions and Europe between 2014 and 2017.
A literature search in 2018 was performed using PubMed to identify relevant peer-reviewed articles and the internet to identify national and European grey reports as well as AMU/AMR databases. The terms used for the search are “antimicrobial resistance”, “antimicrobial use”, “Spain”, “Germany”, “UK”, “United Kingdom”, “Scotland”, “Wales”, “England”, “Northern Ireland”, “Netherlands”, “France”, “Norway”, “Europe”, “food”, “human”, “animal”, “surveillance”, “system” and “monitoring”. Additionally, a questionnaire asking for detailed information on any available AMR and AMU database in each country was developed and sent to all collaborating institutes for completion (Supplementary materials).
A detailed systems' description by country and sector has been performed in order to detect and define the lack of harmonization and standardization on AMU and AMR.
Results
Antimicrobial Resistance Surveillance and Monitoring Systems
A general overview on AMR monitoring and surveillance systems is provided in . The variables collected in the table are the country/region, database name, data type, data origin, unit, interpretation standard, evaluation criteria, public data, published report, report language, submitting data to Europe, laboratory method and set-up year of the database. Additionally, –, showing AMR systems reporting and not reporting to EU per country and sector, are provided.
Table 1 Features of AMR Databases in Human, Food and Animal Sectors by Region
Figure 1 Overview on AMR systems in livestock in six European countries. Inner ring systems (dotted sections) report AMR data to EFSA while outer ring systems not. For details on the systems and their relationship, see the body of the text.
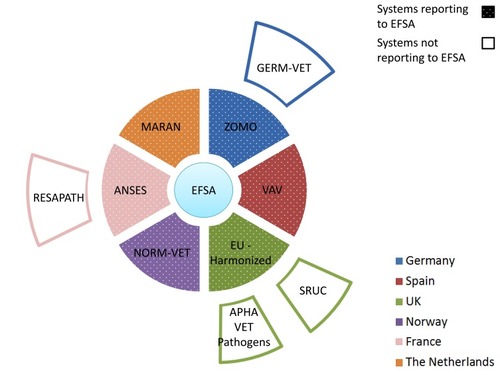
Figure 2 Overview on AMR systems in humans in six European countries. Inner ring systems report (dotted sections) AMR data to EARS-NET while outer ring systems not. For details on the systems and their relationship, see the body of the text.
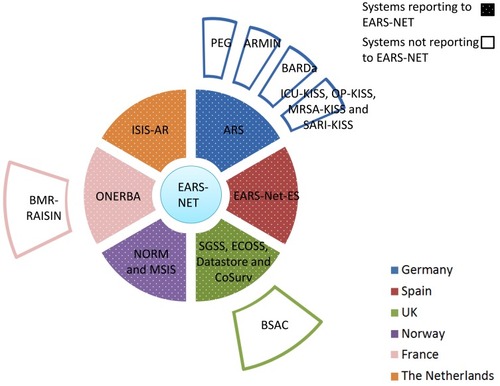
Figure 3 Overview on AMR systems in food in six European countries. Ring systems (dotted sections) report AMR data to EFSA. For details on the systems and their relationship, see the body of the text.

Europe
The European Food Safety Authority (EFSA)Citation11 is responsible for providing independent scientific advice and communication on food chain risks to risk managers and the public. EFSA together with the European Centre for Disease Control and Prevention (ECDC) collect annually AMR data on humans, food and healthy animals from the EU Member States (MS) and some associated countries. EFSA publishes “The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food”.Citation12 EFSA also publishes annually the “Trends and sources of zoonoses and zoonotic agents in foodstuffs, animals and feeding stuffs”Citation13 report (“EFSA report” onwards) for those countries which have not published this information.
The European Animal Health Study Center (CEESA)Citation14 is a non-governmental organization financed by the veterinary pharmaceutical industry doing research on AMR. Two relevant CEESA subsystems for this review are the VetPath monitoring system and the European Antimicrobial Susceptibility Surveillance in Animals (EASSA). CEESA monitors the antimicrobial susceptibility of major disease-causing bacterial pathogens in food animals (VetPath), and of foodborne and commensal bacteria in food animals (EASSA).
The European Antimicrobial Resistance Surveillance Network (EARS-Net)Citation15 is an AMR surveillance network in accordance with the legislationCitation16 for Europe and for the European Economic Area members.Citation17 Through EARS-Net, the ECDC collects AMR data from the EU Member States and publishes the annual EARS-Net report “Surveillance of antimicrobial resistance in Europe” which presents resistance percentages and trends for key resistant bacteria. Data are based on blood and cerebrospinal fluid isolates from humans.Citation18
The European Medicines Agency (EMA), EFSA and ECDC have produced two joint inter-agency antimicrobial consumption and resistance analysis (JIACRA) reportsCitation19 attempting to compare antimicrobial use in animals and humans to AMR in the sectors and to assess potential effects of AMU and AMR in animals on the situation in humans.
Spain
The Spanish Veterinary Antimicrobial Resistance Surveillance Network (VAV)Citation14,Citation20 was created to monitor AMR. It consists of three programs dealing with healthy animals, sick animals and with food and it is performed by the Ministry of Agriculture, Fisheries and Food (MAPAMA). VAV submits non-clinical data to the EFSA that are included in the annual EFSA reports.
The MAPAMA publishes the annual zoonoses and antimicrobial resistance report.Citation21 This report is based on the annual EFSA report and informs on zoonotic pathogens and diseases in animals, humans and food in addition to data on AMR in some zoonotic bacteria and indicator bacteria according to the EU legislation.Citation22,Citation23
The Spanish national plan to tackle and reduce AMR (PRAN)Citation24 has been set up by the Spanish Agency for Consumer Affairs, Food Security and Nutrition (AECOSAN). PRAN publishes a simplified report on zoonoses and antimicrobial resistance of chickens and turkeys for poultry professionals and a simplified report on zoonoses and antimicrobial resistance of laying hens for professionals in the laying hen sector.Citation21 In addition, PRAN publishes the JIACRA Spain reportCitation25 which assesses the relationship between AMU and AMR in humans and animals in Spain.
On the medical side, the national center of Carlos III Institute (ISCIII) coordinates and manages the national AMR database (EARS-Net-ES) submitting the data to the EARS-Net.Citation25
The Netherlands
The AMR monitoring system on animals and food in the Netherlands is the “Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands” (MARAN) bringing together the AMR food database of the Food and Consumer Product Safety Authority (NVWA).Citation26
The Netherlands publishes data on the resistance of foodborne pathogens and of commensal indicators from livestock and food in the annual report also referred to as MARAN.Citation27 The report is produced in collaboration with the NVWA, the National Institute for Public Health and the Environment (RIVM), the Netherlands Veterinary Medicines Institute (SDa), the University of Utrecht and Wageningen University and Research.
In the human sector, the Infectious Disease Surveillance Information System on Antibiotic Resistance (ISIS-AR)Citation28,Citation29 aims at monitoring AMR in major pathogens. The Dutch Foundation of the Working Party on Antibiotic Policy (SWAB) publishes the annual report “Consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands” (NethMap).Citation26 This report provides resistance data for outpatients, inpatients and care in nursing homes. It reports on several surveillance programs such as the ISIS-AR and Mycobacterium tuberculosis surveillance program and others. It has been created by the Ministry of Health, Welfare and Sport and the Dutch Society of Medical Microbiology (NVMM) and coordinated by the RIVM.
United Kingdom
Several systems and reports coexist to monitor AMR in animals in the UK, which is based on the EU decision.Citation30 The EU harmonized surveillance system (a native UK system) collects mandatory AMR data on indicator commensal Escherichia coli and/or Campylobacter spp. from meat and faecal/caecal content of healthy animals (chicken, cattle/beef, turkey and pigs) in the UK. There are also UK National Control Programs for Salmonella in layers, broilers and turkeys, which are hosted in the EU harmonized surveillance system.
In England and Wales the scanning surveillance system Vet Pathogens APHACitation31 provides AMR data from diseased animals provided for diagnostic services on a voluntary basis by veterinarians covering all relevant bacteria and animal species.
In Scotland, a surveillance system carried out by the Scotland’s Rural College Veterinary Services and Capital Diagnostics (SRUC) collects clinical isolates from animals.
In Northern Ireland, an AMR surveillance system performed by the Agri-Food Biosciences Institute (AFBI) collects livestock clinical data from post-mortem investigation of colibacillosis or similar diseases. E. coli isolates mainly originate from samples coming from less than 2-week old calves and animals with bovine mastitis.Citation32
The annual report “UK-Veterinary Antibiotic Resistance and Sales Surveillance” (UK-VARSS) promoted by the UK government and produced by the Veterinary Medicine Directorate (VMD) provides details on veterinary AMR and AMU data in the UK.Citation32
On the human side, the Resistance Surveillance Program of the British Society for Antimicrobial Chemotherapy (BSAC)Citation33 publishes antibiotic resistance data from participating laboratories in the UK and Ireland for a range of clinically significant bacteria from respiratory infections from the community (since 1999), hospitals (since 2008) and bloodstream infections (since 2001).
Public Health England’s Second Generation Surveillance System (SGSS) captures routine laboratory surveillance data on infectious diseases and antimicrobial resistance from 98% of National Health Service (NHS) laboratories across England. SGSS data are reported annually in the English surveillance program for antimicrobial utilization and resistance (ESPAUR) report.Citation34,Citation35
The Electronic Communication of Surveillance in Scotland (ECOSS) database collects AMR data from participating NHS and reference laboratories in Scotland.Citation36 The data are published together with the AMR data on animals from the SRUC and several AMU data sources from humans in the Scottish One Health Antimicrobial Use and Antimicrobial Resistance (SONAAR) report.Citation37
The medical AMR data in Northern Ireland are collected on a voluntary basis by the CoSurv database. These data are published annually in the “Surveillance of Antimicrobial Use and Resistance in Northern Ireland” report (“NI report”). This report was published for the first time in 2017 by the Public Health Agency (PHA).
Finally, the DataStore is an open-access database that collects on a voluntary basis AMR data from Wales covering all hospital labs. The DataStore data are annually published in the “Antibacterial Resistance in Wales” report.
Norway
The Norwegian Surveillance System for Communicable Diseases (MSIS)Citation38 together with the Norwegian Surveillance System for Antimicrobial Drug Resistance (NORM) and Norwegian Veterinary Antimicrobial Resistance Monitoring (NORM-VET) system are the three AMR surveillance programs in Norway.Citation39 These systems publish their data in the “Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway” (NORM/NORM-VET) report.Citation40 This annual report provides updated data on the occurrence and distribution of AMU and AMR in the human, animal and food sectors.
France
The French Agency for Food, Environmental and Occupational Health & Safety (ANSES) monitors AMR data associated with farming, food and the environment.Citation43 This institution coordinates the French surveillance network for antimicrobial resistance in pathogenic bacteria of animal origin (RESAPATH) and the Salmonella network. In addition, ANSES collects AMR non-clinical data from the national programs and the Salmonella network to be submitted to the EFSA.
The Salmonella network is a surveillance system set up to control non-human Salmonella throughout the food chain. Samples are collected from healthy animals, food and the environment.
The passive voluntary surveillance system RESAPATH provides in the annual RESAPATH report the AMR data compilation for the primary bacterial species and general isolates from sick animals from each animal sector.Citation41 This surveillance system started in 1982 under the name of RESABO (only for bovine species). In 2000, it was extended to pigs and poultry and in 2007 to other species including small ruminants, horses and companion animals. This network collaborates with the National Observatory of the Epidemiology of Bacterial Antibiotic Resistance (ONERBA).
On the medical side, ONERBA is the annual French report on AMU and AMR as well as the main AMR network collecting data from a complex network of subsystems. Currently (2019), the French health system (SPF) is taking over the ONERBA network and results are reported in the new tool ConsoRes.Citation42 This tool has been set up by the support centers for the prevention of healthcare-associated infections (CPias) Great East and New Aquitaine. Additionally, the AMR community network driven by SPF reports results using the MedqualCitation43 tool coordinated by the CPias Pays de la Loire. Finally, the Alert, Investigation and Surveillance of Nosocomial Infection Network (RAISIN)Citation44,Citation45 coordinates nationally the nosocomial infection surveillance coordination centers (CCLIN), now CPias. The RAISIN network includes several surveillance system modules. The private RAISIN module for multi-drug resistant bacteria BMR-RAISIN reports on AMR data in the community. However, it will be replaced shortly by the tool ConsoRes (2019).Citation46
Germany
The German veterinary monitoring system (GERM-VET) collects clinical AMR data in Germany from companion and food-producing animals. These data are published in detail by the Federal Office of Consumer Protection and Food Safety (BVL) in a report with the same denomination.Citation47
AMR-testing in the Zoonosis-Monitoring system (ZOMO) is carried out by the Federal Institute for Risk Assessment (BfR). The results are published in the annual zoonosis monitoring report by the BVL.Citation48 The report contains data about zoonotic and commensal bacteria in diverse food chains that are also reported to the EFSA together with AMR-data on Salmonella from the national control programs.
Antimicrobial Resistance Surveillance (ARS)Citation49 is the national AMR surveillance system in human medicine. Established by the Robert Koch Institute, it collects routine susceptibility data for all bacterial species from any kind of sample site from hospital care as well as from outpatient care institutions by an increasing number of laboratories participating on a voluntary basis. Results for main pathogens are published via an interactive database on the ARS website.
Besides the national surveillance ARS, the federal state Lower Saxony sets up a similar system (ARMIN).Citation50 A further system has recently been set up in Bavaria (BARDa).Citation51
The Hospital Infection Surveillance System (KISS) is the nosocomial infection surveillance system in hospitals formed by several sub-systems collecting AMU and AMR data.Citation52 This network assimilated the Surveillance of Antimicrobial Use and Bacterial Resistance in Intensive Care Units (SARI).Citation53,Citation54 SARI collected on a voluntary basis aggregated data on antimicrobial sensitivity for selected pathogenic bacteria and AMU-AMR development. The project is organized by the Institute for Hygiene and Environmental Medicine of the Charité, Berlin.Citation55 Patient-based and unit-based AMR data (MRSA, VRE, ESBL) are collected as well.
Finally, the Paul Ehrlich Society for Chemotherapy (PEG)Citation56 is a society that conducts studies on antimicrobial resistance in human pathogens as part of a longitudinal study in both hospital and community sectors. Results are presented as an interactive database on the PEG website. The report on Antibiotic Consumption and the Spread of Antibiotic Resistance in Human and Veterinary Medicine in Germany (GERMAP),Citation55 a joint work of the PEG, the BVL and Infectiology Freiburg, is published on a regular basis. This report provides AMU and AMR data and trends in human and veterinary medicine in Germany since 2008. GERMAP publishes AMR-data mainly from GERM-VET, ARS and data of the PEG. The report publishes antibiotic consumption data from the community analyzed by the Research Institute of the largest German public non-private Health Insurance AOK (WIdO).
Antimicrobial Consumption Surveillance and Monitoring Systems
A general overview on AMU monitoring and surveillance systems is provided in . The variables collected in the AMU table are country/region, database name, data origin, unit, public data published report, report language, data source, submitting data to Europe and set-up year of the database. Additionally, and show AMU systems reporting and not reporting to EU per country and sector.
Table 2 Features of AMU Databases in Human and Animal Sectors by Region
Figure 4 Overview on AMU systems in livestock in six European countries. Inner ring systems (dotted sections) report AMU data to ESVAC while outer ring systems not. For details on the systems and their relationship, see the body of the text.
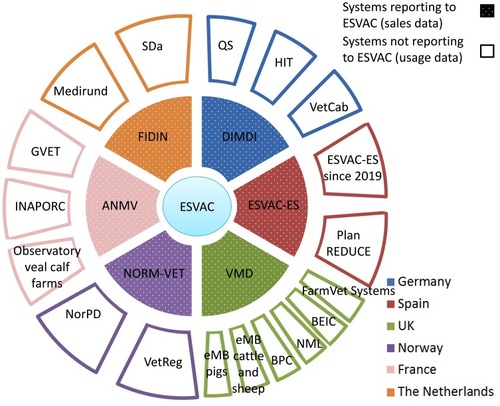
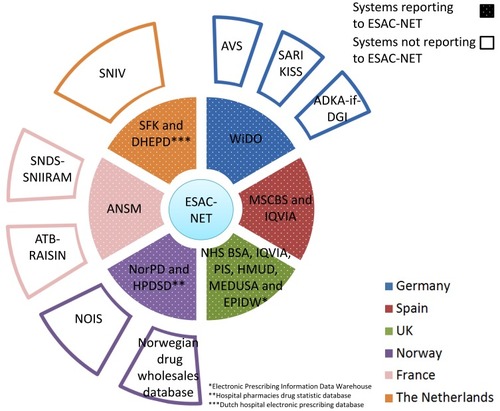
Figure 5 Overview on AMU systems in humans in six European countries. Inner ring systems (dotted sections) report AMU data to ESAC-NET while outer ring systems not. For details on the systems and their relationship, see the main text.
The term “prescription data” has been used in the veterinary field in the later table as “usage data” covering what is prescribed by the veterinarian, supplied by the veterinarian under veterinary prescription, or administered by the farmer under veterinary prescription.
Europe
EMA monitors overall AMU in livestock through sales data in the European Surveillance of Veterinary Antimicrobial Consumption (ESVAC) project.Citation57,Citation58 Since 2010, AMU is provided as an overall consumption (ie overall sales corrected for the animal population present in the country) in mg/kg. Since 2019, European countries are required to set up a data collection system in order to provide antibiotic consumption per species to ESVAC from 2022 onwards.
The Network on quantification of veterinary Antimicrobial usage at herd level and Analysis, CommunicaTion and benchmarkING to improve responsible usage (AACTING) provides in its website guidelines and information on farm level AMU collection systems from AACTING members, mostly European countries.Citation10
Similar to ESVAC, on the medical side, European AMU data from the community and hospital sector are collected by the European Surveillance of Antimicrobial Consumption Network (ESAC-Net),Citation59 that is coordinated by ECDC. AMU data are collected by MS in the community and the hospital sector or both (total care). In addition, ECDC coordinates the Healthcare-Associated Infections Surveillance Network (HAI-Net) since the coordination of the Improving Patient Safety in Europe network (IPSE) was transferred to ECDC in 2008. The HAI-Net supports MS in the prevention and control of healthcare-associated infections (HAI) and coordinates the European point prevalence survey of HAI and AMU in acute care hospitals, the European surveillance of surgical site infections (HAI-Net SSI), the European surveillance of Healthcare-Associated Infections in intensive care units and the repeated prevalence surveys of HAI and AMU in European long-term care facilities.
Spain
The ESVAC-ES is a project from PRAN and carried out by the Spanish Agency of Medicaments and Sanitary Products (AEMPS).Citation60 It collects animal AMU data on a voluntary basis and reports annually these sales data from the veterinary sector to ESVAC. Since 2019, ESVAC-ES additionally collects prescription data. Moreover, since 2016, several collaboration programs between PRAN, MAPAMA and the animal production sectors have been implemented. In these programs that are called “Plan REDUCE” the animal sectors provide AMU data on a voluntary basis to PRAN. These data together with the prescription data that ESVAC-ES collects will be submitted to ESVAC.
On the medical side, the Ministry of Health, Consumption and Social Welfare (MSCBS) database collects community reimbursement data on antimicrobials dispensed from only official prescriptions in the public system. The system is run by the General Directorate of Basic Services of the National Health and Pharmacy System.Citation61 The database for Pharmacoepidemiological Research in Primary Care (BIFAP) and the Primary Care Clinical Database (BDCAP) provide primary care data integrated into the MSCBS database. Spain is able to provide primary and secondary care sales data through The Human Data Science Company (IQVIA) database, formerly Quintiles and IMS Health.Citation24,Citation62
The PRAN website provides charts based on data collected from the latter databases with estimates on AMU in hospitals since 2012 and in the community (national and regional) since 2014.Citation63
The Netherlands
The MARAN report includes AMU data from two sourcesCitation26:
The Federation of the Dutch veterinary pharmaceutical industry (FIDIN) provides antibiotic sales data on the major livestock farming sectors.Citation27
The Netherlands Veterinary Medicines Institute (SDa) is an independent institute that promotes responsible drug consumption. It hosts a mandatory delivery records AMU database on the main livestock sectors and publishes it in the SDa report.Citation64 Similarly, Medirund,Citation65 the central database for the mandatory registration of antibiotics in cattle in the Netherlands, reports AMU data quarterly.Citation66
On the medical side, the NethMap report publishes AMU data from electronic antibiotic prescriptions on patient level.Citation26 These are extracted from the Dutch hospital electronic prescribing system for hospitals by SWAB and from the Foundation for Pharmaceutical Statistics (SFK) system for the community. This report also assimilates data from the national sentinel surveillance network for infectious diseases in nursing homes (SNIV).Citation67
United Kingdom
The VMD collates and analyses overall sales data from marketing authorization holders and aggregated usage data by species provided on a voluntary basis by several industry-based databases. Both sales and usage data are published in the UK-VARSS report. The industry-based databases on pig, cattle and poultry are
The Electronic Medicine Book for Pigs (eMB pigs),Citation68 launched by the Agriculture and Horticulture Development Board Pork (AHDB-Pork), collects usage data at farm level from the pig industry in the UK, covering around 90% of production.
The British Poultry Council (BPC) StewardshipCitation69 provides meat poultry usage data (for chickens, turkeys and ducks) in the UK, covering 90% of UK poultry meat production.
The British Egg Industry Council (BEIC) organizes the collection of antibiotic usage data for the laying hen industry. The Lion Scheme, representing over 90% of the UK laying hen industry, requires sharing usage data with BEIC.
FarmVet Systems is a private company, which collects usage data from veterinary practice and this data is published for cattle (dairy, around 30% UK coverage, and beef, around 5% UK coverage). This represents a convenience sample and so may not be representative of the UK cattle industry.Citation32
Additionally, the National Milk Laboratories (NML) database collects AMU data at farm level in dairy cattle.Citation70 However, this data source is currently at an early stage. Likewise, the new eMB cattle and sheep databaseCitation71 has been set up during 2018 as a pilot project collecting usage data at farm level and it is still in pilot stage.
On the medical side, the NHS Digital database in England displays antibiotic prescribing and AMR indicators from general practice (GP). Additional primary care sources such as hospices, nursing homes, police custody (among others) are displayed in the ESPAUR report. The IQVIA database shares secondary care data with Public Health England (PHE). However, these data are not openly available.
In Scotland, the Information Service Division (ISD) holds the Prescribing Information System (PIS) database that provides AMU data from primary care. The data are supplied by the Practitioner and Counter Fraud Services (P&CFS) of the National System Scotland (NSS). This system is responsible for the processing and pricing of all prescriptions dispensed in Scotland. Data from secondary care are provided by the Hospital Medicines Utilisation Database (HMUD) that is also held by the ISD. Primary and secondary care data are published in the SONAAR report.
In Northern Ireland, the Electronic Prescribing Database and the JAC Medicines Management Systems are the two datasets on AMU data from primary and secondary care, respectively. Data are published in the NI report.
In Wales, the AMU data are collected from the prescribing Information Data Warehouse for primary care provided by the Prescribing Services Unit (PSU)Citation72 and the Medusa datasetCitation73 for secondary care. Both are managed by Public Health Wales (PHW) and they cover 100% of Welsh dispensing contractors and hospital pharmacies.
Norway
The NORM-VET monitoring system collects AMU data from the Norwegian drug wholesales statistics database and the Veterinary Prescription Register (VetReg).Citation40,Citation74 The latter register is owned by the Norwegian Food Safety Authority (NFSA) and applies to veterinarians, pharmacies and feed mills. There, prescription data on farmed fish (since 2011) and terrestrial animals (since 2012) are stored.
On the medical side, the NORM surveillance program gathers AMU data in humans from the following surveillance systems:
The Hospital Pharmacies Drug Statistics Database.
The national prescription database (NorPD)Citation75 that contains dispensed drugs in hospitals and nursing homes from pharmacies in Norway.
The Norwegian drug wholesales statistics database that contains all sales data in Norway provided by the Norwegian Institute of Public Health (NIPH).
Additionally, the Norwegian Surveillance System for Antibiotic Consumption and Healthcare-Associated Infections (NOIS) is a nationwide mandatory system administrated by the NIPH. It is largely based on hospital automated data extraction on AMU.
France
The French Agency for Veterinary Medicinal Products (ANMV)Citation76 within ANSES collects annually sales data on veterinary drugs containing antibiotics in France, and also reports these data stratified by animal species. ANMV uses marketing authorisation holder estimates on the proportion sold per target species. In addition, The Permanent Observatory of Antibiotics in Veal Calf FarmsCitation77 is a voluntary system collecting AMU data in veal calves. Data are collected and analyzed by ANSES and the French Livestock Institute (IDELE).
The French Pork Interprofessional Organization (INAPORC)Citation78 is a voluntary system gathering consumption data on pig categories (sows, fatteners, weaners and sucklers) from 160 randomly chosen pig farms (approximately 1% of French farms). This system is run by ANSES, the swine industry’s French technical institute (IFIP) and stakeholders. The results are delivered to each farmer by the end of the study. Similar to INAPORC, the GVETCitation79 system is a voluntary pig register run by ANSES and IFIP. The latter system also collects AMU data from the same pig categories adopted by INAPORC providing an online result access to farmers.
On the medical side, the national health insurance cross-schemes information system (SNIIRAM) is a large French healthcare database which covers around 99% of the French population. SNIIRAM was extended with outpatient data through the National health data system (SNDS) by French lawCitation80 in 2016. The SNIIRAM-SNDS dataset provides prescription data covering primary and secondary care in the ambulatory and hospital sectors. It includes systems such as CNAMTS (for employed workers), RSI (for independent workers) and MSA (for farmers) among others. Through CNAMTS, the MEDIC’AM spreadsheetCitation81,Citation82 provides all medication reimbursement data and also the costs to the system (overall and reimbursement) and packages sold. SPF will take over shortly SNIIRAM-SNDS (2019).
In parallel, The Antibiotic Consumption Monitoring RAISIN module (ATB RAISIN) provides AMU data collected on a voluntary basis from the hospital sector. It is connected to the CPIAS network. It will be replaced shortly by the tool ConsoRes (2019).Citation46
The French National Agency for Medicine and Health Products Safety (ANSM) publishes on a regular basis the antibiotic consumption trends in France report.Citation83 It includes data on outpatient and hospital AMU in humans, critical antibiotics and AMU in Europe. The data in the report are extracted from the following databases: ANSM, IQVIA, Permanent Sample of Medical Prescription (EPPM), OPEN-MEDIC and ESAC-Net.
Germany
Several initiatives collect AMU data in Germany. The animal antimicrobial sales data are reported annually by the industry and wholesalers to the German Institute for Medical Documentation and Information (DIMDI).Citation84
The industry-based system run by Quality and Safety GmbH (QS)Citation85 carries out an antibiotic monitoring program on AMU data in broilers, turkeys, ducks, veal and pork productions. Some QS data are transferred to the Hi-Tier (HIT)Citation27 database. The HIT database is hosted by the Bavarian Ministry for the Food Chain, Agriculture and Forestry. The data collection includes treatment data on pigs, turkeys, broilers and cattle. This AMU database receives data from farmers and vets including the antimicrobial product, treatment days and number of treated animals. From the data, benchmarks for AMU in the different livestock sectors are calculated twice a year and published by the BVL. These inform farmers on the necessity to reduce AMU, ie farmers with a use above the third quartile of all reporting farms of their sector need to take action.
The sentinel project Veterinary Consumption of Antibiotics (VetCab)Citation27,Citation86 is carried out by the Institute for Biometry, Epidemiology and Information Processing of the Hanover University of Veterinary Medicine Foundation (IBEI-TIHO). VetCab aims to describe and assess AMU in farm animals in Germany and includes data on pigs, cattle and broilers.
In the human sector, two national surveillance systems are in place for hospital data: AVS (RKI) and ADKA-if-DGI.
The Federal Association of German Hospital Pharmacists (ADKA) created together with the infectious disease department of the University Freiburg the ADKA-if project in 2007. Since 2015, the network supported by the German Society for Infectious Diseases (DGI) is called ADKA-if-DGI.Citation87
The AVSCitation87,Citation88 housed by the RKI with technical support of the Charité collects data from German and Austrian hospitals on antimicrobial consumption for individual substances and groups of substances in acute care hospitals and rehabilitation centers since 2015 (2014 pilot study) according to the German Infection Protection Act.
On behalf of the SHI (Statutory Health Insurance Funds), the WIdOCitation55,Citation89 collects all antimicrobial prescriptions from mandatory health-insured patients (totaling about 89% of the German population). Only reimbursement data from the ambulatory sector (about 85–90%) are included in the WIdO dataset. Since 2001, all prescription data have become available. WIdO data are yearly reported to ESAC-Net via the RKI. In addition, the ZI, a research institute of the Federal Association of Statutory Health Insurance Physicians collects AMU data of ambulant patients.
Discussion
Harmonization and Interpretation of the AMU and AMR Data
A wide variety of AMU and AMR monitoring and surveillance systems and reports were identified at country and regional levels in the six countries. Funding of the systems is mostly public, but may also be private.
Monitoring and surveillance databases are mostly not freely accessible. Some databases do not report to the public on a regular basis. Even when most of the reports like NORM/NORM-VET, UK-VARSS, ESPAUR, MARAN/NETHMAP, ARS, EARS-Net among others, which publish aggregated data, are freely accessible others like Medirund or EASSA reports are not. This lack of free access to the available information may contribute to the existence of overlap between systems, reports and databases that may duplicate efforts and economic resources.
A further potential overlap source may be that the development of the different systems is frequently due to specific interests that are not fully covered by earlier systems leading to a substantial diversity in objectives and procedures. Therefore, it seems essential to reduce the number of overlapping systems joining forces, promoting synergies and planning the systems properly. Note that some overlap between systems may also contribute to validate system results. Some examples of overlapping are presented in . In addition, often newer systems are easier to use with the possibility of web-based reporting and feedback.
Table 3 Complementary Systems with Some Overlap
The presence of overlap between systems also translates to the existence of different reports that provide information on the same type of data generated in different systems. Furthermore, reports not based on a specific system may also produce overlaps (eg GERMAP resistance data on animals are also published by GERM-VET). However, the level of reported information may be different.
National AMU and AMR reports are published in different languages (see and ). Annual publication of these reports in an international agreed language would facilitate access to published data.
AMR Surveillance and Monitoring Systems
AMR surveillance and monitoring systems vary substantially between sectors and across the countries in the type of data collected and reported. Besides the human, animal or food population studied, main sources of variability include the type of samples collected (clinical vs non-clinical samples) and the sample collection basis (voluntary, sentinel or mandatory). Both define the bacterial population that the isolates may be representative for and influence the degree of representativeness of the data.
Diversity was also observed regarding the laboratory methods (eg micro broth dilution, disk diffusion or other, automatic systems) and the reported result type (minimum inhibition concentration (MIC), inhibition zone (IZ) or susceptible-intermediate-resistant (SIR)). The laboratory method selected may affect final results. As an example, colistin, a key antimicrobial in human and animal health, diffuses poorly into the agar medium. Therefore, disk diffusion results from colistin are not reliable.Citation90 Quantitative data allow for interpretation using different clinical breakpoints (CBP) or epidemiological cut-offs (ECOFF) as provided by EUCAST, CLSI or other, sometimes national institutions. SIR data can only be validly compared to other SIR data, if the methodology used is standardized. This includes both laboratory methods as well as the ECOFFS or breakpoints used for the categorization of the isolate populations to SR or SIR. Therefore, AMR reports should capture quantitative data rather than qualitative values (SIR or SR) to allow for interpretation of data using different thresholds. However, the comparability of quantitative data from different laboratory methodologies remains as an issue.
Data collection systems often adopt a specific standard. However, most standards and their corresponding evaluation criteria do not cover all drug/bug combinations. In that case, different standards and/or evaluation criteria may be used for different drug/bug combinations in the same data collection system. Therefore, AMR collecting systems should have a similar approach (ie standard, evaluation criteria, antibiotic panel, unit and data type (clinical-non clinical)) so that data comparison, evaluation and analyses across countries and sectors were valid.
In addition, ECOFFs and CBPs are regularly revised, so their threshold values may vary over time. Differences between ECOFFs and CBPs are frequently underlined in literature.Citation91,Citation92 ECOFFs identify the wild-type (those assumed to have no acquired/mutational resistance) from non-wild-type populations (those that show a degree of acquired/mutational resistance) while CBPs define clinically a microorganism as “sensitive”, “intermediate” or “resistant” in relation to the likelihood of therapeutic success. CBPs take into account information such as the infection site, ability of the antimicrobial to reach the infection site, dosage regimens and formulations available to determine the effectiveness against the pathogen. Therefore, interpretation of results between countries may not be directly comparable as different dose regimens are used.Citation93 However, in most instances, the differences between published ECOFF and CBP values are limited, given that one dilution step is the tolerance of microdilution in both systems. Moreover, values for both evaluation criteria are constantly evolving when new data become available and those with the greatest differences (eg ciprofloxacin in E. coli) tended to converge over time.Citation94
The main task of surveillance systems is to provide an overview of patterns and trends, however some systems may provide additional useful information for risk factor analysis.
A further source of variability identified on AMR systems is the collection of sample results from diverse laboratories, using different diagnostic methods and interpretation standards. Antimicrobial panels to be tested in laboratories against zoonotic and commensal bacteria are standardized in the livestock sector by Commission Implementing Decision 2013/652/EU. This is not the case in the medical sector and for the testing of clinical isolates from animals. However, EU institutions clearly indicate that the antimicrobial panel described for indicator bacteria on livestock by Decision 2013/652/EU takes human relevance into consideration.Citation95 Therefore, reporting part of the data adopting this standardized panel would help minimize current standardization and harmonization issues.
AMU Surveillance and Monitoring Systems
AMU collection systems are based on a variety of data sources ranging from overall national sales data to individual prescription or treatment data.Citation7 Data are displayed in very diverse units (such as weight of active ingredient, therapy frequency, mg/Population Correction Unit (PCU), Defined Daily Dose (DDD)/1000 inhabitants/days, DDD/1000 Specific Therapeutic Group Age-sex weightings Related Prescribing Units (STAR-PU) among others) hampering the comparison of data from different sources.
Differences in dosage regimes and treatment durations between hospitals and countries might result in an erroneous assessment of the treatment numbers if they are deduced from the amount of drug sold. For these reasons, any evaluation and comparison of AMU data from different sources should be done carefully.
A consensus has been reached to report AMU data to European level (ESVAC and ESAC-Net systems) adopting the unit DDD/1000 inhabitants/days on the medical side and antibiotic weight per population correction unit (mg/PCU) on the animal side. However, as with breakpoints in AMR, these units have drawbacks and the consensus is a compromise that is continuously under debate.
The weight of the active ingredient as collected for the ESVAC project does not account for its potency, ie the amount needed to treat 1 kg of animal. Moreover, most antimicrobials may be used in several animal species and may also be licensed at a different dosage for different and sometimes even for the same animal species. Sales data on the veterinary side therefore only provide a general overview, but for further analyses, farm-level data are needed. These are frequently collected on the regional or national level as shown in , but at a very low level of harmonization. At best they allow for assessing trends within the system, but between systems analyses are very challenging. This also applies for comparisons to the medical side. The differences between the systems have repeatedly been described and critically reviewed.Citation27,Citation96
The DDD/1000 inhabitants/day is widely used as a standard for monitoring antimicrobial consumption for the human sector. However, it does not necessarily reflect the dose prescribed to the individual patient. This particularly plays a role for special patient populations (eg children or patients with renal insufficiency). The same issue applies for defined daily doses for animals. Dosing of drugs for systemic use ideally should be done giving the amount of drug needed per kg of treated individual. If the weight of the treated animal is not accounted for and DDDs are calculated from the amount of drug used alone, substantial miscalculations are possible. In broilers 1-day-old chicks weigh about 50 g and 1-month-old broilers around 2 kg, ie, that is 40 times more. If 1 kg of drug dosed at 20mg/kg/day can be used to treat 1,000,000 1-day-old chicks, it will only serve 25,000 1-month-old broilers. Using a standard weight for broilers at about 1 kg (average weight at the time of treatment applied by ESVAC) to calculate a DDD would result in 50,000 DDDs which neither reflects the exposure of 1-day-old chicks nor the exposure of 1-month-old broilers.
Therapy frequency, used for farm animals in Germany, on the other hand, has the drawback that it does not account for dosing as it only considers the number of animals that were treated with the drug, assuming that this happens at a standard dose. DDDs, in case the DDD are equivalent to the prescribed daily dose (PDD), and therapy frequency both may represent days under treatment, but the results may differ substantially when describing the same population. This is because one is based on counted treatment days and therefore the amount of active substance used cannot be deduced from the figure. DDD, on the other hand, is deduced from used amounts of drugs and therefore does not have to be equivalent to real treatment days because of the issues explained above.
In summary, regarding antimicrobial use in animals, there is need for a measure that includes the name of the active ingredient, the amount of active ingredient, the number of treated animals, the population at risk, the weight of treated animals, the time under treatment and the duration of the therapeutic effect of the active ingredient in the body. If those are collected, most of the units that are currently in use should be deducible from the information with a reasonable accuracy. Likewise, besides DDDs, additional metrics should be collected in order to describe the different aspects of AMU in the human sector (eg days of treatment, number of prescriptions).
However, the EU agreed data type is not always provided in the national or regional surveillance and monitoring system reports but other units such as DDD/1000 STAR-PU are.Citation97
Those datasets that do not report their data at European level may have different units than the agreed ones, such as HIT or the SNIIRAM-SNDS system.
The health-care system implemented in each country is of great relevance to understand the data collection. As an example, in England, it is common to dispense outpatient medications by hospitals, whereas in Northern Ireland these are usually prescribed by the GP at the request of secondary care specialists.Citation98 Thus, there may be significant dissimilarities in the data collected across countries from homologous databases.
The usage data per animal species is a more useful source than sales data; however, it is not consistently collected by all countries and also not provided to ESVAC yet. Collections of these data are laborious if they are not available in electronic formats. However, in most countries, prescription data are collected at least from a part of the animal population such as in the UK (collecting prescription data on a voluntary basis from pigs through eMB pigs and meat poultry through BPC stewardship), among others or Norway (mandatory data collection of prescription data from food-producing animals in VetReg and on a voluntary basis from companion animals). However, the VetReg system has been compared to the sales data and there is a proportion of underreporting among the prescription data recorded since the registry started.Citation99
Tools for Comparison
JIACRA analyses comparing AMU in animals and humans to AMR in the sectors are accompanied by a long list of disclaimers but provide a valuable general overview. Major progress could further be improved by including prescription data, or at least use data by animal species.
Diverse tools have been developed to assess correlations and associations between AMU and the development of AMR. As an example, the hospital-based ARVIACitation100 and Conso-Res are similar initiatives under development in the human sector launched by Germany and France, respectively. These efforts supplement the JIACRA reports and address the issue at the hospital level.
Conclusions
AMU and AMR Systems and Reports Need Further Harmonization to Support the One Health approach.
Availability of prescription data or similar for animals would allow a more detailed analysis of antimicrobial treatment and resistance data, and enhance interpretation of the findings published in the JIACRA reports by EFSA, EMA and ECDC.
In addition, major challenges need to be addressed in order to harmonize AMU and AMR data in the animal sector through uniform and robust standards that are either fully harmonized or allow for conversion of data to different units. To this end, for AMU, the name of the active ingredient, amount of active ingredient, number of treated animals, the population at risk, weight of treated individual, treatment duration and the duration of the therapeutic effect of the active ingredient in the body are needed.
AMR collecting systems should have as far as possible a similar approach (ie standard, evaluation criteria, antibiotic panel, unit and data type (clinical-non clinical)) to be compared, evaluated and analyzed across countries and sectors. Otherwise, the data may not be directly assessed. Additionally, reports on AMR should capture quantitative values rather than data on the SIR level to allow for interpretation of data using different thresholds. However, there will be still an issue with comparability of quantitative data from different methodologies.
ECDC, EMA and EFSA indicate that the antimicrobial panel described for livestock by Decision 2013/652/EU takes human relevance into consideration. Reporting at least part of the data adopting this standardized panel would ensure uniformity.
Currently, there is some overlap between national and international systems (see ). Therefore, it seems essential to join forces, promote synergies and plan the systems properly in order to avoid overlapping and address potential gaps making better use of the available resources. A first step to achieve the latter goal is to address the system harmonization that will substantially increase data sharing with the EU. It seems that some resources could be used more efficiently by reducing the number of overlapping systems. However, note that some overlap between systems may be useful for system and data validation.
Preferably national AMU and AMR reports should be published annually and provided in one international agreed language (eg English) to facilitate access to published data.
Abbreviations
AACTING, Network on quantification of veterinary Antimicrobial usage at herd level and Analysis, CommunicaTion and benchmarkING to improve responsible usage; ADKA, Federal Association of German Hospital Pharmacists; AECOSAN, Spanish Agency for Consumer Affairs, Food Safety and Nutrition; AEMPS, Spanish Agency of Medicaments and Sanitary Products; AHDB, Agriculture and Horticulture Development Board; AMR, Antimicrobial Resistance; AMU, Antimicrobial Use; ANMV, National Agency for Veterinary Medicines; ANSES, French Agency for Food, Environmental and Occupational Health & Safety; ANSM, French National Agency for Medicine and Health Products Safety; APHA, Animal and Plant Health Agency; ARDIG, Antibiotic Resistance Dynamics: the influence of geographic origin and management systems on resistance gene flows within humans, animals and the environment; ARMIN, Antibiotic Resistance Monitoring in Lower Saxony; ARS, Antibiotics Resistance Surveillance; AST, Antibiotic Susceptibility Testing; ATB RAISIN, Antibiotic Consumption Monitoring programme; AVS, Antibiotic Consumption Surveillance; BDCAP, Primary Care Clinical Database; BfR, Federal Institute for Risk Assessment; BEIC, British Egg Industry Council; BIFAP, Database for Pharmacoepidemiological Research in Primary Care; BPC, British Poultry Council Stewardship; BSAC, British Society for Antimicrobial Chemotherapy; BVL, Federal Office of Consumer Protection and Food Safety; CAESAR, Central Asian and Eastern European Surveillance of Antimicrobial Resistance network; CA-SFM, Antibiogram Committee of the French Society for Microbiology; CBP, Clinical Breakpoints; CCLIN, Nosocomial Infection Surveillance Coordination Centers in France; CEESA, European Animal Health Study Center; CLSI, Clinical Laboratory Standard Institute; CPIAS, National Network for the Prevention of Care-Related Infections; DARC, Defra Antibiotic Resistance Coordination; DART, German antibiotic resistance strategy; DCD, Defined Course Dose; DDD, Defined Daily Dose; DGI, German Society for Infectious Diseases; DIMDI, German Institute for Medical Documentation and Information; DIN, German Institute for Standardization; EARS-Net, European Antimicrobial Resistance Surveillance Network; EASSA, European Antimicrobial Susceptibility Surveillance in Animals; ECDC, European Center for Disease Prevention and Control; ECOFF, Epidemiological Cut-Off; EFFORT, Ecology from Farm to Fork Of microbial drug Resistance and Transmission; ECOSS, Electronic Communication of Surveillance in Scotland; EFSA, European Food Safety Authority; EJP, European Joint Programme; EMA, European Medicines Agency; eMB cattle and sheep, Cattle and sheep Electronic Medicine Book; eMB pigs, Pig Electronic Medicine Book; EPPM, Permanent Sample of Medical Prescription; ESAC-Net, European Surveillance of Antimicrobial Consumption Network; ESPAUR, English Surveillance Programme for Antimicrobial Utilisation and Resistance; ESVAC, European Surveillance of Veterinary Antimicrobial Consumption; EU, European Union; EUCAST, European Committee on Antimicrobial Susceptibility Testing; FIDIN, Federation of the Dutch veterinary pharmaceutical industry; GAP, Global Action Plan; GERMAP, Antibiotic Consumption and the Spread of Antibiotic Resistance in Human and Veterinary Medicine in Germany; GERM-VET, German Veterinary Monitoring System; GP, General Practice; HIT, German animal movement and information system; IBEI-TIHO, Institute for Biometry, Epidemiology and Information Processing of the Hanover University of Veterinary Medicine Foundation; IDELE, French Livestock Institute; IFIP, Swine Industry’s French Technical Institute; INAPORC, French Pork Interprofessional Organisation; IQVIA, Human Data Science Company; ISCIII, Carlos III Health Institute; ISD, Information Service Division; ISIS-AR, Infectious Disease Surveillance Information System on Antibiotic Resistance; IZ, Inhibition Zone; JIACRA, Joint Interagency Antimicrobial Consumption and Resistance Analysis; KISS, Hospital Infection Surveillance System; MABUSE, Medical Antimicrobial Use Surveillance and Evaluation; MAPAMA, Ministry of Agriculture, Fisheries and Food; MARAN, Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands; Medirund, Central Database for the Mandatory Registration of Antibiotics in Cattle in Netherlands; MIC, Minimum Inhibitory Concentration; MS, Member State; MSIS, Norwegian Surveillance System for Communicable Diseases; NethMap, Consumption of Antimicrobial Agents and Antimicrobial Resistance Among Medically Important Bacteria in the Netherlands; NFSA, Norwegian Food Safety Authority; NHS, National Health Service; NIPH, Norwegian Institute of Public Health; NOIS, Norwegian Surveillance System for Antibiotic Consumption and Healthcare-Associated Infections; NORM, surveillance programme for antimicrobial resistance in human pathogens; NORM-VET, Monitoring Programme for Antimicrobial Resistance in the Veterinary and Food Production sectors; NorPD, Norwegian Prescription Database; NSS, National System Scotland; NVI, Norwegian Veterinary Institute; NVMM, Ministry of Health, Welfare and Sport and the Dutch Society of Medical Microbiology; NVWA, Netherlands Food and Consumer Product Safety Authority; ONERBA, National Observatory of the Epidemiology of Bacterial Antibiotic Resistance; P&CFS, Practitioner and Counter Fraud Services; PEG, Paul Ehrlich Society for Chemotherapy; PHE, Public Health England; PHW, Public Health Wales; PIS, Prescribing Information System; PRAN, National Antibiotic Resistance Plan; PSU, Prescribing Services Unit; QS, Quality and Safety GmbH; RAISIN, Alert, Investigation and Surveillance of Nosocomial Infection Network; RDD, Recommended Daily Dose; RESAPATH, French Surveillance Network for Antimicrobial Resistance in Pathogenic Bacteria of Animal Origin; RIVM, National Institute for Public Health and the Environment; RKI, Robert Koch Institute; SARI, Surveillance of Antimicrobial Use and Bacterial Resistance in Intensive Care Units; SPF, French Health System; SDa, Netherlands Veterinary Medicines Institute; SGSS, Second Generation Surveillance System; SHI, Statutory Health Insurance; SIR, Sensible, Intermediate, Resistant; SNDS, French National Health Data System; SNIIRAM, French National Health Data System; SNIV, National sentinel surveillance network for infectious diseases in nursing homes; SONAAR, Scottish One Health Antimicrobial Use and Antimicrobial Resistance; SRUC, Scotland’s Rural College Veterinary Services and Capital Diagnostics; STAR-PU, Specific Therapeutic Group Age-sex weightings Related Prescribing Units; SWAB, Dutch Foundation of the Working Party on Antibiotic Policy; UK, United Kingdom; UK-VARSS, UK-Veterinary Antibiotic Resistance and Sales Surveillance; VAV, Spanish Veterinary Antimicrobial Resistance Surveillance Network; VetCab, Sentinel project Veterinary Consumption of Antibiotics; VetReg, Veterinary Prescription Register; VMD, Veterinary Medicines Directorate; WHO, World Health Organization; WIdO, Scientific Institute of the AOK; ZOMO, German Zoonosis Monitoring.
Acknowledgments
This work was carried out within the framework of the ARDIG project, the European Joint Programme (EJP) on AMU and AMR in humans, food and animals. ARDIG project has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 773830.
We thank Mike Brouwer, Jean-Yves Madec, Philippe Glaser, Ines Noll, Karin Gröschner, Alexandra Hoffmann, Robby Markwart (RKI) for their input and collaboration.
Disclosure
The authors declare grants from European Union’s Horizon 2020 research and innovation programme during the conduct of the study. The authors report no other possible conflicts of interest in this work.
References
- Zhang D, Cui Y, Zhang X. Estimating factors related to fluoroquinolone resistance based on one health perspective: static and dynamic panel data analyses from Europe. Front Pharmacol. 2019;10:1145. doi:10.3389/fphar.2019.0114531636561
- Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–596. doi:10.1128/CMR.00064-1628275006
- Global Action Plan on Antimicrobial Resistance. World Health Organization (WHO). Available from: http://www.who.int/antimicrobial-resistance/global-action-plan/en/. Accessed 913, 2018.
- A European One Health Action Plan against Antimicrobial Resistance (AMR). EU Commission. Available from: https://ec.europa.eu/health/amr/sites/amr/files/amr_action_plan_2017_en.pdf. Accessed 403, 2019.
- Central Asian and Eastern European Surveillance of Antimicrobial Resistance (CAESAR). World Health Organization (WHO). Available from: http://www.euro.who.int/en/health-topics/disease-prevention/antimicrobial-resistance/surveillance/central-asian-and-european-surveillance-of-antimicrobial-resistance-caesar. Accessed March 31, 2020.
- ORION Knowledge Hub Catalogue. One Health EJP.ORION. Available from: https://foodrisklabs.bfr.bund.de/ohejp-glossary/. Accessed 1030, 2019.
- Interagency Coordination Group on Antimicrobial Resistance (IACG). Surveillance and monitoring for antimicrobial use and resistance; 2018 Available from: http://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_Surveillance_and_Monitoring_for_AMU_and_AMR_110618.pdf. Accessed 1012, 2018.
- The Ecology from Farm to Fork Of microbial drug Resistance and Transmission (EFFORT). EFFORT. Available from: http://www.effort-against-amr.eu/page/project.php. Accessed 71, 2019.
- Antibiotic resistance dynamics: the Influence of Geographic origin and management systems on resistance gene flows within humans, animals and the environment (ARDIG). One Health EJP. Available from: https://onehealthejp.eu/jrp-ardig/. Accessed 1012, 2018.
- Network on quantification of veterinary antimicrobial usage at herd level and Analysis, CommunicaTion and benchmarkiNG to improve responsible usage (AACTING). AACTING consortium. Available from: http://www.aacting.org/monitoring-systems/. Accessed 71, 2019.
- European Food Safety Authority (EFSA). Available from: https://www.efsa.europa.eu/. Accessed 222, 2019.
- European Food Safety Authority (EFSA). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019;17(2):5598.
- Biological hazards reports. European Food Safety Authority (EFSA). Available from: https://www.efsa.europa.eu/en/biological-hazards-data/reports. Accessed 1012, 2018.
- Schrijver R, Stijntjes M, Rodriguez-Bano J, Tacconelli E, Babu Rajendran N, Voss A. Review of antimicrobial resistance surveillance programmes in livestock and meat in EU with focus on humans. Clin Microbiol Infect. 2018;24(6):577–590. doi:10.1016/j.cmi.2017.09.01328970159
- European Antimicrobial Resistance Surveillance Network (EARS-Net). European Centre for Disease Prevention and Control (ECDC). Available from: https://ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/ears-net. Accessed 907, 2018.
- European parliament. Decision No 2119/98/EC of the European Parliament and of the Council of 24 September 1998 setting up a network for the epidemiological surveillance and control of communicable diseases in the Community. Off J Eur Union. 2019 Available from https://publications.europa.eu/en/publication-detail/-/publication/13a83657-97b6-4a80-aa32-3b335bdf80be/language-en. Accessed March 26, 2020.
- About the EARS Network. European Centre for Disease Prevention and Control (ECDC). Available from: https://ecdc.europa.eu/en/about-us/networks/disease-networks-and-laboratory-networks/ears-net-about. Accessed 402, 2019.
- European Centre for Disease Prevention and Control (ECDC). Surveillance of antimicrobial resistance in Europe 2017 Available from: https://ecdc.europa.eu/sites/portal/files/documents/AMR-surveillance-EARS-Net-2017-updated-dec-18.pdf. Accessed 914, 2018.
- European Medicines Agency (EMA). Analysis of antimicrobial consumption and resistance (‘JIACRA’ reports) Available from: https://www.ema.europa.eu/en/veterinary-regulatory/overview/antimicrobial-resistance/analysis-antimicrobial-consumption-resistance-jiacra-reports#report-on-2013%E2%80%9315-(jiacra-ii)-section. Accessed 219, 2019.
- Red de Vigilancia Veterinaria de Resistencias a Antibióticos (VAV). [Veterinary Antibiotic Resistance Surveillance Network]. Available from: https://www.visavet.es/data/VAV_vigilancia_veterinaria_resistencias_antibioticos_06.pdf. Accessed 907, 2018. Spanish.
- Resistencias antimicrobianas. Ministerio de Agricultura pesca y alimentación (MAPAMA). [Antimicrobial Resistance Surveillance. Ministry of Agriculture Fisheries and Food]. Available from: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&cad=rja&uact=8&ved=2ahUKEwjMn8DdpoTjAhVGLFAKHUAXA_kQFjAAegQIABAB&url=https%3A%2F%2Fwww.mapa.gob.es%2Fca%2Fganaderia%2Ftemas%2Fsanidad-animal-higiene-ganadera%2Fsanidad-animal%2Fzoonosis-resistencias-antimicrobianas%2Fresistencias_antimicrobianas.aspx&usg=AOvVaw2SbcAVpbgP9R3TFhECViAb. Accessed 1102, 2018. Spanish.
- European parliament. Directive 2003/99/EC of The European Parliament and of the Council of 17 November 2003 on the monitoring of zoonoses and zoonotic agents, amending Council Decision 90/424/EEC and repealing Council Directive 92/117/EEC. Off J Eur Union. Available from: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32003L0099&from=EN. Accessed October, 12 2018.
- European parliament. Regulation (EC) No 2160/2003 of the European Parliament and of the Council of 17 November 2003 on the control of salmonella and other specified food-borne zoonotic agents. Off J Eur Union. Available from https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32003R2160&from=EN. Accessed October, 12 2018.
- Plan Nacional frente a la Resistencia a los Antibióticos (PRAN). [Spanish Action Plan against Antimicrobial Resistance]. Available from: http://www.resistenciaantibioticos.es/es/publicaciones. Accessed 1012, 2018. Spanish.
- Informe JIACRA España. Primer análisis integrado del consumo de antibióticos y su relación con la aparición de resistencia. Plan Nacional de Resistencia a Antibióticos (PRAN). [JIACRA SPAIN report. First integrated analysis of antibiotic consumption and its relationship to the emergence of resistance. Spanish Action Plan against Antimicrobial Resistance]. Available from: http://www.resistenciaantibioticos.es/en/system/files/field/files/informe_jiacra-espana.pdf?file=1&type=node&id=410&force=0. Accessed 1012, 2018. Spanish.
- National Institute for Public Health and the Environment; Netherlands Food and Consumer Product Safety Authority; Wageningen university; Utrech university and SDa. Consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands in 2018. NethMap 2019/Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2018.MARAN 2019. 2018 Available from: https://www.rivm.nl/bibliotheek/rapporten/2019-0038.pdf. Accessed 719, 2019.
- Werner N, Kreienbrock L, Kreienbrock L. Monitoring antimicrobial drug usage in animals: methods and applications. Microbiol Spectrum. 2018;6(4):ARBA-0015-2017 2017. doi:10.1128/microbiolspec.ARBA-0015-2017
- ISISweb. Nederlandse Vereniging voor Medische Microbiologie (NVMM). [Dutch Association for Medical Microbiology]. Available from: https://isis-web.nl/. Accessed 907, 2018. Dutch.
- Altorf-van der Kuil W, Schoffelen AF, de Greeff SC, et al. National laboratory-based surveillance system for antimicrobial resistance: a successful tool to support the control of antimicrobial resistance in the Netherlands. Eurosurveillance. 2017;22(46):17–00062. doi:10.2807/1560-7917.ES.2017.22.46.17-00062
- COMMISSION IMPLEMENTING DECISION of 12 November 2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria (notified under document C(2013) 7145) (Text with EEA relevance) (2013/652/EU). The European commission. Available from: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32013D0652&qid=1539349584328&from=EN. Accessed 1012, 2018.
- Scanning surveillance - How is it delivered? Animal and Plant Health Agency. Available from: http://apha.defra.gov.uk/vet-gateway/surveillance/scanning/index.htm. Accessed 907, 2018.
- Veterinary Antibiotic Resistance and Sales Surveillance. UK Government. Available from: https://www.gov.uk/government/collections/veterinary-antimicrobial-resistance-and-sales-surveillance. Accessed 503, 2019.
- The BSAC Resistance Surveillance Programme is a well-established initiative designed to provide long-term surveillance of antibiotic resistance.The British Society for Antimicrobial Chemotherapy (BSAC). Available from: http://www.bsacsurv.org/. Accessed 402, 2019.
- Laboratory reporting to Public Health England. A guide for diagnostic laboratories. Public Health England (PHE). Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/545183/PHE_Laboratory_Reporting_Guidelines.pdf. Accessed 907, 2018.
- English Surveillance Programme for Antimicrobial Utilisation and Resistance (ESPAUR). England PH. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/759975/ESPAUR_2018_report.pdf. Accessed 1205, 2018.
- Surveillance Data and Systems. Protection Scotland. Available from: https://www.hps.scot.nhs.uk/data/. Accessed 907, 2018.
- Scottish one health antimicrobial use and antimicrobial resistance. Report in 2017. Health Protection Scotland. Available from: https://www.hps.scot.nhs.uk/resourcedocument.aspx?id=6971. Accessed 201, 2019.
- MSIS-Statistikk. Folkehelseinstituttet (FHI). [MSIS Statistics. Norwegian Institue of Public Health (NIPH)]. Available from: http://www.msis.no/. Accessed 907, 2018.
- Antibiotic resistance in Norway. Norwegian Institue of Public Health (NIPH). Available from: https://www.fhi.no/en/id/antimicrobial/. AccessedMarch 31, 2020.
- NORM/NORM-VET 2018. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Public Health Institute and Veterinary institute. Available from: https://norsvin.no/wp-content/uploads/2019/10/NORM-NORM-VET-2018.pdf. Accessed 326, 2020.
- French surveillance network for antimicrobial resistance in pathogenic bacteria of animal origin (RESAPATH 2017). French Agency for Food Environmental and Occupational Health & Safety (ANSES) Available from: https://resapath.anses.fr/resapath_uploadfiles/files/Documents/2017_RESAPATH%20annual%20report.pdf. Accessed 625, 2019.
- ConsoRes. CPias Grand.Est. Available from: https://www.consores.net/. Accessed 403, 2019. French.
- MedQual. Santé Publique France (SPF). French Health System. Available from: http://www.medqual.fr/. Accessed 217, 2019. French.
- The RAISIN Working Group C. “RAISIN” – a national programme for early warning, investigation and surveillance of healthcare-associated infection in France. Eurosurveillance. 2009;14(46):19408.19941798
- de Léotoing L, Barbier F, Dinh A, et al. French hospital discharge database (PMSI) and bacterial resistance: is coding adapted to hospital epidemiology? Med Mal Infect. 2018;48(7):465–473. doi:10.1016/j.medmal.2018.03.00729673880
- Surveillance et prévention de la RATB en établissements de santé. CPIAS. [Surveillance and prevention of RATB in health institutions]. Available from: https://cpias-occitanie.fr/surveillance-et-prevention-de-la-ratb-en-etablissements-de-sante/. Accessed 1029, 2019. French.
- Bericht zum GERM-Vet Monitoring programm 2016 Bundesamt für Verbraucherschutz und Lebensmittelsicherkeit (BVL). [Report on the GERM-Vet Monitoring Programme 2016. Federal Office of Consumer Protection and Food Safety]. Available from: https://www.bvl.bund.de/DE/09_Untersuchungen/03_Fachmeldungen/2018/2018_10_09_Fa_GERM-Vet-2016.html. Accessed 403, 2019. German.
- Zoonosen-Monitoring. Bundesamt für Verbarucherschutz und Lebensmittelsicherheit. [Zoonosis Monitoring. Federal Office of Consumer Protection and Food Safety]. Available from: https://www.bvl.bund.de/DE/08_PresseInfothek/04_Publikationen/03_Berichte/infothek_berichte_node.html#doc1401838bodyText4. Accessed 1012, 2018. German.
- Resistenzübersicht. Robert Koch Institut (RKI). [Resistance overview]. Available from: https://ars.rki.de/Content/Database/ResistanceOverview.aspx. Accessed 907, 2018. German.
- Antibiotika-Resistenz-Monitoring in Niedersachsen (ARMIN). Niedersächsisches Landesgesundheitsamt. [Antibiotic resistance monitoring in lower Saxony. Office. LSSH]. Available from: https://www.nlga.niedersachsen.de/infektionsschutz/armin_resistenzentwicklung/antibiotika-resistenz-monitoring-in-niedersachsen-armin-19418.html. Accessed 1012, 2018. German.
- Die Bayerische Antibiotikaresistenz-Datenbank (BARDa). Bayerisches Landesamt für Gesundheit und Lebensmittelsicherheit. [The Bavarian Antibiotic Resistance Database. Safety. BSOfHaF]. Available from: https://www.lgl.bayern.de/gesundheit/infektionsschutz/barda/index.htm. Accessed 907, 2018. German.
- KISS (Krankenhaus-Infektions-Surveillance-System) Projektbeschreibung. Nationales Referenzzentrum für Surveillance von nosokomialen Infektionen. [Hospital Infection Surveillance System. Infections. NRCfSoN]. Available from: http://www.nrz-hygiene.de/surveillance/kiss/. Accessed 1012, 2018. German.
- Surveillance der Antibiotika-Anwendung und bakteriellen Resistenzen auf Intensivstationen (SARI). Institut für Hygiene und Umweltmedizin. [Surveillance of antibiotic use and bacterial resistance in intensive care units. Institute for Hygiene and Environmental Medicine]. Available from: http://sari.eu-burden.info/. Accessed 326, 2020. German.
- Remschmidt C, Schneider S, Meyer E, Schroeren-Boersch B, Gastmeier P, Schwab F. Surveillance of Antibiotic Use and Resistance in Intensive Care Units (SARI). Dtsch Arztebl Int. 2017;114(50):858–865.29271345
- GERMAP. Paul-Ehrlich-Gesellschaft (PEG). Available from: https://www.p-e-g.org/germap-47.html. Accessed 1012, 2018.
- Paul-Ehrlich-Gesellschaft (PEG). Resistenzdaten [resistance data]. Available from: https://www.p-e-g.org/resistenzdaten-46.html. Accessed 907, 2018. German.
- European Surveillance of Veterinary Antimicrobial Consumption (ESVAC). European Medicines Agency (EMA). Available from: http://www.ema.europa.eu/ema/index.jsp?curl=pages/regulation/document_listing/document_listing_000302.jsp. Accessed 907, 2018.
- Silley P, Simjee S, Schwarz S. Surveillance and monitoring of antimicrobial resistance and antibiotic consumption in humans and animals. Rev Sci Tech. 2012;31(1):105–120. doi:10.20506/rst.31.1.210022849271
- European Surveillance of Antimicrobial Consumption Network (ESAC-Net). European Centre for Disease Prevention and Control (ECDC). Available from: https://ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/esac-net. Accessed 907, 2018.
- Agencia Española de Medicamentos y Productos Sanitarios (AEMPS). Ministerio de Sanidad Consumo y Bienestar Social. [Spanish Agency for Medicines and Health Products Ministry of Health Consumer Affairs and Social Welfare]. Available from: https://www.aemps.gob.es/home.htm. Accessed 913, 2018. Spanish.
- Funciones Dirección General de Cartera Básica de Servicios del Sistema Nacional de Salud y Farmacia. Ministerio de Sanidad Consumo y Bienestar Social. [Functions General Directorate of Basic Portfolio of Services of the National Health and Pharmacy System. Ministry of Health CAaSW]. Available from: https://www.mscbs.gob.es/organizacion/ministerio/organizacion/sgralsanidad/dgcbssnsyfF.htm. Accessed 1012, 2018. Spanish.
- IQVIA. IQVIA. Available from: https://www.iqvia.com/locations/. Accessed 907, 2018.
- Mapa de Consumo. Plan Nacional de Resistencia a los Antibióticos (PRAN). [Consumption map. Spanish Action Plan against Antimicrobial Resistance]. Available from: http://www.resistenciaantibioticos.es/es/profesionales/vigilancia/mapas-de-consumo. Accessed 1012, 2018. Spanish.
- Netherlands Veterinary Medicines Institute (SDa). Netherlands Veterinary Medicines Institute (SDa). Available from: http://www.autoriteitdiergeneesmiddelen.nl/. Accessed 1012, 2018.
- Database Diergeneesmiddelen runderen. MediRund. [Database Veterinary medicines bovine animals]. Available from: https://www.medirund.nl/. Accessed 1012, 2018. Dutch.
- Mills HL, Turner A, Morgans L, et al. Evaluation of metrics for benchmarking antimicrobial use in the UK dairy industry. Vet Rec. 2018;182(13):379. doi:10.1136/vr.10470129476032
- SNIV. National Institute of Public Health and the Environment. Available from: https://www.rivm.nl/sniv. Accessed 917, 2018. Dutch.
- Electronic medicine book to record antibiotic usage data. Agriculture and Horticulture Development Board (AHDB). Available from: https://pork.ahdb.org.uk/news/news-releases/2016/april/electronic-medicine-book-to-record-antibiotic-usage-data/. Accessed 907, 2018.
- Welcome to the British Poultry Council. British Poultry Council. Available from: https://www.britishpoultry.org.uk/. Accessed 907, 2018.
- NML. National Milk Laboratories (NML). Available from: https://www.nationalmilklaboratories.co.uk/vets/farm-assist. Accessed 716, 2019.
- eMB-Cattle. Agriculture and Horticulture Development Board (AHDB). Available from: http://beefandlamb.ahdb.org.uk/research/animal-health-and-welfare-beef/cattle-emb-phase-ii/. Accessed 1012, 2018.
- Antibacterial Usage in Primary Care in Wales 2013–2017. Public Health Wales (PHW). Available from: http://www.wales.nhs.uk/sitesplus/documents/888/Antibacterial%20Usage%20in%20Primary%20Care%20in%20Wales%202013%2D2017%20%28financial%20years%29.pdf. Accessed 222, 2019.
- Antimicrobial Usage in Secondary Care in Wales 2007–2016. Public Health Wales. Available from: http://www.wales.nhs.uk/sitesplus/documents/888/All%20Wales%20Antimicrobial%20Usage%20in%20Secondary%20Care%20FINAL.pdf. Accessed 222, 2019.
- Lillehaug A, Bornes C, Grave K. A pharmaco-epidemiological study of antibacterial treatments and bacterial diseases in Norwegian aquaculture from 2011 to 2016. Dis Aquat Organ. 2018;128(2):117–125. doi:10.3354/dao0321929733026
- Berg C, Red BH, Fenne O, et al. Drug consumption statistics 2018:2 Norwegian Prescription Database 2013–2017. 2018 Available from: https://www.fhi.no/en/publ/2018/legemiddelstatistikk-20182-reseptregisteret-20132017/. Accessed 210, 2019.
- Monitoring sales of veterinary antimicrobials. French Agency for Food Environmental and Occupational Health & Safety (ANSES). Available from: https://www.anses.fr/en/content/monitoring-sales-veterinary-antimicrobials. Accessed 1012, 2018.
- Chanteperdrix M, Chevance A, Orlianges M, Moulin G, Urban D, Parois A. Observatoire pérenne du suivi des usages d’antibiotiques en production de veaux de boucherie: Résultats de l’année 2016 et évolution par rapport à 2013. [Permanent observatory monitoring the use of antibiotics in the veal calves production: 2016 results and evolution compared to 2013]. 24ème ed. Journées 3R, Rencontres, recherches, ruminants; 2018 French.
- Hémonic A, Chauvin C, Delzescaux D, Verliat F, Corrégé I. French Working Group ‘antimicrobials in the swine i. Reliable estimation of antimicrobial use and its evolution between 2010 and 2013 in French swine farms. Porcine Health Manag. 2018;4:8. doi:10.1186/s40813-018-0084-729692925
- Moulin G, Orand J-P. Le suivi des antibiotiques en élevages. [Monitoring of antibiotics on farms]. Les cahiers de la Recherche Santé, Environnement, Travail. 2017;10:59–67. French.
- Loi no◦ 2016-41 du 26 janvier 2016 de modernisation de notre système de santé. The French National Assembly and the Senate. [Law no◦ 2016-41 of 26 January 2016 on the modernisation of our health system]. Available from: https://www.legifrance.gouv.fr/affichTexte.do?cidTexte=JORFTEXT00031912641. Accessed 222, 2019. French.
- Open Medic: base complète sur les dépenses de médicaments interrégimes. French goverment. [Open Medic: complete database on inter-scheme drug expenditures]. Available from: https://www.data.gouv.fr/en/datasets/open-medic-base-complete-sur-les-depenses-de-medicaments-interregimes/. Accessed 218, 2019. French.
- Bezin J, Duong M, Lassalle R, et al. The national healthcare system claims databases in France, SNIIRAM and EGB: powerful tools for pharmacoepidemiology. Pharmacoepidemiol Drug Saf. 2017;26(8):954–962. doi:10.1002/pds.423328544284
- L’ANSM publie un rapport sur la consommation des antibiotiques en France en 2016 - Point d’Information. Agence Nationale de Sécurité du Médicament et des produits de santé (ANSM). [ANSM publishes a report on the consumption of antibiotics in France in 2016 - Information Point. National Agency for the Safety of Medicines and Health Products]. Available from: https://ansm.sante.fr/S-informer/Points-d-information-Points-d-information/L-ANSM-publie-un-rapport-sur-la-consommation-des-antibiotiques-en-France-en-2016-Point-d-Information. Accessed 1029, 2019. French.
- Abgabemengen von Tierarzneimitteln werden beim DIMDI zentral erfasst. Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL). [Dispensing quantities of veterinary drugs are centrally recorded at DIMDI. Office for Consumer Protection and Food Safety]. Available from: https://www.bvl.bund.de/DE/08_PresseInfothek/01_FuerJournalisten_Presse/01_Pressemitteilungen/05_Tierarzneimittel/2010/2010_11_18_pi_abgabemengenregister.html;jsessionid=9D2FE13408BFF8D5F5E702CD3A473318.1_cid332. Accessed 907, 2018 German.
- Schaekel F, May T, Seiler J, Hartmann M, Kreienbrock L. Antibiotic drug usage in pigs in Germany—are the class profiles changing? PLoS One. 2017;12(8):e0182661. doi:10.1371/journal.pone.018266128841685
- Hemme M, Ruddat I, Hartmann M, et al. Antibiotic use on German pig farms - A longitudinal analysis for 2011, 2013 and 2014. PLoS One. 2018;13(7):e0199592. doi:10.1371/journal.pone.019959229969477
- Noll I, Schweickert B, Tenhagen B-A, Käsbohrer A. Antibiotikaverbrauch und Antibiotikaresistenz in der Human- und Veterinärmedizin [Antimicrobial use and antimicrobial resistance in human and veterinary medicine]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2018;61(5):522–532. doi:10.1007/s00103-018-2724-0. German.29633033
- Birgitta Schweickert MF, Schneider M, Willrich N, et al. Antibiotic consumption in Germany: first data of a newly implemented web-based tool for local and national surveillance. J Antimicrob Chemother. 2018;73(12):3505–3515. doi:10.1093/jac/dky34530239728
- Arzneiverordnungs-Report. WIdO. Wissenchaftliches Institut der AOK. [Prescription report. WIdO. AOK Scientific Institute]. Available from: https://www.wido.de/publikationen-produkte/buchreihen/arzneiverordnungs-report/. Accessed 913, 2018. German.
- Ezadi F, Ardebili A, Mirnejad R. Antimicrobial susceptibility testing for polymyxins: challenges, issues, and recommendations. J Clin Microbiol. 2019;57(4):e01390–e01318. doi:10.1128/JCM.01390-1830541939
- Toutain P-L, Bousquet-Mélou A, Damborg P, et al. En route towards European clinical breakpoints for veterinary antimicrobial susceptibility testing: a position paper explaining the VetCAST approach. Front Microbiol. 2017;8:2344. doi:10.3389/fmicb.2017.0234429326661
- Hombach M, Courvalin P, Böttger EC. Validation of antibiotic susceptibility testing guidelines in a routine clinical microbiology laboratory exemplifies general key challenges in setting clinical breakpoints. Antimicrob Agents Chemother. 2014;58(7):3921–3926. doi:10.1128/AAC.02489-1324777093
- European Food Safety Authority (EFSA). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019;17(2):5598.
- O’Halloran C, Walsh N, O’Grady MC, et al. Assessment of the comparability of CLSI, EUCAST and Stokes antimicrobial susceptibility profiles for Escherichia coli uropathogenic isolates. Br J Biomed Sci. 2018;75(1):24–29. doi:10.1080/09674845.2017.139273629210602
- European Center for Disease prevention and Control (ECDC), European Food Safety Authority (EFSA) and European Medicices Agency (EMA). ECDC, EFSA and EMA Joint Scientific Opinion on a list of outcome indicators as regards surveillance of antimicrobial resistance and antimicrobial consumption in humans and food-producing animals. EFSA J. 2017;15(10):e05017.
- Merle R, Robanus M, Hegger-Gravenhorst C, et al. Feasibility study of veterinary antibiotic consumption in Germany–comparison of ADDs and UDDs by animal production type, antimicrobial class and indication. BMC Vet Res. 2014;10:7. doi:10.1186/1746-6148-10-724401194
- Antimicrobial Usage in Primary Care in Wales 2013/14–2017/18. Public Health Wales. Available from: http://www.wales.nhs.uk/sitesplus/documents/888/Antibacterial%20Usage%20in%20Primary%20Care%20in%20Wales%202013%2D2017%20%28financial%20years%29.pdf. Accessed 1030, 2019.
- Surveillance of Antimicrobial Use and Resistance in Northern Ireland, Annual Report, 2018. Public Health Agency. Available from: https://www.publichealth.hscni.net/sites/default/files/2019-02/AMR%20annual%20report%20final%202018.pdf. Accessed 1029, 2019.
- Veterinært legemiddelregister (VetReg) - datakvalitet for antibakterielle midler. Norwegian Veterinary Institute (NVI). [Veterinary drug register (VetReg) - data quality for antibacterial agents]. Available from: https://www.mattilsynet.no/om_mattilsynet/annual_report_the_norwegian_food_safety_authority_2018.35406/binary/Annual%20report%20The%20Norwegian%20Food%20Safety%20Authority%202018. Accessed March 131, 2020.
- Hoffmann A, Schneider MJ, Zacher B, Krings A, Eckmanns T. ARVIA “ARS und AVS Integrierte Analyse. Epidemiologisches Bull. 2019;6:46–53. German.
