Abstract
Background
Bloodstream infection (BSI) caused by carbapenem-resistant Enterobacteriaceae are potentially life-threatening related to poorer outcomes. Colistin is considered one of the last-resort treatments against human infections caused by multidrug-resistant (MDR) Gram-negative bacteria. Therefore, emergence of strains from the blood that co-harboring mcr and carbapenem resistance genes were considered as a serious problem.
Purpose
In this study, two mcr-9-harboring MDR Enterobacter cloacae isolates BSI034 and BSI072 recovered from BSI patients were identified, one of which co-harbored mcr-9 and blaNDM-1. The genetic characteristics of the MDR plasmid needed to be clarified.
Methods
S1-PFGE and Southern blotting were conducted to determine the location of mcr-9. Whole-genome sequencing was performed to obtain the complete genome and plasmid sequences. The resistome and virulence genes of the strains, accompanied by the genetic characteristics of mcr-9- and blaNDM-1-harboring plasmids, were analyzed.
Results
Whole-genome sequencing showed that BSI034 harbored mcr-9-carrying IncHI2-type pBSI034-MCR9 and blaNDM-1-carrying IncX3-type pBSI034-NDM1. The 278,517 bp pBSI034-MCR9 carried mcr-9 along with the other 19 resistance genes. mcr-9 was flanked by IS903B (1057 bp) and IS26 (820 bp) in the same orientation. In addition to resistance genes, strain BSI034 also carried a chromosome-located Yersinia high-pathogenicity island, which harbored genes of yersiniabactin biosynthesis operon ybtSXQPAUTE, irp1/2, and fyuA.
Conclusion
We described the complete genome and mcr-9/blaNDM-1-co-harboring plasmid of E. cloacae from a BSI patient. Notable differences were observed within mosaic modules between pBSI034-MCR9 and other mcr-9-harboring plasmids due to extensive recombination via horizontal gene transfer.
Keywords:
Introduction
Colistin is considered one of the last-resort treatments against human infections caused by multidrug-resistant Gram-negative bacteria.Citation1 The first plasmid-mediated colistin resistance gene mcr-1 was identified on an IncI2 plasmid from Escherichia coli and Klebsiella pneumoniae in China.Citation2 While mcr-1 remains the predominant plasmid-mediated colistin resistance gene, mcr-2 to −8Citation3,Citation4 have been identified in various species from humans and animals. Recently, mcr-9 was identified from a colistin-susceptible Salmonella enterica serotype Typhimurium isolate recovered from a human patient in the USA in 2010. Induced expression of mcr-9 in E. coli conferred MIC of colistin at 2.5 mg/L.Citation5 mcr-9 shared 65% and 63% amino acid identity with the most closely related MCR-3 and MCR-7 enzymes, respectively, and between 33% and 45% with the other MCR-like enzymes.Citation6 mcr-9 has also been identified among ESBL-producing Enterobacteriaceae isolates from horses in Sweden, including Enterobacter cloacae, E. coli, Klebsiella oxytoca and Citrobacter freundii.Citation7
Carbapenem-resistant Enterobacteriaceae (CRE), which is a member of the ESKAPE group of pathogens, is an emerging pathogen and a leading cause of nosocomial infectionsCitation8 associating with great difficulty of clinical treatment.Citation9 Bloodstream infection (BSI) caused by CRE are potentially life-threatening related to poorer outcome and longer hospital stays.Citation10 To better understand the characteristic of mcr-9-harboring plasmid, the mcr-9-positive isolates from BSI patients in China were identified.
Materials and Methods
Bacterial Strains
We collected 188 Enterobacteriaceae from the blood of BSI patients in Guangdong province, China. The isolation of those strains was part of the routine hospital laboratory procedure. Preliminary species identification was achieved by MALDI-TOF MS (BrukerDaltonik GmbH, Bremen, Germany) and 16s rRNA sequencing, and the identification of E. cloacae species was confirmed by WGS. The mcr-9 gene was detected by using WGS data in two E. cloacae isolates BSI034 and BSI072 from two patients.
Antimicrobial Susceptibility Testing
MICs were determined for colistin, tigecycline, cefotaxime, ceftazidime, cefepime, gentamicin, amikacin, imipenem, ertapenem, meropenem, ciprofloxacin, fosfomycin, trimethoprim-sulfamethoxazole, piperacillin-tazobactam, aztreonam for both mcr-9-carrying isolates, using the agar dilution method except for colistin, for which used the broth dilution method was used, in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines. The E. coil ATCC25922 was the quality control strain used for the MIC measurement. The results interpreted according to CLSI instructions, while colistin and tigecycline resistance was defined according to EUCAST clinical breakpoints.
S1-PFGE and Southern Blotting
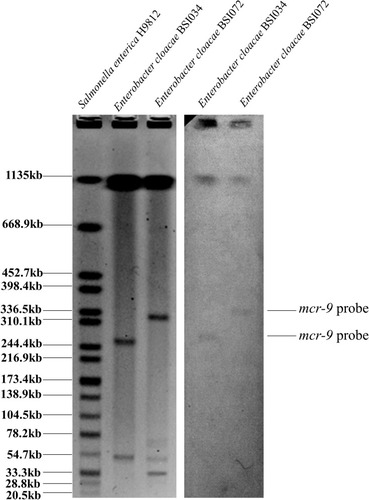
The plasmid and/or chromosomal locations of mcr-9 were determined by S1-PFGE, followed by Southern blotting hybridizations.Citation11 Southern blotting hybridizations of plasmid DNA were performed with a digoxigenin-labeled mcr-9 probe, according to the manufacturer’s instructions (Roche Diagnostics, Germany).
Whole-Genome Sequencing and Analysis
The five E. cloacae isolates were subjected to genomic DNA extraction using the Qiagen Blood & Tissue kit (Qiagen, Hilden, Germany) and whole-genome sequencing. DNA libraries were constructed with 350-bp paired-end fragments and sequenced using an Illumina HiSeq 2000 platform. The sequencing reads were assembled into contigs using SPAdes version 3.10.Citation12 The transferrable antimicrobial resistance genes and virulence genes were identified using ABRicate version 0.5 (https://github.com/tseemann/abricate). In silico multilocus sequence typing (MLST) was performed by MLST 1.8 (https://cge.cbs.dtu.dk/services/MLST/). Considering strain BSI034 co-harboring mcr-9 and blaNDM-1 and other MDR features, we further used the long-read MinION sequencer (Oxford Nanopore Technologies, Oxford, UK) to sequencing this strain. De novo hybrid assembly both of short Illumina reads and long MinION reads was performed using Unicycler v.0.4.3.Citation13 Complete circular contigs were then corrected using Pilon v.1.22 with Illumina reads. The complete nucleotide sequence of pBSI034-MCR9 and pBSI034-NDM1 reported in this study has been submitted to the NCBI database and assigned accession numbers MN937241 and MN937240.
Results and Discussion
Characteristics of Patients Infected by Both Isolates
The ST114 strain BSI034 was isolated from a 35-year-old male patient in September 2013, who was suffering the renal calculus and urinary tract infection. After receiving the percutaneous nephrolithotomy, the man developed fever and bloodstream infection. Then, he was treated with meropenem and discharge from the hospital alive. While the ST190 strain BSI072 was recovered from a 3-year-old boy in June 2018, who was suffering a Burkitt lymphoma and acute kidney failure in a pediatric ICU. After receiving chemotherapy, the boy developed myelosuppression and bloodstream infection, which the source was considered as catheter-associated. Before infection, the patient was treated with piperacillin-tazobactam and imipenem. He then received the therapy of meropenem, cefoperazone sulbactam, levofloxacin, and tigecycline subsequently after infection. The boy eventually developed septic shock and died four days later.
Antimicrobial Susceptibility and Location of mcr-9
MICs of 15 antimicrobial agents for strain BSI034 and BSI072 were determined (Table S1). Except for colistin, strain BSI034 was resistant to all the tested antimicrobial agents. While strain BSI072 was susceptible to colistin, tigecycline, and amikacin (Table S1). Both BSI034 and BSI072 did not show resistance to colistin, with 0.25 μg/mL and 0.5 μg/mL, respectively. This further support that mcr-9 only causes colistin resistance under induction expression.Citation6
S1-PFGE and Southern blotting hybridization revealed that mcr-9 located on plasmids in size of ~270 Kb and ~320 Kb in BSI034 and BSI072, respectively ().
Resistome and Virulence Genes in E. cloacae Isolates
It showed that BSI034 co-harbored mcr-9 and blaNDM-1genes, and the other 24 antimicrobial resistance genes mediating resistance to aminoglycosides (aac(6ʹ)-IIc, aadA5, aph(3ʹʹ)-Ib, aph(3ʹ)-Ia, aph(6)-Id, aph(3ʹ)-Ia, armA), β-lactams (blaACT-16, blaDHA-1, blaOXA-1, blaSHV-12), chloramphenicol (catA2, catB3), fosfomycin (fosA), macrolide (msr(E), mph(E), ere(A)), quinolones (aac(6ʹ)-Ib-cr, oqxB, oqxA, qnrB4), sulphonamides (sul1), and trimethoprim (dfrA1, dfrA19). By contrast, BSI072 harbored mcr-9 along with other 18 resistance genes mediating resistance to aminoglycosides (ant(2ʹʹ)-Ia, aadA16), β-lactams (blaACT-7, blaCTX-M-65, blaOXA-10), chloramphenicol (catA1, catB8), fosfomycin (fosA), quinolones (oqxB, oqxA, aac(6ʹ)-Ib-cr, qnrB6), rifampicin (ARR-3), sulphonamides (sul1), tetracycline (tet(A), tet(G)), and trimethoprim (dfrA27).
In addition to resistance genes, both strains carried many virulence genes. A total of 35 different virulence genes were identified among these mcr-9 positive isolates. Both isolates carried virulence genes including acrB, fepA, fepC, fepD, fepG, ybdA, entE, entA, entB, ompA, gnd, galF, rcsB, vipA/tssB. acrB, part of the AcrAB-TolC tripartite system, belonging to the RND family, have been proved to be associated with increasing antibiotic resistance and virulence in E.cloacae.Citation14 Besides the virulence genes above, BSI034 also carried a chromosome-located Yersinia high-pathogenicity island (HPI) which harbored yersiniabactin biosynthesis operon ybtSXQPAUTE, irp1/2, and fyuA. This HPI was inserted at the 3ʹ-end of a tRNA gene (tRNA-Asn) and has no other mobile elements.
Genetic Characteristics of the Plasmid-Borne mcr-9
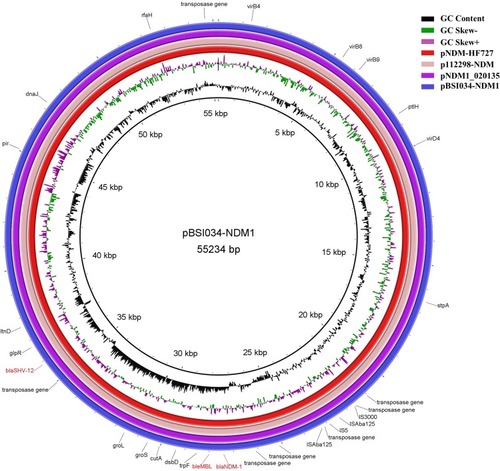
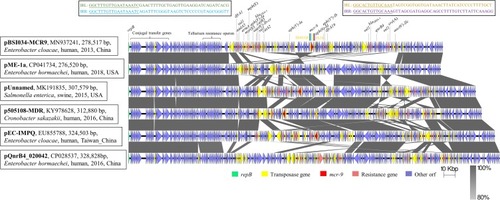
The mcr-9-carrying pBSI034-MCR9, which has IncHI2 and IncHI2A replicons, was found to encode 297 open reading frames (ORFs) (). In pBSI034-MCR9, mcr-9 was flanked by two insertion sequences (IS) in the same orientation. With 132 bp upstream of mcr-9, an IS903B (1057 bp, IS6 family) was identified. This element included the intact IRL and IRR sequences but lacked the 8 bp direct repeat (DR) sequence surrounding it. Besides, downstream of mcr-9 with 655 bp interval is an IS26 (820 bp, IS6 family). Similar to the conclusion in another study, IS903 and IS903-like have been found upstream of mcr-9, and IS1R, IS26-like, IS15DII have been found downstream of mcr-9.Citation6,Citation15 The consistent adjacency of mcr-9 to IS903-like element suggests the acquisition of mcr-9 by an IS903-dependent mechanism. The major differences of the genetic context of mcr-9 were various IS elements and ORFs downstream of mcr-9, thus forming diverse mcr-9 gene contexts. Aside from mcr-9, pBSI034-MCR9 harbored other 22 resistance genes including four copies of sul1 to form a large MDR plasmid, including those involved in resistance to aminoglycoside, β-lactams, chloramphenicol, macrolide, quinolones, sulphonamides, and trimethoprim. Those resistance genes were surrounded by various IS elements or transposons. A BLAST comparison of pBSI034-MCR9 screened five most close plasmids (). These included a newly identified mcr-9 and blaVIM-4-co-harboring plasmid pME-1a (CP041734) in an E. hormaechei isolate from a pediatric patient in the USA;Citation16 pUnnamed (MK191835) from Salmonella enterica subsp. enterica serovar Braenderup strain from swine in the USA;Citation17 p505108-MDR (KY978628) in Cronobacter sakazakii isolated from a patient in China;Citation18 pEC-IMPQ (EU855788) in a blaIMP-8-carrying E. cloacae from a patient in Taiwan;Citation19 non-mcr-9-carrying plasmid pQnrB4_020042 (CP028537) in a clinical E. hormaechei in China. They all carried a backbone related to a conjugal transfer locus (~30 Kb), a tellurium resistance operon (ter locus, 16785 bp) and parAB and parMR for partition. Additionally, pUnnamed, p505108-MDR, and pEC-IMPQ carried a mercuric resistance operon (merEDACPTR, 3976 bp). However, the accessory modules exhibited some similarities but also notable differences across those plasmids, which are likely to contain several MDR modules to form large MDR plasmids due to extensive recombination mediated by multiple mobile elements.
Figure 2 Schematic presentation of major structural features of pBSI034-MCR9 in comparison with five reference plasmids. Areas shaded in gray indicate homologous regions of ≥80% nucleotide sequence identity in the plasmid scaffold regions. ORFs are portrayed by arrows to indicate the direction of transcription and colored based on their predicted gene functions. The figure was drawn to scale.
Analysis of the Plasmid Carrying blaNDM-1
blaNDM-1 was located on the IncX3 plasmid pBSI034-NDM1. The blaNDM-1 gene accompanied by bleMBL (bleomycin resistance gene) located downstream of an ISAba125 element (1092 bp). This ISAba125 was disrupted by the insertion of IS5 (1195 bp) at the 917 bp position site. Furthermore, an IS3000 (3236 bp) and a truncated Tn2 (580 bp) element were found upstream of the ISAba125 element. Indeed, blaNDM-1 has always been found in association with an upstream insertion sequence ISAba125 which provides the −35 promoter sequence.Citation20 pBSI034-NDM1 also harbored blaSHV-12, which was bounded by two IS26 (820 bp) elements. pBSI034-NDM1 is organized very similarly (100% query coverage, 99% identity) to that of plasmids pNDM1_020135 (K. pneumoniae, CP037965), p112298-NDM (Citrobacter freundii, KP987216), and pNDM-HF727 (E. cloacae, KF976405) (). The main difference between them is pBSI034-NDM1 carries an additional segment that contains an IS5 (1195 bp).
Conclusion
BSI caused by carbapenem-resistant and colistin-resistant Enterobacteriaceae differs from most other multidrug-resistant bacterial pathogens in that there is no reliable treatment. In summary, we reported the complete sequence of plasmid co-harboring the mcr-9 and other resistance genes in NDM-1-producing E. cloacae. The linkage is a matter of concern since it could herald the possibility of a co-spread of the two genes, both involved in resistance to last-resort agents. Screening for the mcr-9 gene in other species in animals, the environment, and humans is necessary to understand its dissemination throughout the world.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81830103, 81722030, and 81902123), National Science and Technology Key Projects for Major Infectious Diseases (2017ZX10302301), China Postdoctoral Science Foundation (2019M653192), the Guangdong Natural Science Foundation (2017A030306012), the Science and Technology Planning Project of Guangdong (2017A020215017), Science, Technology & Innovation Commission of Shenzhen Municipality (JCYJ20190807151601699), Open project of Key Laboratory of Tropical Disease Control (Sun Yat-sen University), Ministry of Education, Supported by the 111 Project, Grant No. B12003.
Disclosure
The authors report no conflicts of interest in this work.
References
- Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:1–18. doi:10.3389/fmicb.2014.0064324478763
- Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-726603172
- Nang SC, Li J, Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol. 2019;45(2):131–161. doi:10.1080/1040841X.2018.149290231122100
- Wang XM, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi:10.1038/s41426-018-0124-z29970891
- Carroll LM, Gaballa A, Guldimann C, et al. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. mBio. 2019;10(3):e00853–e00819. doi:10.1128/mBio.00853-1931064835
- Kieffer N, Royer G, Decousser JW, et al. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother. 2019;63(9):e00965–19. doi:10.1128/AAC.00965-1931209009
- Borjesson S, Greko C, Myrenas M, et al. A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying blaSHV-12 from horses in Sweden. J Glob Antimicrob Resist. 2019. doi:10.1016/j.jgar.2019.08.007
- Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016:2475067.27274985
- Bonomo RA, Burd EM, Conly J, et al. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis. 2017;66(8):1290–1297. doi:10.1093/cid/cix893
- Stewardson AJ, Marimuthu K, Sengupta S, et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis. 2019;19(6):601–610. doi:10.1016/S1473-3099(18)30792-831047852
- Sirichote P, Hasman H, Pulsrikarn C, et al. Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar choleraesuis isolates from patients in Thailand and Denmark. J Clin Microbiol. 2010;48(3):883–888. doi:10.1128/JCM.01792-0920032253
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi:10.1089/cmb.2012.002122506599
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.100559528594827
- Guerin F, Lallement C, Isnard C, Dhalluin A, Cattoir V, Giard JC. Landscape of resistance-nodulation-cell division (RND)-type efflux pumps in Enterobacter cloacae complex. Antimicrob Agents Chemother. 2016;60(4):2373–2382. doi:10.1128/AAC.02840-1526856831
- Yuan Y, Li Y, Wang G, et al. Coproduction of MCR-9 and NDM-1 by colistin-resistant Enterobacter hormaechei isolated from bloodstream infection. Infect Drug Resist. 2019;12:2979–2985. doi:10.2147/IDR.S21716831571950
- Chavda KD, Westblade LF, Satlin MJ, et al. First report of bla VIM-4 and mcr-9 coharboring enterobacter species isolated from a pediatric patient. mSphere. 2019;4(5):e00629–00619. doi:10.1128/mSphere.00629-1931511372
- Elnekave E, Hong SL, Lim S, et al. Circulation of plasmids harboring resistance genes to quinolones and/or extended-spectrum cephalosporins in multiple Salmonella enterica serotypes from swine in the United States. Antimicrob Agents Chemother. 2019;63(4):e02602–e02618. doi:10.1128/AAC.02602-1830745386
- Shi L, Liang Q, Zhan Z, et al. Co-occurrence of 3 different resistance plasmids in a multi-drug resistant Cronobacter sakazakii isolate causing neonatal infections. Virulence. 2018;9(1):110–120. doi:10.1080/21505594.2017.135653728771073
- Chen YT, Liao TL, Liu YM, Lauderdale TL, Yan JJ, Tsai SF. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob Agents Chemother. 2009;53(3):1235–1237. doi:10.1128/AAC.00970-0819075060
- Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents. 2010;36(Suppl 3):S8–S14. doi:10.1016/S0924-8579(10)70004-2