Abstract
Objective
A colistin-resistant Escherichia coli strain isolated from dog feces was characterized in this study.
Methods and Results
A multiplex PCR assay was used to detect the presence of colistin-resistant mcr genes; it was found that E. coli QDFD216 co-harbored the mcr-1 and mcr-3 genes. Whole-genome sequencing and further bioinformatics analysis revealed that E. coli QDFD216 belonged to serotype O176:H11, fimH1311 type and ST132. The resistance genes blaCTX-M-14, mdfA, dfrA3, acrA, acrB, tolc, and sul3 were present in the chromosome. The mcr-1.1 and mcr-3.7 genes were located in two plasmids of different incompatibility groups. mcr-1.1 was carried by a IncX4-type plasmid within an typical IS26-parA-mcr-1.1-pap2 cassette, while mcr-3.7 was encoded by an IncP1-type plasmid with a genetic structure of TnAs2-mcr-3.7-dgkA-IS26. No additional antibiotic resistance genes were carried by either plasmid.
Conclusion
This is the first report of an E. coli isolate co-harboring a mcr-1.1-carrying IncX4 plasmid and a mcr-3.7-carrying IncP1 plasmid. The evolution and mechanism of mcr gene co-existence need further study to assess its impact on public health.
Introduction
Colistin (polymyxin E) is a cyclic polypeptide antibiotic produced by Bacillus polymyxa.Citation1 This antibiotic is lethal to Gram-negative bacteria by targeting lipopolysaccharides (LPSs) and phospholipids in the cell membrane, inducing alterations in the cell permeability and thus leading to leakage of cellular contents and cell death.Citation2 Colistin has been considered as one of the last-resort options for the treatment of severe human infections caused by multidrug-resistant Gram-negative organisms.Citation3 It was thought that chromosomal mutation was the only mechanism to acquire resistance to colistin in bacteria.Citation4 However, the plasmid-mediated transferable colistin-resistance determinant MCR-1 was first reported in 2016 in Escherichia coli and Klebsiella pneumonia strains isolated from China.Citation5 The mcr-1 gene has been detected worldwide in recent years in dozens of bacterial species from multiple origins, which raises great clinical concerns.Citation6–Citation9 MCR proteins function as phosphoethanolamine (PEA) transferases that catalyze the addition of a PEA moiety from phosphatidylethanolamine to the head group of lipid A through a ping-pong mechanism.Citation10,Citation11 Modification of lipid A with PEA is thought to reduce the affinity of colistin to LPS, and thus confers bacterial resistance to the antibiotic.Citation11 To date, 10 mcr genes (mcr-1 to mcr-10) carried by plasmids with varied replicon types have been identified in various bacterial species.Citation5,Citation12–Citation20 Each mcr gene has multiple variants. For example, 27 mcr-1 and 30 mcr-3 variant sequences have been reported in the NCBI database.
It is common for one bacterial isolate to harbor a single mcr determinant. However, recent studies have revealed the co-existence of the mcr-1 and mcr-3 genes in E. coli and K. pneumonia strains isolated from different countries.Citation21–Citation23 These observations depict the complexities of colistin resistance dissemination mediated by MCR determinants. Here we characterized one E. coli strain isolated from dog feces that co-harbors mcr-1.1 and mcr-3.7 in different plasmids.
Materials and Methods
Bacterial Isolates and Antimicrobial Susceptibility Testing
We collected fecal samples from dogs in pet hospitals during the period from June to August 2019 in Jilin Province, China. The feces were resuspended in brain-heart infusion broth (Hopebio, Qingdao, China) followed by centrifugation at 300 rpm for 5 min at 4°C. The supernatants were plated on the MacConkey agar (Hopebio, Qingdao, China) plates containing 2 mg/L of colistin (Sigma-Aldrich, St Louis, MO, USA). Strain identification was confirmed using 16S rRNA gene sequencing. The presence of mcr genes in the bacterial isolates was tested by an established multiplex-PCR assay.Citation24 The susceptibility of the isolates to 30 antibiotics (Table S1) was measured by the Kirby-Bauer disk diffusion method. The data were analyzed according to the protocol of Clinical and Laboratory Standards Institute (CLSI) guidelines. The MICs of colistin, tigecycline and meropenem were analyzed according to the standard of the European Committee on Antibiotic Susceptibility Testing.Citation25 Multidrug resistances were defined as resistance to at least three antibiotic classes. The E. coli strain ATCC 25922 was used as the control.
Whole-Genome Sequencing, Assembly and Bioinformatics Analysis
Total genomic DNA of the isolate was prepared using Qiagen’s DNA Mini Kit with the manufacturer’s instructions (Qiagen, Hilden, Germany). The genomic DNA extracted from the isolate was qualified and quantified with a Qubit™ 3.0 Fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Whole-genome sequencing was performed using PacBio single-molecule real-time (SMRT) and Illumina Novaseq sequencing technology. PacBio RS II and Illumina Novaseq 6000 sequencer were used, respectively. The PacBio library (10 kb) and Illumina PE150 library (350 bp) were constructed. The low-quality and short reads were filtered out with the SMRT portal (version 5.0.1). The filtered high-quality reads were assembled de novo using SOAPdenovo to generate the complete chromosome and plasmids without gaps. Genome annotation was performed using GeneMarkS version 4.17 to predict the related genes. Serotypes, sequence types, FimH types and resistance genes were identified using SerotypeFinder 2.0, Multi-Locus Sequence Typing (MLST) v.2.0, FimTyper 1.0 and ResFinder v.3.2 databases from the Center for Genomic Epidemiology (CGE). In addition, antimicrobial resistance genes were confirmed using the Comprehensive Antibiotic Resistance Database (CARD) and the Antibiotic Resistance Genes Database (ARDB).
Plasmid Analysis
The plasmid replicon genotype, resistance genes and insertion sequence (IS) elements of the plasmid were identified using PlasmidFinder 2.1, ResFinder 3.2 and ISfinder. Comparative analysis of plasmids was performed and visualized using Easyfig version 2.2.3 and BLAST Ring Image Generator (BRIG).
Nucleotide Sequence Accession Numbers
The sequences of the chromosome and plasmids have been submitted in the NCBI database under the accession numbers: CP053211, CP053212 and CP053213.
Plasmid Conjugation
Conjugation experiment was performed between the colistin-resistant E. coli QDFD216 (donor) and the rifampin-resistant E. coli strain C600 (recipient) as described previously.Citation26 Transconjugants were selected on MacConkey agar plates containing 2 mg/L of colistin and 512 mg/L of rifampin. The presence of the mcr-1 and mcr-3 in the transconjugants was analysed by PCR with specific primers.
Results and Discussion
Antibiotic Resistance Profile of the E. coli Strain QDFD216 That Co-Harbors mcr-1 and mcr-3
A total of 513 colistin-resistant isolates were obtained from 91 samples of dog feces in Jilin Province, China, in 2019. All colistin-resistant isolates were screened by a multiplex PCR assay to detect the presence of mcr genes. Interestingly, PCR and sequencing showed that one isolate (designated E. coli QDFD216, isolated from one sample collected at the Veterinary Teaching Hospital of Jilin University) co-harbored the mcr-1 and mcr-3 genes. Antimicrobial susceptibility testing against a panel of antibiotics revealed that E. coli QDFD216 was resistant to penicillin, oxacillin, ampicillin, carboxypenicillin, piperacillin, cefazolin, cephalexin, cefradine, ceftriaxone, cefuroxime, cefoperazone, ceftazidime, amikacin, gentamicin, kanamycin, maddie mycin, clindamycin and colistin (MIC=16 mg/L), but remained susceptible to minocycline, norfloxacin, ofloxacin, ciprofloxacin, chloramphenicol, tigecycline, meropenem and rifampin (Table S1).
Genome Analysis of the E. coli Strain QDFD216
Next, the E. coli strain QDFD216 was subjected to whole-genome sequencing by the PacBio and Illumina platforms. The de novo assembly resulted in a circular chromosome and two circular plasmids (designated pQDFD216.1 and pQDFD216.2). The chromosome of E. coli QDFD216 was 4,577,885 bp in length and had a GC content of 50.91%. Genome annotation by GeneMarkS resulted in the identification of 4543 coding sequences. MLST, SerotypeFinder and FimTyper analyses showed that E. coli strain QDFD216 belonged to ST132, serotype O176:H11and fimH1311 (Table S2). Resistance determinants were determined by the CARD and ARDB; blaCTX-M-14, mdfA, dfrA3, acrA, acrB, tolc, and sul3 were present in the chromosome. mcr-1.1 and mcr-3.7 were found to be located on two different plasmids (pQDFD216.1 and pQDFD216.2, respectively) (Table S2).
The Genetic Context of mcr-1.1 on pQDFD216.1
The mcr-1.1 was located on pQDFD216.1 which was 33,308 bp in length and had an average GC content of 41.83%. PlasmidFinder analysis indicated that pQDFD216.1 belonged to the IncX4 incompatibility group (Table S2). pQDFD216.1 had a typical IncX4 plasmid structure, including plasmid replication (pir-type replicon), maintenance (H-NS and topB), toxin-antitoxin genes (hicA-hicB), insertion sequence elements (IS26), a partition gene (parA) and the conjugative transfer system (TraG, VirB1, VirB2, VirB4, VirB5, VirB6, VirB8-11, VirD2 and VirD4) ( and ).
Figure 1 Comparison of mcr-carrying plasmids and other closely related plasmids. (A) Comparative analysis of pQDFD216.1 with five closely related mcr-1.1-carrying plasmids including pLD93-1-MCR1 (GenBank accession: CP047664.1), p2HS-C-1 (GenBank accession: CP038181.1), p787-MCR (GenBank accession: MG825367.1), pE13-43-mcr-1 (GenBank accession: LT838201.1) and pWI2-mcr (GenBank accession: LT838201.1). pQDFD216.1 was used as the reference plasmid for BRIG. (B) Comparative analysis of pQDFD216.2 with five closely related mcr-3-carrying plasmids, pMCR3_WCHEC1943 (GenBank accession: MF678351.1), pGDZJ003-1-MCR-3 (GenBank accession: MH043625.1), pMCR3_WCHEC-LL123 (GenBank accession: MF489760.1), pZR12 (GenBank accession: MF455227.1) and pMCR_WCHEC1622 (GenBank accession: KY463452.1). The plasmid pQDFD216.2 was used as the reference plasmid for BRIG.
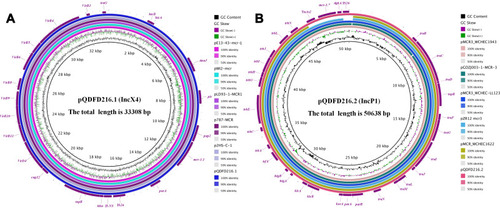
Figure 2 A linear depiction of the genetic configuration of mcr-carrying plasmids compared to other closely related plasmids. (A) Structure comparisons of pQDFD216.1, pmcr1_IncX4 (GenBank accession: KU761327.1) and pCSZ4 (GenBank accession: KX711706.1). (B) Structure comparisons of pQDFD216.2, pGDZJ003-1-MCR-3 (GenBank accession: MH043625.1) and pMCR3_WCHEC-LL123 (GenBank accession: MF489760.1). The map was generated with Easyfig.
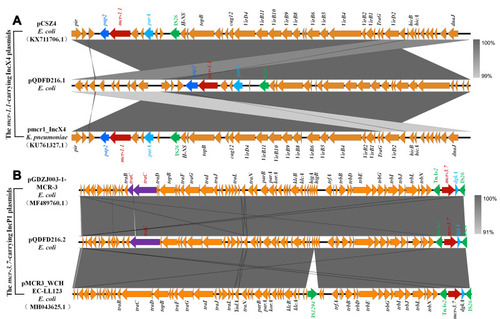
BLASTn analysis of pQDFD216.1 with the current database showed that this plasmid was highly similar to pLD93-1-MCR1 (GenBank accession: CP047664.1), p2HS-C-1 (GenBank accession: CP038181.1), p787-MCR (GenBank accession: MG825367.1), pE13-43-mcr-1 (GenBank accession: LT838201.1) and pWI2-mcr (GenBank accession: LT838201.1) (). These mcr-1-carrying plasmids showed 99.91–99.95% nucleotide sequence identity with pQDFD216.1. The mcr-1.1 gene in pQDFD216.1 is carried by the genetic structure IS26-parA-mcr-1.1-pap2 ( and ), which is nearly identical to the mcr-1.1-carrying IncX4 plasmids pmcr1_IncX4 (GenBank accession: KU761327.1) and pCSZ4 (GenBank accession: KX711706.1) ().
Similar to other mcr-1.1-carrying IncX4-type plasmids, the typical IS26-parA-mcr-1.1-pap2 cassette was identified in pQDFD216.1 ().Citation27 The ISApl1 insertion sequence (IS30 family) was lost in the mcr-1 gene cassette.Citation28 Generally, ISApl1 is a highly active insertion element and a key component required for the mobilization of the gene-cassette containing the mcr-1 gene.Citation29,Citation30 The absence of ISApl1 along mcr-1 genes could reduce further chromosomal integration.Citation28 Previous studies have shown that IncX4-type plasmids play a vital role in increasing the spread of plasmid-mediated mcr genes.Citation31,Citation32 Remarkably, in all the mcr-1-carrying IncX4-type plasmids, ISApl1 in front of mcr-1 was lost.Citation33 Therefore, the loss of the composite transposon ISApl1 might increase the stability of the mcr gene in IncX4 plasmids, and promote the widespread dissemination of the mcr gene.
The Genetic Contexts of mcr-3.7 on pQDFD216.2
The mcr-3.7-bearing plasmid pQDFD216.2 was 50,638 bp in size and had a GC content of 47.08%. PlasmidFinder analysis revealed that pQDFD216.2 belonged to the IncP1 type plasmid (Table S2). The IncP1 plasmids have a broad host range and are widespread in Gram-negative organisms, including E. coli, K. pneumonia and S. enteric.Citation34–Citation36 The association of mcr-3.7 with the IncP plasmid is worrisome because it may have the potential to promote the dissemination of mcr-3 in Gram-negative pathogens.
The plasmid pQDFD216.2 did not carry any other resistance gene besides mcr-3.7. The pQDFD216.2 plasmid has the conjugative transfer/type IV secretion system (tra and trb), replication initiator (trfA), replication and partition genes (parA and parB), toxin-antitoxin genes (higA-higB), host-lethal genes (klc) and their regulators (kor) ( and ). BLASTn analysis showed that plasmids pMCR3_WCHEC1943 (GenBank accession: MF678351.1), pGDZJ003-1-MCR-3 (GenBank accession: MH043625.1), pMCR3_WCHEC-LL123 (GenBank accession: MF489760.1), pZR12 (GenBank accession: MF455227.1) and pMCR_WCHEC1622 (GenBank accession: KY463452.1) were most similar to pQDFD216.2 (). These plasmids showed 99.96–99.98% nucleotide sequence identity with pQDFD216.2. The mcr-3.7 gene was located in pQDFD216.2 with the structure TnAs2-mcr-3.7-dgkA-IS26, which was also found in the mcr-3.7-carrying IncP1 plasmids pMCR3_WCHEC-LL123 and pGDZJ003-1-MCR-3 (). However, unlike pQDFD216.2, pGDZJ003-1-MCR-3 partially lacked the conjugal transfer gene TraC of the IncP-type plasmid (). Furthermore, when compared with pMCR3_WCHEC-LL123, the mobile genetic element IS1294 was absent in pQDFD216.2 ().
Plasmid Conjugation
Conjugation experiment was performed to determine the transferability of the pQDFD216.1 and pQDFD216.2. The mcr-harboring plasmids in the E. coli strain QDFD216 can be transferred to the recipient E. coli strain C600 at a frequency of 1.2x10−3 per donor cell. One hundred transconjugants were selected for PCR to characterize the presence of mcr genes. Thirty-two transconjugants contained only one mcr gene (19 for mcr-1 and 13 for mcr-3). Strikingly, 68 out of the 100 transconjugants were found to harbor both mcr-1 and mcr-3, indicating the co-transfer of pQDFD216.1 and pQDFD216.2.
The rapid dissemination of mcr genes in bacteria has raised great public concerns. Recent publications described the co-occurrence of different mcr alleles within a single bacterial strain of diverse species isolated from various regions,Citation21,Citation22,Citation37,Citation38 adding complexity and flexibility to the spread of colistin resistance. Based on the available literatures, mcr-1/mcr-3 are the most frequently detected combination of co-existing mcr genes,Citation21–Citation23,Citation39 while at a lower detection frequency, the co-occurrence of mcr-3/mcr-8 as well as mcr-1/mcr-5 has also been described.Citation37,Citation38 Further molecular surveillance of colistin resistance may reveal more combinations of co-existing mcr genes, and their influences on public health require further assessment. The presence of mcr genes in companion animals has been reported previously, which may cause possible transmission of mcr-harboring bacterial strains between companion animals and humans.Citation40,Citation41 To the best of our knowledge, our study reports the first bacterial strain isolated from companion animals that co-harbors mcr-1 and mcr-3, adding another layer of threat of colistin-resistant bacteria from companion animals to human health.
Conclusions
In this study, we identified the co-occurrence of a mcr-1.1-carrying IncX4 plasmid and a mcr-3.7-carrying IncP1 plasmid in E. coli strain QDFD216 isolated from dog feces in China. Compared to other studies worldwide, our data provide insight into the dissemination routes of plasmid-mediated colistin resistance. Furthermore, the co-existence of different mcr genes in the same isolate presents a great challenge for infection control in multidrug-resistant Gram-negative organisms.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Acknowledgments
We thank Dr. Xiangru Wang at College of Animal Science and Veterinary Medicine, Huazhong Agricultural University for providing us the rifampin-resistant E. coli strain C600. This work was supported by the Thousand Young Talents Program of the Chinese government (JZQ) and startup fund from Jilin University (JZQ).
Disclosure
The authors declare no conflicts of interest.
References
- Falagas ME, Kasiakou SK, Saravolatz LD. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clinical Infectious Diseases. 2005;40(9):1333–1341. doi:10.1086/42932315825037
- Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther. 2012;10(8):917–934. doi:10.1586/eri.12.7823030331
- Panknin HT. [Multidrug-resistant bacteria require new treatment options]. Med Monatsschr Pharm. 2015;38(12):517–521. German.26837160
- Anjum MF, Duggett NA, AbuOun M, et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother. 2016;71(8):2306–2313. doi:10.1093/jac/dkw14927147305
- Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-726603172
- Anyanwu MU, Jaja IF. Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: a review. Int J Environ Res Public Health 2020;17:3. doi:10.3390/ijerph17031028
- Gröndahl-Yli-Hannuksela K, Lönnqvist E, Kallonen T, et al. The first human report of mobile colistin resistance gene, mcr-1, in Finland. APMIS. 2018;126(5):413–417. doi:10.1111/apm.1283429696722
- Robert M, Renaud A, Hendricx S, Beyrouthy R, Blanckaert K. Premier cas de transmission croisée de K. pneumoniae BLSE mcr-1 en France. Médecine et Maladies Infectieuses. 2018;48(3):226–229. doi:10.1016/j.medmal.2018.01.00629422424
- Wang W, Baloch Z, Zou M, et al. Complete genomic analysis of a salmonella enterica serovar typhimurium isolate cultured from ready-to-eat pork in china carrying one large plasmid containing mcr-1. Front Microbiol. 2018;9:616. doi:10.3389/fmicb.2018.0061629755416
- Gao R, Hu Y, Li Z, et al. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 2016;12(11):e1005957. doi:10.1371/journal.ppat.100595727893854
- Zhang H, Hou M, Xu Y, et al. Action and mechanism of the colistin resistance enzyme MCR-4. Communications Biol. 2019;2:36. doi:10.1038/s42003-018-0278-1
- Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveillance European Communicable Disease Bulletin. 2016;21:27.
- Yin W, Li H, Shen Y, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in escherichia coli. mBio. 2017;8:3.
- Carattoli A, Villa L, Feudi C, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveillance European Communicable Disease Bulletin. 2017;22:31.
- Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317–3324. doi:10.1093/jac/dkx32728962028
- AbuOun M, Stubberfield EJ, Duggett NA, et al. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2017;72(10):2745–2749. doi:10.1093/jac/dkx28629091227
- Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(7):1791–1795. doi:10.1093/jac/dky11129912417
- Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae.Emerg Microbes Infect.2018;7(1):122. doi:10.1038/s41426-018-0124-z
- Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. mBio. 2019;10:3. doi:10.1128/mBio.00853-19
- Wang C, Feng Y. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect 2020;9(1):508–516. doi:10.1080/22221751.2020.1732231
- Li R, Zhang P, Yang X, et al. Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain. J Antimicrob Chemother. 2019;74(6):1517–1520. doi:10.1093/jac/dkz05830793748
- Creighton J, Anderson T, Howard J, Dyet K, Ren X, Freeman J. Co-occurrence of mcr-1 and mcr-3 genes in a single Escherichia coli in New Zealand. J Antimicrob Chemother. 2019;74(10):3113–3116. doi:10.1093/jac/dkz31131339995
- Yu Y, Andrey DO, Yang RS, et al. A Klebsiella pneumoniae strain co-harbouring mcr-1 and mcr-3 from a human in Thailand. J Antimicrob Chemother. 2020.
- Lescat M, Poirel L, Nordmann P. Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn Microbiol Infect Dis. 2018;92(4):267–269. doi:10.1016/j.diagmicrobio.2018.04.01030220493
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 10.0, 2020 http://www.eucast.org/clinical_breakpoints/. Accessed 102, 2020.
- Peng Z, Li XS, Hu ZZ, et al. Characteristics of carbapenem-resistant and colistin-resistant escherichia coli co-producing NDM-1 and MCR-1 from Pig Farms in China. Microorganisms. 2019;7:11. doi:10.3390/microorganisms7110482
- Veldman K, van Essen-zandbergen A, Rapallini M, et al. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J Antimicrobial Chemotherapy. 2016;71(8):2340–2342. doi:10.1093/jac/dkw181
- Sun J, Fang L-X, Wu Z, et al. Genetic analysis of the IncX4 plasmids: implications for a Unique Pattern in the mcr-1 Acquisition. Sci Rep. 2017;7(1):424.28336940
- Snesrud E, Ong A, Corey A. Analysis of serial isolates of mcr-1- positive escherichia coli reveals a highly active ISApl1 transposon. Antimicrob Agents Chemother. 2017;61(5):AAC.00056–00017. doi:10.1128/AAC.00056-17
- Li R, Xie M, Zhang J, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72(2):393–401. doi:10.1093/jac/dkw41128073961
- Migura-Garcia L, González-López JJ, Martinez-Urtaza J, et al. mcr-colistin resistance genes mobilized by IncX4, IncHI2, and IncI2 plasmids in escherichia coli of pigs and white stork in Spain. Front Microbiol. 2020;10:3072. doi:10.3389/fmicb.2019.0307232010114
- Ageevets V, Lazareva I, Mrugova T, Gostev V, Sidorenko S. IncX4 plasmids harboring mcr-1 genes: further dissemination. J Global Antimicrobial Resistance. 2019;18.
- Sun J, Fang LX, Wu Z, et al. Genetic analysis of the incx4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep. 2017;7(1):424. doi:10.1038/s41598-017-00095-x28336940
- Lalaoui R, Djukovic A, Bakour S, et al. Detection of plasmid-mediated colistin resistance, mcr-1 gene, in Escherichia coli isolated from high-risk patients with acute leukemia in Spain. J Infection Chemotherapy. 2019;25:605–609. doi:10.1016/j.jiac.2019.03.007
- Kieffer N, Aires-de-Sousa M, Nordmann P, Poirel L. High Rate of MCR-1-producing escherichia coli and klebsiella pneumoniae among pigs, Portugal. Emerg Infect Dis. 2017;23(12):2023–2029. doi:10.3201/eid2312.17088329148380
- Lu X, Hu Y, Luo M, et al. MCR-1.6, a New MCR variant carried by an Incp plasmid in a colistin-resistant salmonella enterica serovar typhimurium isolate from a healthy individual. Antimicrob Agents Chemother. 2017;61(5):e0263202616. doi:10.1128/AAC.02632-16
- Hadjadj L, Baron SA, Olaitan AO, Morand S, Rolain JM. Co-occurrence of variants of mcr-3 and mcr-8 genes in a klebsiella pneumoniae isolate from laos. Front Microbiol. 2019;10:2720. doi:10.3389/fmicb.2019.0272031849875
- Fukuda A, Sato T, Shinagawa M, et al. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int J Antimicrob Agents. 2018;51(1):163–164. doi:10.1016/j.ijantimicag.2017.11.01029180277
- Sun D, Jin S, Wang J, et al. Multidrug-resistant escherichia coli strain isolated from swine in china harbors mcr-3.1 on a Plasmid of the IncX1 Type That Cotransfers with mcr-1.1. Foodborne Pathog Dis. 2020. doi:10.1089/fpd.2019.2769
- Zhang XF, Doi Y, Huang X, et al. Possible transmission of mcr-1-harboring escherichia coli between companion animals and human. Emerg Infect Dis. 2016;22(9):1679–1681. doi:10.3201/eid2209.16046427191649
- Lei L, Wang Y, Schwarz S, et al. mcr-1 in enterobacteriaceae from companion animals, Beijing, China, 20122016. Emerg Infect Dis. 2017;23(4):710–711. doi:10.3201/eid2304.16173228322714
