Abstract
Background and Aim
The association of microbiota changes with sensitive skin remains controversial until now. Although a strong correlation is detected between skin microbiota distribution and biophysical parameters, there is little knowledge on the link between sensitive skin and skin microbiota in Chinese women. This study aimed to unravel the correlation between facial skin microbiota distribution and skin barriers in Chinese women with sensitive skin.
Materials and Methods
In total, 34 volunteers were enrolled, including 24 subjects with sensitive skin (SS group) and 10 subjects with non-sensitive skin (NS group). The cuticle moisture content, transepidermal water loss (TEWL), and facial skin sebum secretion were measured, and the facial skin surface morphology was evaluated. Sensitive skin samples were collected from the facial (SS-F group) and chest skin of subjects in the SS group (SS-C group), while non-sensitive skin samples were collected from the facial skin of subjects in the NS group (NS-F group). All skin samples were subjected to 16S rRNA sequencing.
Results
16S rRNA sequencing detected Actinobacteria, Firmicutes, and Proteobacteria as the three most common microbiota phyla and Propionibacterium, Paracoccus, and Corynebacterium as the three most common microbiota genera, and there were no significant differences in the relative frequency of Actinobacteria, Firmicutes, or Proteobacteria, or Propionibacterium, Paracoccus, or Corynebacterium among the SS-F, SS-C, and NS-F groups (P>0.05). We detected no significant difference in the diversity of bacterial communities among the SS-F, SS-C, and NS-F groups; however, the Shannon’s diversity index was significantly higher in the NS-F group than in the SS-C group. In addition, Spearman correlation analysis showed a correlation between the microbiota genera and skin physiological parameters (P<0.05).
Conclusion
This study preliminarily unravels the skin microbiota of sensitive skin using a high-throughput tool, and there are no microbiota genera with strong associations with skin physiological parameters.
Introduction
As the first line of defense against pathogens, skin protects organisms from external physical, chemical, and microbial invasions,Citation1 and maintains a steady state of skin barriers, including physical barrier, chemical barrier, and more importantly, microbial barrier.Citation2 Previous studies have demonstrated the diversity of bacteria on the skin surface and in superficial skin layers,Citation3–Citation5 and have identified microbes with diverse relative abundances as a cause of skin disorders.Citation6–Citation10
Sensitive skin syndrome is a subjective discomfort, and a hyper-responsiveness to external stimuli, sometimes accompanied by erythema and objective manifestations.Citation11 Sensitive skin condition is not exactly the same as skin diseases; however, multiple skin disorders present a sensitive condition.Citation12 To date, the pathological and physiological mechanisms of sensitive skin syndrome are not fully clear; however, the hyper-responsiveness of nerves and blood vessels associated with abnormal skin barriers has been demonstrated to contribute to the pathogenesis of sensitive skin.Citation11,Citation13,Citation14 In addition, the involvement of microbiota changes with sensitive skin remains in debate until now.Citation15,Citation16 Our previous study detected a strong correlation between skin microbiota distribution and skin biophysical parameters, including transepidermal water loss (TEWL), pH, skin scaliness and roughness, sebum and hydration levels, at different body sites and under diverse skin environments among Chinese women living in Shanghai, China.Citation17 Nevertheless, there is still little knowledge pertaining to the link between sensitive skin and skin microbiota in Shanghai women. The present study was therefore designed with aims to unravel the microbiota difference between subjects showing a positive lactic acid sting test and with self-perceived sensitive skin and controls, and examine the correlation between microbiota distribution and physiological parameters of skin in a Chinese female population.
Materials and Methods
Study Subjects
Chinese women that went to Shanghai Skin Disease Hospital (Shanghai, China) for counseling of skin problems since February 2018 were recruited. The inclusion criteria involved 1) Chinese women showing positive in lactic acid sting test,Citation18 with self-perceived sensitive skin (confirmed in the detailed inquiry of a medical history and recorded in medical records), and at ages of 18–60 years, which were assigned as the sensitive skin (SS) group; 2) Chinese women showing negative in lactic acid sting test, with self-perceived non-sensitive skin (confirmed in the detailed inquiry of a medical history and recorded in medical records), and at ages of 18–60 years, which were categorized as the non-sensitive skin (NS) group; 3) subjects in both the SS group and the NS group had a habit of using facial skin care products, but without skin diseases; 4) all subjects read and signed the informed consent form prior to the enrollment, participated in the study as required, and agreed to maintain the habit of using facial skin care products during the study period (face washing was no longer allowed 12 hours prior to collection of microbes in the formal study). Those who met the following criteria were excluded from the study: 1) lactating or pregnant women; 2) current use of corticosteroids, angiotonics, antibiotics, anti-inflammatory drugs, or systemic or local use of these agents within 2 weeks prior to the study; 3) confirmed skin disorders on the face or at other sites; 4) participation in other systemic drug trials within 2 weeks prior to the study, current use, or use of anti-histamines within 1 week prior to the study, a medical history of acute or chronic skin diseases or drug allergy, or physical status which may interfere with the use of the test product and/or affect study results.
Finally, a total of 34 volunteers were enrolled in this study, including 24 subjects in the SS group and 10 subjects in the NS group. The women had a mean age of 42.21±9.86 years (range= 19–58 years) in the SS group and 46.3±12.18 years (range=18–58 years) in the NS group, and the age distribution was comparable between the two groups (P>0.05).
Measurement of Facial Skin Conditions
Face washing was no longer allowed 12 hours prior to the study and after participation in the study. After resting for 20–30 minutes, subjects’ facial skin conditions were measured with noninvasive instruments. The cuticle moisture content was measured using a Corneometer CM825 cuticle moisture content tester (Courage & Khazaka Electronic GmbH; Cologne, Germany), and the TEWL was determined with an AquaFluxAF200 TEWL tester (Biox Systems, Ltd.; London, UK). In addition, the facial skin sebum secretion was measured with a Sebumeter SM810 tester (Courage & Khazaka Electronic GmbH; Cologne, Germany), and the facial skin surface morphology was evaluated using a VC98 skin scanner (Courage & Khazaka Electronic GmbH; Cologne, Germany). All measurements were performed in a random left–right manner according to the randomized table.
Detection of Facial Skin Surface Microbiota
Following noninvasive detection of skin, facial skin microbes were collected from the other side of the face. Sensitive skin samples were collected from the facial (SS-F group) and chest skin of subjects in the SS group (SS-C group), while non-sensitive skin samples were collected from the facial skin of subjects in the NS group (NS-F group). Then, all skin samples were subjected to 16S rRNA sequencing performed by the Chinese National Human Genome Center at Shanghai (Shanghai, China). The differences of facial skin microbiota were compared among the three groups through sample pretreatment, operational taxonomic units (OUT) analysis, and taxonomic analysis.
Ethical Statement
This study was approved by the Ethics Review Committee of Shanghai Skin Disease Hospital (approval no.: 2018–16). All participants were informed of the purpose, experimental procedures and potential risks of the study, and signed an informed consent form. All experiments were performed in accordance with the Declaration of Helsinki and National Regulations for Ethics of Biological Medical Sciences on Human Studies released by the Ministry of Health, People’s Republic China.
Statistical Analysis
All measurement data were described as mean±standard deviation (SD), and all categorical data were expressed as proportions. The STAMP software was employed to calculate means and SD, and compute the skin microbial differences among different groups at phylum and genus levels.Citation19 Differences of means were tested for statistical significance with paired t-test between groups and with one-way analysis of variance (ANOVA) among groups. The correlation between facial skin microbiota and skin barriers was evaluated using a Spearman correlation analysis, and a correlation coefficient of >0.35 or <‒0.35 was defined as significantly correlated (weak correlation, correlation coefficient of <0.35 or >‒0.35; medium correlation, correlation coefficient of >0.35 and <0.68 or >‒0.68 and <‒0.35; strong correlation, correlation coefficient of >0.68 or <‒0.68). All statistical analyses were performed using the software SPSS version 21.0 (SPSS, Inc.; Chicago, IL, USA), and a P-value of <0.05 was considered statistically significant.
Results
Comparison of Physiological Parameters Between Sensitive and Non-Sensitive Facial Skin Surface
The sting score and abnormal sensation score (total score of sting, pruritus, burning heat sensation, and other abnormal sensations) were both significantly greater in the SS group than in the NS group (P<0.01); however, no significant differences were detected between the two groups in terms of TEWL, sebum secretion, or cuticle moisture content (P>0.05) (). These data indicate that there are significant differences in skin barrier-associated physiological parameters between the SS group and the NS group.
Table 1 Comparison of Physiological Parameters Between Sensitive and Non-Sensitive Facial Skin Surface
Skin Microbiota Characteristics
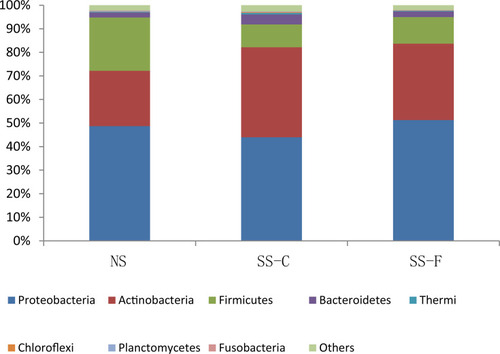
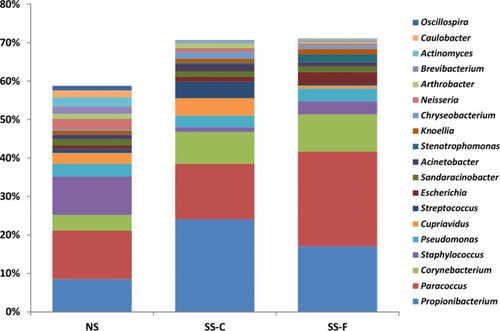
To unravel the correlation between microbiota distribution and physiological parameters of skin, we first investigated the skin microbiota characteristics. 16S rRNA sequencing showed Actinobacteria, Firmicutes, and Proteobacteria as the three most common microbiota phyla; however, there was no significant difference detected in the relative frequency of Actinobacteria, Firmicutes, or Proteobacteria among the SS-F, SS-C, and NS-F groups (P>0.05). The highest frequency of Actinobacteria was detected in the SS-C group, with the lowest distribution seen in the NS-F group, and the greatest frequency of Firmicutes was found in the NS-F group, with the lowest detected in the SS-C group, while the highest frequency of Proteobacteria was identified in the SS-F group, with the lowest detected in the SS-C group ().
Figure 1 16S rRNA sequencing detects phylum-wide distribution of microbes on the facial and chest skin of subjects with sensitive skin and facial skin of subjects without sensitive skin.
16S rRNA sequencing detected 19 microbiota genera with high contents, with a total content of 58–70%, and the three most common microbiota genera included Propionibacterium, Paracoccus, and Corynebacterium; however, there was no significant difference in the relative frequency of Propionibacterium, Paracoccus, or Corynebacterium among the SS-F, SS-C, and NS-F groups (P>0.05). The highest relative frequency of Propionibacterium was detected in the SS-C group, with the lowest seen in the NS-F group, and the greatest frequency of Paracoccus was found in the SS-F group, with the lowest seen in the SS-C group, while the highest frequency of Corynebacterium was identified in the SS-F group, with the lowest in the NS-F group ().
Figure 2 16S rRNA sequencing detects genus-wide distribution of microbes on the facial and chest skin of subjects with sensitive skin and facial skin of subjects without sensitive skin.
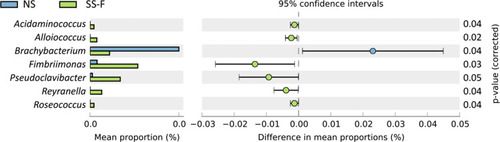
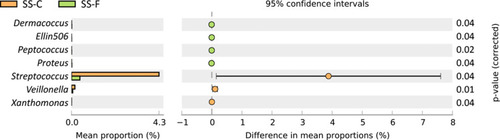
Then, we compared the difference in the frequency of microbiota between the SS-F and NS-F groups and between the SS-F and SS-C groups at a genus level. The frequency of Acidaminococcus, Alloiococcus, Fimbriimonas, Pseudoclavibacter, Reyranella, and Roseococcus was significantly lower in the NS-F group than in the SS-F group (P<0.05), while a higher frequency of Bradyrhizobium was seen in the NS-F group than in the SS-F group (P<0.05) ( and ). In addition, the frequency of Dermacoccus, Ellin506, Peptococcus, and Proteus was lower in the SS-C group than in the SS-F group (P<0.05), while a higher frequency of Streptococcus, Veillonella, and Xanthomonas was seen in the SS-C group than in the SS-F group (P<0.05) ( and ). Our data showed that all microbiota genera with intergroup statistical significance in the relative freqeuncy were rare bacteria in skin microbiota.
Table 2 Comparison of the Relative Frequency of Microbe Genus Between the NS-F and SS-F Groups
Table 3 Comparison of the Relative Frequency of Microbe Genus Between the SS-C and SS-F Groups
Skin Microbiota Diversity
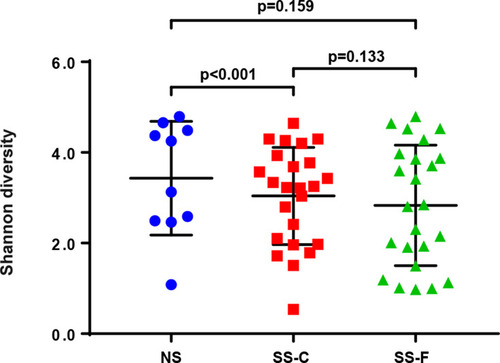
Shannon’s diversity index was employed to compare the diversity of bacterial communities among the three groups, and a greater Shannon’s index means a higher diversity. The greatest Shannon’s index was detected in the NS-F group, and the lowest was seen in the SS-F group (). We detected no significant difference in the diversity of bacterial communities among the three groups; however, the Shannon’s diversity index was significantly higher in the NS-F group than in the SS-C group (P<0.001).
Correlation Between Skin Microbiota and Physiological Properties of Skin
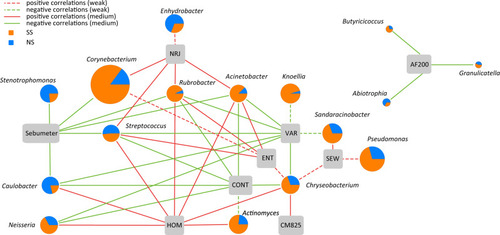
Spearman correlation analysis showed that the microbiota genera significantly correlated with the physiological parameters of the skin (P<0.05) ().
Figure 6 Correlation between skin microbiota and physiological parameters of skin. Dashed lines indicate a weak correlation (correlation coefficient of <0.35 or >‒0.35), and solid lines denote a medium correlation (correlation coefficient of >0.35 and <0.68 or >‒0.68 and <‒0.35). Sebumeter, skin sebum secretion; NRJ, energy; HOM, homology; CONT, contrast; ENT, entropy; VAR, variety; CM825, cuticle moisture content; SEW, surface evaluation of wrinkles; AF200, transepidermal water loss.
Discussion
Our previous study detected a strong correlation between biophysical properties of skin and the distribution of common microbes at different body sites and skin environments among Shanghai women, and TEWL, pH, skin scaliness and roughness, sebum, and hydration levels were found to pose a great impact on the distribution of microbial colonies.Citation17 In addition, exposed and non-exposed sites also have large effects on microbiota distribution.Citation20,Citation21 In this study, we, therefore, aimed to investigate the biophysical parameters which have proven useful in describing sensitive skin, to see if they had effects on skin microbiota. Besides the face (a typical site to study sensitive skin) that was selected as an exposed site, we also selected the chest as a non-exposed control.
Although sensitive skin is pretty similar to non-sensitive skin in appearance, non-invasive physiological parameters are effective to distinguish between sensitive and non-sensitive skin.Citation12 There are some differences between sensitive skin and non-sensitive skin in specific biochemical compositions of the skin surface.Citation22 For instance, lower pyrrolidone carboxylic acid, bleomycin hydrolase, and transglutaminase activities, and smaller and less mature keratinocytes are seen in sensitive skin than in non-sensitive skin, and all these factors contribute to the high permeability of surface microbiota, and aggravate the symptoms of sensitive skin.Citation22 It is therefore hypothesized that the microbiota of sensitive skin has its specific characteristics.
In this study, 16S RNA sequencing detected that Actinobacteria, Firmicutes, and Proteobacteria were the three most common microbiota phyla on the facial skin surface. Although a mild variation was seen in the relative frequency of Actinobacteria, Firmicutes, or Proteobacteria among the SS-F, SS-C, and NS-F groups; however, there was no significant difference detected among these three groups (P>0.05). Similarly, we observed a variation in the relative frequency of Propionibacterium, Paracoccus, and Corynebacterium among the SS-F, SS-C, and NS-F groups; however, there was no significant difference detected among these three groups (P>0.05). Our findings uncover a slight difference in the microbiota changes of resident bacteria. Further studies including much more study samples were required to validate the conclusion from this study.
In a previous large-scale study on bacterial strain culture, non-sensitive skin and sensitive skin were compared in terms of skin microbial colonies, and gender-specific differences were identified.Citation21 However, results from a recent study showed no significant correlation between non-sensitive and sensitive skin phenotypes in terms of bacterial phyla, bacterial genera, or dominant bacterial species.Citation23 In this study, we detected that the bacteria that showed significant differences in the relative frequency between the sensitive skin and non-sensitive skin were all rare bacteria, and most of these rare bacteria were found to colonize in sensitive skin. Such a finding is not contradictory to the higher microbiota diversity in non-sensitive skin. This is because the remarkable decline of the microbiota diversity of sensitive skin is relative to non-sensitive skin creates the opportunity for specific rare bacteria to colonize on sensitive skin. Thus, the significant increase of rare bacteria is probably a secondary phenomenon of the lower microbiota diversity of sensitive skin.
In the present study, we examined the correlation between skin microbiota and skin physiological parameters; however, we found no bacterial genus that had strong associations with skin physiological parameters. Skin hydration level, TEWL, and skin surface topography are considered as indirect parameters that are used to describe skin microbiota, and those showing significant associations with skin microbiota include the mobile water on skin surface, the sebum level on each type of skin, and the pH value of the skin surface.Citation3 Notably, the mobile water has shown a critical role in microbial colonization on skin surface. Traditional moisturizers improve the hydration level of the skin surface through combining water with skin cells, without increasing the mobile water content. Further sensitive skin studies introducing more direct parameters that may describe sensitive skin seem justified.
It has been reported that intestinal microbiota disorders may promote the development and deterioration of the symptoms of irritable bowel syndrome,Citation24 and intestinal microbiota may affect the two-way communication between the enteric nervous system and central nervous system.Citation25 Previous studies have demonstrated that immune system activation and proinflammatory cytokines may indirectly affect skin.Citation26,Citation27 This may be the possible mechanism underlying the indirect impact of intestinal skin microbiota on sensitive skin.Citation28 For this reason, an important key to further understanding of the microbiota changes of sensitive skin is to investigate the intestinal or microbiota status of sensitive skin and examine the immune status.
Previous clinical studies have demonstrated that plant-derived ingredients or oils remarkably improve inflammatory skin disorders, including acne.Citation29,Citation30 However, whether the ingredients of the skin care products improve the rash in patients with acne through altering the skin microbiota remains unknown until now, which deserves further investigations.
Conclusions
The results of the present study demonstrate that Actinobacteria, Firmicutes, and Proteobacteria are the three most common microbe phyla and Propionibacterium, Paracoccus, and Corynebacterium are the three most common microbe genera, and there were no significant differences in the relative frequency of Actinobacteria, Firmicutes, or Proteobacteria, or in the relative frequency of Propionibacterium, Paracoccus, or Corynebacterium among the SS-F, SS-C, and NS-F groups (P>0.05). Our data show that the microbes with significant differences in the relative frequency between the sensitive skin and non-sensitive skin are all rare bacteria in skin microbiota. Although no significant difference is detected in the diversity of bacterial communities among the SS-F, SS-C, and NS-F groups, there is a higher Shannon’s diversity index in the NS-F group than in the SS-C group. In addition, we did not identify the microbe genera that had strong associations with the physiological parameters of the skin. This study preliminarily unravels the skin microbiota of sensitive skin using a high-throughput technique, which provides insights into the management of sensitive skin.
Disclosure
The authors declare no conflicts of interest for this work.
References
- Hwa C, Bauer EA, Cohen DE. Skin biology. Dermatol Ther. 2011;24(5):464–470. doi:10.1111/j.1529-8019.2012.01460.x22353152
- Jensen JM, Proksch E. The skin’s barrier. Giornale italiano di dermatologia e venereologia. 2009;144(6):689–700.19907407
- Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi:10.1038/nrmicro253721407241
- Grice EA, Kong HH, Conlan S, NISC Comparative Sequencing Program, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192.19478181
- Egert M, Simmering R. The Microbiota of the human skin. Adv Exp Med Biol. 2016;902:61–81.27161351
- Ellis SR, Nguyen M, Vaughn AR, et al. The skin and gut microbiome and its role in common dermatologic conditions. Microorganisms. 2019;7(11):550. doi:10.3390/microorganisms7110550
- Picardo M, Ottaviani M. Skin microbiome and skin disease: the example of rosacea. J Clin Gastroenterol. 2014;48(Suppl 1):S85–S86. doi:10.1097/MCG.000000000000024125291137
- Kandil A, Hanora A, Azab M, Enany S. Proteomic analysis of bacterial communities associated with atopic dermatitis. J Proteomics. 2020;229:103944. doi:10.1016/j.jprot.2020.10394432822829
- Paller AS, Kong HH, Seed P, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):26–35. doi:10.1016/j.jaci.2018.11.01530476499
- Shi B, Bangayan NJ, Curd E, et al. The skin microbiome is different in pediatric versus adult atopic dermatitis. J Allergy Clin Immunol. 2016;138(4):1233–1236. doi:10.1016/j.jaci.2016.04.05327474122
- Berardesca E, Farage M, Maibach H. Sensitive skin: an overview. Int J Cosmet Sci. 2013;35(1):2–8. doi:10.1111/j.1468-2494.2012.00754.x22928591
- Richters R, Falcone D, Uzunbajakava N, Verkruysse W, van Erp P. van de Kerkhof P. What is sensitive skin? A systematic literature review of objective measurements. Skin Pharmacol Physiol. 2015;28(2):75–83. doi:10.1159/00036314925322670
- Farage MA, Maibach HI. Sensitive skin: closing in on a physiological cause. Contact Dermatitis. 2010;62(3):137–149. doi:10.1111/j.1600-0536.2009.01697.x20565500
- Primavera G, Berardesca E. Sensitive skin: mechanisms and diagnosis. Int J Cosmet Sci. 2005;27(1):1–10. doi:10.1111/j.1467-2494.2004.00243.x18492176
- Seite S, Misery L. Skin sensitivity and skin microbiota: is there a link? Exp Dermatol. 2018;27(9):1061–1064. doi:10.1111/exd.1368629781556
- Misery L, Weisshaar E, Brenaut E, et al. Special Interest Group on sensitive skin of the International Forum for the Study of Itch (ISFI). Pathophysiology and management of sensitive skin: position paper from the special interest group on sensitive skin of the International Forum for the Study of Itch (IFSI). J Eur Acad Dermatol Venereol. 2020;34(2):222–229.31660659
- Li X, Yuan C, Xing L, Humbert P. Topographical diversity of common skin microflora and its association with skin environment type: an observational study in Chinese women. Sci Rep. 2017;7(1):18046. doi:10.1038/s41598-017-18181-529273721
- Chen SY, Yin J, Wang XM, Liu YQ, Gao YR, Liu XP. A new discussion of the cutaneous vascular reactivity in sensitive skin: A sub-group of SS? Skin Res Technol. 2018;24(3):432–439. doi:10.1111/srt.1244629396896
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi:10.1093/bioinformatics/btu49425061070
- Raj N, Voegeli R, Rawlings AV, et al. A fundamental investigation into aspects of the physiology and biochemistry of the stratum corneum in subjects with sensitive skin. Int J Cosmet Sci. 2017;39(1):2–10. doi:10.1111/ics.1233427079667
- Hillion M, Mijouin L, Jaouen T, et al. Comparative study of normal and sensitive skin aerobic bacterial populations. Microbiologyopen. 2013;2(6):953–961. doi:10.1002/mbo3.13824151137
- Buhé V, Vié K, Guéré C, et al. Pathophysiological study of sensitive skin. Acta Derm Venereol. 2016;96(3):314–318. doi:10.2340/00015555-223526337000
- Keum HL, Kim H, Kim HJ, et al. Structures of the skin microbiome and mycobiome depending on skin sensitivity. Microorganisms. 2020;8(7):1032. doi:10.3390/microorganisms8071032
- Takakura W, Pimentel M. Small intestinal bacterial overgrowth and irritable bowel syndrome - an update. Front Psychiatry. 2020;11:664. doi:10.3389/fpsyt.2020.0066432754068
- Kuwahara A, Matsuda K, Kuwahara Y, Asano S, Inui T, Marunaka Y. Microbiota-gut-brain axis: enteroendocrine cells and the enteric nervous system form an interface between the microbiota and the central nervous system. Biomed Res. 2020;41(5):199–216. doi:10.2220/biomedres.41.19933071256
- Yu SH, Bordeaux JS, Baron ED. The immune system and skin cancer. Adv Exp Med Biol. 2014;810:182–191. doi:10.1007/978-1-4939-0437-2_1025207366
- Nosenko MA, Ambaryan SG, Drutskaya MS. Proinflammatory cytokines and skin wound healing in mice. Mol Biol (Mosk). 2019;53(5):741–754. doi:10.1134/S002689331905012131661475
- Ma YF, Yuan C, Jiang WC, Wang XL, Humbert P. Reflectance confocal microscopy for the evaluation of sensitive skin. Skin Res Technol. 2017;23(2):227–234. doi:10.1111/srt.1232727782310
- Mazzarello V, Donadu MG, Ferrari M, et al. Treatment of acne with a combination of propolis, tea tree oil, and Aloe vera compared to erythromycin cream: two double-blind investigations. Clin Pharmacol. 2018;10:175–181. doi:10.2147/CPAA.S18047430588129
- Mazzarello V, Gavini E, Rassu G, et al. Clinical assessment of new topical cream containing two essential oils combined with tretinoin in the treatment of acne. Clin Cosmet Investig Dermatol. 2020;13:233–239. doi:10.2147/CCID.S236956