Abstract
Introduction
We assessed the effect of Mycobacterium tuberculosis (MTB) bacterial load on Xpert MTB/RIF accuracy for detection of rifampicin (RIF)-resistant MTB in bronchoalveolar lavage fluid (BALF) specimens obtained at a national tuberculosis (TB) specialized hospital in Beijing, China.
Methods
A retrospective study was conducted at Beijing Chest Hospital. Patients with symptoms suggestive of pulmonary TB who provided BALF specimens for routine MTB detection between June 2019 and July 2020 were enrolled in the study. Chi-square test and Student’s t-test were used to compare results across groups stratified according to BALF bacterial load.
Results
In total, 1125 patients with positive Xpert results who were enrolled in final analysis, 263 provided BALF specimens that tested positive for RIF-resistant MTB via Xpert MTB/RIF. The RIF-resistance rate of specimens with very low MTB bacterial load was 30.9%, a resistance rate significantly greater than rates obtained for groups with high (25.0%), medium (17.3%) and low (19.2%) MTB loads (P<0.01). Notably, false-positive results obtained for the very low bacterial load group led to markedly reduced positive predictive value of Xpert MTB/RIF to provide correct RIF-resistance predictions for that group (67.1%, 95% CI: 56.1%–78.1%5) relative to the predictive value obtained for all other groups combined (about 90%, P<0.05). Sanger sequencing data obtained for 20 (32.8%) MTB isolates deemed RIF-resistant via Xpert (Probe E) lacked rpoB RRDR mutations. Meanwhile, of another group of 23 isolates deemed RIF-susceptible via DST but RIF-resistant via Xpert MTB/RIF, 20 isolate sequences (87.0%) lacked rpoB RRDR mutations, while sequences of the remaining 3 isolates harbored single rpoB RRDR mutations predicted to cause amino acid substitutions.
Conclusion
Xpert MTB/RIF assay performed alarmingly poorly when used to detect RIF-resistant MTB in BALF specimens with very low bacterial loads. A high rate of Xpert probe E hybridization failure was the main driver of false-positive RIF-resistant results.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB) complex, is among the major global causes of morbidity and mortality.Citation1,Citation2 According to World Health Organization (WHO) estimates, over 10.0 million persons worldwide have developed incident TB and 1.4 million died from this disease in 2019.Citation1 Meanwhile, an ongoing epidemic of drug-resistant tubercle bacilli has undermined gains made in TB prevention and treatment.Citation3 Early diagnosis of drug-resistant TB is important for reducing TB morbidity and mortality,Citation4 prompting use of sputum culture and drug susceptibility testing (DST) as key tools for diagnosing drug-resistant TB.Citation5 However, few tuberculosis control programs in low-income countries can utilize testing services available through phenotypic DST facilities, due to high testing costs and technical constraints.Citation6 Moreover, culture-based acid-fast bacilli (AFB) testing and drug susceptibility testing (DST) are time consuming activities that require at least 8 weeks to yield interpretable results. Such prolonged test time to diagnosis often delays initiation of appropriate anti-TB treatment.Citation7 Therefore, implementation of rapid and accurate laboratory diagnostic testing would likely lead to improved TB treatment outcomes and reduce the global burden of drug-resistant tuberculosis disease.Citation8
Nucleic acid amplification technologies, such as polymerase chain reaction (PCR) amplification, could potentially revolutionize rapid detection of MTB from clinical samples.Citation9 Xpert MTB/RIF (Xpert, Cepheid, USA), an automated cartridge-based molecular assay, can detect MTB and rifampicin (RIF) resistance within two hours and has been endorsed by WHO to achieve rapid diagnosis of RIF-resistant TB.Citation10 Although Xpert provides high overall sensitivity and specificity for detecting RIF-resistant MTB in various types of clinical samples,Citation10 accumulating evidence obtained from case reports and small studies indicates that false-positive results supporting RIF resistance are frequently obtained when Xpert MTB/RIF is used to test sputum specimens with very low bacterial loads.Citation11,Citation12 Thus, concerns about how to correctly interpret such Xpert results are growing, since Xpert results may greatly impact the clinical management of TB patients.
Bronchoscopy is commonly performed on smear-negative patients with presumed pulmonary TB and patients who are unable to produce adequate sputum.Citation13,Citation14 As a consequence, bronchial lavage fluid (BALF) can serve as an alternative to sputum for increasing the diagnostic yield of laboratory examinations used to diagnose pulmonary TB cases.Citation13 However, BALF samples are generally paucibacillary in nature and thus require sensitive techniques for MTB detection and characterization. Consequently, Xpert was initially embraced as a highly sensitive method and suitable method for testing of specimens with very low bacterial loads, in spite of concerns associated with false-positive Xpert results. To address such concerns, here we assessed the effect of bacterial load on the Xpert accuracy for detection of RIF-resistant MTB in BALF specimens collected at a national TB specialized hospital in Beijing, China.
Materials and Methods
Study Design
This study was retrospectively conducted at Beijing Chest Hospital, a 612-bed tertiary hospital providing health care for patients afflicted with tuberculosis and other chest diseases. Patients with symptoms suggestive of pulmonary TB who provided BALF specimens for routine MTB detection between June 2019 and July 2020 were enrolled in this retrospective study. After 20 mL of normal saline was instilled into patient airways through bronchoscopy, a minimum of 10 mL of BALF was obtained. BALF specimens were subjected to laboratory testing that included Xpert MTB/RIF assays and phenotypic DST that were performed according to manufacturers’ instructions.Citation13
Laboratory Examination
One milliliter of fresh BALF specimen was mixed with 2 mL Xpert sample agent and the mixture was incubated for 15 min at room temperature with intermittent shaking. After completion of pretreatment, 2 mL of sample was added to the Xpert G4 cartridge for automatic processing. The results were automatically generated after a 2-hour amplification reaction. The bacterial load was categorized by the Xpert system semi-quantitatively in relation to sample positivity as high (Ct≤16), medium (Ct>16–22), low (Ct>22–28), and very low (Ct>28–38). Five probes were preloaded in the Xpert cartridge, including ProbeA, ProbeB, ProbeC, ProbeD, and ProbeE, the latter of which covered the 81-bp rifampicin resistance determining region (RRDR).
The remainder of the BALF specimen was decontaminated by addition of N-acetyl-L-cysteine-NaOH-Na citrate and the sample was vortexed for 30 seconds. Following 15 min of incubation at room temperature, the decontaminated sample was neutralized by addition of phosphate buffer (pH=7.4) then centrifuged at 4000×g for 15 min. The supernatant was discarded and the sediment was resuspended in 2 mL PBS buffer; then, 0.5 mL of the suspension was inoculated into a MGIT tube supplemented with 10% OADC and PANTA. The tube was then loaded into the Bactec MGIT 960 system where it remained for 6 weeks or until it was flagged as positive by the machine readout. Rapid identification of positive mycobacterial cultures was performed using a TibiliaTB Rapid Test kit (Chuangxin, Hangzhou). Briefly, 0.1 mL of liquid culture was transferred to the sample well. After incubation at room temperature for 15 min, a pink band in the T region was interpreted as a positive result for MPT64 Ag to demonstrate the presence of MTB in the positive culture.
MTB-positive cultures were subcultured on Löwenstein-Jensen (L-J) medium for 4 weeks. Colonies were harvested and then subjected to phenotypic DST using a commercial microdilution method.Citation15 Growth of MTB at a concentration of RIF of 1.0 mg/L was considered to be a positive test result for RIF resistance. All isolates were stored at −80°C in Middlebrook 7H9 medium supplemented with 10% OADC and 5% glycerol.
DNA Amplification and Sequencing
Prior to DNA extraction, frozen MTB isolates were inoculated on L-J medium; then, 4-week-old bacteria were processed using a boiling method to extract crude genomic DNA as previously described.Citation16 A 688-bp rpoB amplicon containing the RRDR was amplified using published primer sets.Citation15 PCR products were sent to the Tsingke Company (Beijing, China) for DNA sequencing after they were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Automated sequencing was performed by capillary electrophoresis on an ABI3730 (Thermo Fisher Scientific, Waltham, MA). DNA sequences were compared with the corresponding sequence of the reference MTB strain (H37Rv) using BioEdit software version 7.1.11 (http://www.mbio.ncsu.edu/bioedit/bioedit.html).
Statistical Analysis
Indicators used to evaluate Xpert performance related to detection of RIF-resistant MTB included sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), with Xpert results compared to results obtained via pDST, the gold standard. We performed chi-square analysis to compare the performance of Xpert across subgroups based on bacterial load. Additionally, the Student’s t-test was used to assess differences involving continuous variables. Intergroup differences were declared significant if the P value was less than 0.05. All calculations were conducted using SPSS version 20.0 (IBM Corp. Armonk, NY, USA).
Ethics Statement
This study was conducted in accordance with the tenets of the World Medical Association’s Declaration of Helsinki and approved by the Ethics Committee of Beijing Chest Hospital, Capital Medical University (Approval No.:2016KY005). Because this study only included data obtained from clinical isolates and not from other patient record-based data, individual patient consent was waived.
Results
Patients
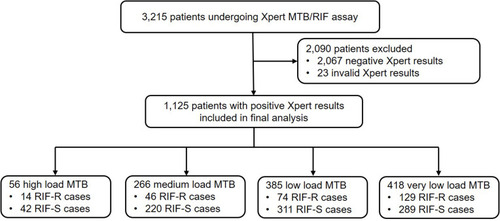
Between June 2019 and July 2020, BALF specimens obtained from a total of 3215 patients with suggestive TB symptoms were subjected to Xpert MTB/RIF testing at Beijing Chest Hospital. Of these cases, Xpert failed to identify the presence of MTB in 2090 patient specimens, while 23 specimens yielded invalid Xpert results. Finally, positive Xpert results from 1125 patients were included in our final analysis. As shown in , numbers of patient BALF specimens with high, medium, low, and very low MTB bacterial loads were 56 (56/1125, 5.0%), 266 (266/1125, 23.6%), 385 (385/1125, 34.2%) and 418 (418/1125,37.2%), respectively. Notably, the RIF-resistance rate, as determined using Xpert for specimens with very low MTB bacterial loads, was 30.9% (129/418), a rate significantly higher than rates obtained for high (14/56, 25.0%), medium (46/266, 17.3%) and low (74/385, 19.2%) bacterial load groups (P<0.01).
Diagnostic Accuracy of Xpert Stratified to Bacterial Load
Next, Xpert MTB/RIF diagnostic accuracy for predicting RIF resistance was analyzed according to bacterial load, with results provided in . Of 1125 enrolled patients, 970 (86.2%) yielded positive MTB cultures, of which 812 (83.7%) yielded in vitro DST results. Based on conventional DST results as a reference, the specificity of Xpert results obtained for MTB-positive specimens with very low bacterial load was only 85.9% (140/163), a rate significantly lower than the overall rate obtained for specimens with medium and low bacterial loads (>98.0%, P<0.01). In addition, significant differences were found among positive predictive values (PPVs) obtained across groups, with PPV obtained for the very low bacterial load group (47/70, 67.1%) markedly lower than PPVs of other groups (about 90.0%). Taken together, these results emphasize the high potential risk of obtaining false-positive RIF-resistance results when testing BALF specimens with very low MTB load (low positivity grade).
Table 1 Diagnostic Accuracy of Xpert MTB/RIF Assay Stratified to Bacterial Load
Interpretation of False-Positive Rifampicin Resistance
In order to explore the distribution of mutations within the rpoB gene for specimens with very low bacterial load, partial DNA fragments containing the rpoB RRDR region were analyzed using Sanger sequencing, with results summarized in . Among the 129 cases with rpoB mutations that were detected using Xpert, 59 were excluded due to negative culture results. Ultimately, sequence results of 70 isolates were included in our analysis, of which 61 isolate sequences (87.1%) possessed Probe E rpoB mutations and 9 (12.9%) had non-ProbeE rpoB mutations. For 9 isolate sequences belonging to the non-Probe-E group, all (100.0%) were shown to harbor mutations that encoded amino acid substitutions within the rpoB RRDR sequence, including 4 with His526Tyr, 2 with Asp516Val, 1 with Leu511Pro, 1 with Gln513Pro, and 1 with His526Leu substitutions. By contrast, RRDR sequences of 20 (32.8%) isolates that had been predicted to possess Probe E-associated rpoB RRDR mutations based on Xpert results tested negative for mutations via sequence analysis. For cases with very low bacterial load, we also analyzed mutation profiles among 23 MTB isolates deemed RIF-susceptible based on DST results that tested as RIF-resistant based on Xpert results. Of 23 false-positive cases, 20 (87.0%) had no mutations within RRDR, while 3 had RRDR mutations encoding amino acid substitutions including 1(4.3%) with Leu511Pro, 1 (4.3%) with His526Tyr, and 1 (4.3%) with Ser531Leu.
Table 2 Sequence Analysis of Cases Detected by Xpert MTB/RIF in BALF Samples with Very Low Bacterial Load
Discussion
The Xpert MTB/RIF assay has greatly shortened the time to diagnosis of TB and rifampicin-resistant TB.Citation17 Since its endorsement by the WHO, many high-burden TB countries have started scaling-up use of this innovative assay; however, concerns about Xpert specificity for assessing RIF resistance have arisen due to silent mutations detected in the rpoB gene and delayed binding of a specific Xpert probe.Citation11,Citation12,Citation18 Here we found that the Xpert assay had alarmingly low positive predictive value (67%) when used to evaluate BALF specimens with very low MTB load for RIF resistance, whereas this assay had an acceptable positive predictive value (>90%) when used to evaluate specimens with greater bacterial loads. Therefore, we hypothesize that false-positive RIF resistance results obtained using Xpert are caused by delayed binding of the rpoB probe to target DNA under low bacterial load conditions as the major explanation for significantly and incorrectly high RIF-resistance rates obtained for samples with very low bacterial loads. It is well recognized that RIF resistance results obtained using Xpert assay are interpreted based on the universal >4 cycle difference in Ct values between probes.Citation15 Thus, the risk of obtaining diagnostic false-negative Xpert RIF-resistance results using any rpoB probe is greater when samples with low MTB load are tested, due to inefficient amplification at low MTB loads. Indeed, false-positive results may have led to incorrect RIF results in a recent population-based study conducted in Rwanda. In that study, 86% of patients with very low bacillary load were falsely diagnosed with RIF-resistant TB based on Xpert results.Citation19 Meanwhile, differences in positive predictive values between studies have reflected the diversity of RIF-resistance rates across regions.
Variations in Xpert performance with respect to clinical interpretation of RIF susceptibility have important implications for testing of BALF samples. On one hand, approximately one-third of BALF specimens have extremely low bacterial loads and thus would be expected to have increased rates of false-positive RIF-resistance results. As a solution, repeat testing of a second specimen would reduce the Xpert false-positive RIF-resistance rate. On the other hand, the relatively high cost of Xpert MTB/RIF cartridges precludes use of repeat testing in low-income countries. However, when the Xpert assay cannot provide a reliable result due to low bacterial load in the specimen, centrifugation could ideally be used to concentrate MTB within the sample.Citation14 Further work is urgently needed to determine if Xpert performance would improve when applied to centrifuged BALF samples obtained from patients with symptoms suggestive of TB.
Notably, it is highly likely that a high frequency of Probe E hybridization failure occurred during Xpert PCR amplification of rpoB regions. In line with this speculation, Ocheretin and colleagues found that a false-positive RIF-resistance result was due to unequal dynamics of probe/wild-type target hybridization between Probe E and other probes after an extended number of PCR cycles.Citation12 Our Sanger sequencing results confirmed that approximately one-third of RIF-resistant cases detected using Xpert with Probe E lacked rpoB RRDR mutations, suggesting that low bacterial load had a pronounced effect on the results by delaying Probe E binding. In other words, RIF-resistant cases that were detected based on Probe E hybridization as based on samples with very low bacterial load had greater odds of generating false-positive results than when other probes were used. Worldwide, the most frequent mutation conferring RIF resistance occurs within the RRDR at rpoB codon 531,Citation20 which specifically binds Xpert Probe E. Therefore, RIF resistance results obtained using Xpert for specimens with very low bacterial load require more careful interpretation that do other specimens, due to the high potential for false positive-results resulting from delayed binding of Probe E.
Our study had several limitations. First, as based on results of previous studies, in addition to Probe E, Probe B has also been associated with false-positive RIF-susceptibility results.Citation11 However, due to the small sample size of cases with mutations in Probe B-binding region of rpoB, we could not verify this result in our cohort. Second, the small number of false-positive RIF resistance cases also made it difficult to obtain statistically unbiased results for these cases. Third, heteroresistance is another potential explanation for discordant RIF susceptibility results between different methods.Citation21 It should be noted that Sanger sequencing was conducted on the genomic DNA of MTB strains isolated from patients. As a consequence, dynamic changes that occur in bacterial populations during subculture may lead to discordance between Xpert and DNA sequencing results, a factor that was not investigated here. Fourth, due to inherent drawbacks associated with retrospective study design and high Xpert testing costs, we did not repeat Xpert testing of BALF samples at very low bacterial loads. Finally, the WHO recently endorsed the use of novel Xpert Ultra, which has greater specificity for diagnosing pulmonary and extrapulmonary TB.Citation22 Therefore, the accuracy of this novel assay for testing of specimens with very low bacillary loads should be assessed in the future.
In conclusion, our data demonstrate that the Xpert MTB/RIF assay has alarmingly low positive predictive value for assessing RIF resistance in BALF specimens with very low bacterial loads. A high frequency of Probe E hybridization failure was the main driver of false-positive RIF resistance results. Therefore, Xpert results obtained for specimens with very low bacterial load require careful interpretation. Further postimplementation field studies are urgently needed to determine if Xpert Ultra would overcome inherent shortcomings of the classic Xpert assay.
Acknowledgments
We would like to thank all the staffs participating this study from Beijing Chest Hospital.
Disclosure
The authors report no conflicts of interest in this work.
References
- World Health Organization. Global Tuberculosis Report 2020. Geneva: World Health Organization; 2020. Licence: CC BY-NC-SA 3.0 IGO.
- Chakaya JM, Harries AD, Marks GB. Ending tuberculosis by 2030-pipe dream or reality? Int J Infect Dis. 2020;92S:S51–S4. doi:10.1016/j.ijid.2020.02.02132114202
- Dheda K, Gumbo T, Maartens G, et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir Med. 2017.
- van Cutsem G, Isaakidis P, Farley J, Nardell E, Volchenkov G, Cox H. Infection control for drug-resistant tuberculosis: early diagnosis and treatment is the key. Clin Infect Dis. 2016;62(Suppl 3):S238–43. doi:10.1093/cid/ciw01227118853
- Siddiqi K, Lambert ML, Walley J. Clinical diagnosis of smear-negative pulmonary tuberculosis in low-income countries: the current evidence. Lancet Infect Dis. 2003;3(5):288–296. doi:10.1016/S1473-3099(03)00609-112726978
- Schon T, Miotto P, Koser CU, Viveiros M, Bottger E, Cambau E. Mycobacterium tuberculosis drug-resistance testing: challenges, recent developments and perspectives. Clin Microbiol Infect. 2017;23(3):154–160. doi:10.1016/j.cmi.2016.10.02227810467
- Pang Y, Xia H, Zhang Z, et al. Multicenter evaluation of genechip for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2013;51(6):1707–1713. doi:10.1128/JCM.03436-1223515537
- Parsons LM, Somoskovi A, Gutierrez C, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. 2011;24(2):314–350. doi:10.1128/CMR.00059-1021482728
- MacLean E, Kohli M, Weber SF, et al. Advances in molecular diagnosis of tuberculosis. J Clin Microbiol. 2020;58(10). doi:10.1128/JCM.01582-19
- World Health Oragnization. Automated Real-Time Nucleic Acid Amplification Technology for Rapid and Simultaneous Detection of Tuberculosis and Rifampicin Resistance: Xpert MTB/RIF Assay for the Diagnosis of Pulmonary and Extrapulmonary TB in Adults and Children. Geneva, Switherland: World Health Organization; 2013. WHO/HTM/TB/2013.16.
- Van Rie A, Mellet K, John MA, et al. False-positive rifampicin resistance on Xpert(R) MTB/RIF: case report and clinical implications. Int J Tuberc Lung Dis. 2012;16(2):206–208. doi:10.5588/ijtld.11.039522236921
- Ocheretina O, Byrt E, Mabou MM, et al. False-positive rifampin resistant results with Xpert MTB/RIF version 4 assay in clinical samples with a low bacterial load. Diagn Microbiol Infect Dis. 2016;85(1):53–55. doi:10.1016/j.diagmicrobio.2016.01.00926915638
- Xu P, Tang P, Song H, et al. The incremental value of bronchoalveolar lavage for the diagnosis of pulmonary tuberculosis in a high-burden urban setting. J Infect. 2019;79(1):24–29. doi:10.1016/j.jinf.2019.05.00931100361
- Theron G, Peter J, Meldau R, et al. Accuracy and impact of Xpert MTB/RIF for the diagnosis of smear-negative or sputum-scarce tuberculosis using bronchoalveolar lavage fluid. Thorax. 2013;68(11):1043–1051. doi:10.1136/thoraxjnl-2013-20348523811536
- Huo F, Ma Y, Liu R, et al. Interpretation of discordant rifampicin susceptibility test results obtained using genexpert vs phenotypic drug susceptibility testing. Open Forum Infect Dis. 2020;7(8):ofaa279. doi:10.1093/ofid/ofaa27932766385
- Pang Y, Liu G, Wang Y, Zheng S, Zhao YL. Combining COLD-PCR and high-resolution melt analysis for rapid detection of low-level, rifampin-resistant mutations in Mycobacterium tuberculosis. J Microbiol Methods. 2013;93(1):32–36. doi:10.1016/j.mimet.2013.01.00823396215
- Lawn SD, Mwaba P, Bates M, et al. Advances in tuberculosis diagnostics: the Xpert MTB/RIF assay and future prospects for a point-of-care test. Lancet Infect Dis. 2013;13(4):349–361. doi:10.1016/S1473-3099(13)70008-223531388
- Mathys V, van de Vyvere M, de Droogh E, Soetaert K, Groenen G. False-positive rifampicin resistance on Xpert(R) MTB/RIF caused by a silent mutation in the rpoB gene. Int J Tuberc Lung Dis. 2014;18(10):1255–1257. doi:10.5588/ijtld.14.029725216843
- Ngabonziza JCS, Decroo T, Migambi P, et al. Prevalence and drivers of false-positive rifampicin-resistant Xpert MTB/RIF results: a prospective observational study in Rwanda. Lancet Microbe. 2020;1(2):e74–e83. doi:10.1016/S2666-5247(20)30007-0
- Jing W, Pang Y, Zong Z, et al. Rifabutin resistance associated with double mutations in rpoB gene in Mycobacterium tuberculosis isolates. Front Microbiol. 2017;8:1768. doi:10.3389/fmicb.2017.0176828959248
- Zhang Z, Wang Y, Pang Y, Liu C. Comparison of different drug susceptibility test methods to detect rifampin heteroresistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58(9):5632–5635. doi:10.1128/AAC.02778-1425022589
- Qi XY, Qiu XS, Jiang JY, Chen YX, Tang LM, Shi HF. Microwaves increase the effectiveness of systemic antibiotic treatment in acute bone infection: experimental study in a rat model. J Orthop Surg Res. 2019;14(1):286. doi:10.1186/s13018-019-1342-331488167