Abstract
Background
Early and accurate diagnosis of rifampicin (RIF)-resistant Mycobacterium tuberculosis (MTB) is essential for controlling community spread of drug-resistant tuberculosis (TB). In order to discover mutations residing outside the rifampicin resistance-determining region (RRDR) of the MTB rpoB gene, we conducted this retrospective study.
Methods
We retrospectively screened patient records to obtain Xpert MTB/RIF assay results for patients who received care at the Beijing Chest Hospital from 2016 to 2019 in order to identify subjects who met study selection criteria. Stored frozen patient isolates were cultured, harvested, and then subjected to drug susceptibility testing. Concurrently, entire rpoB gene DNA of each isolate was amplified and then sequenced to reveal rpoB mutations.
Results
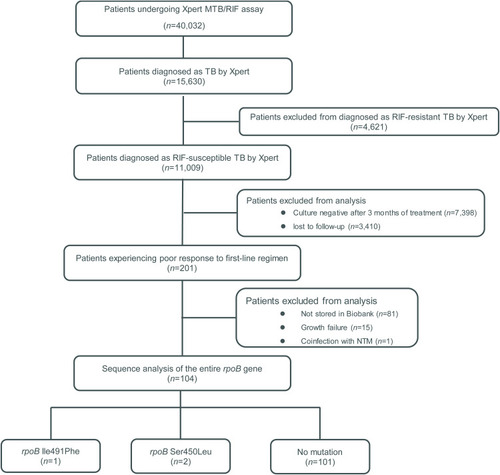
Overall, 104 RIF-susceptible tuberculosis patients who were tested using the Xpert MTB/RIF assay also had poor first-line regimen treatment responses. Isolates obtained from these cases included 101 MTB isolates that possessed wild-type rpoB allelic profiles, as demonstrated using Sanger sequencing. However, sequences from the other three isolates confirmed that rpoB of one isolate harbored a mutation encoding the amino acid substitution Ile491Phe and that rpoB genes of two isolates contained a mutation encoding the amino acid substitution Ser450Leu.
Conclusion
Our data demonstrated that mutations found outside the RRDR of MTB rpoB are rare in Beijing, China, indicating that World Health Organization-approved molecular diagnostics are generally suitable for diagnosing RIF resistance.
Introduction
Tuberculosis (TB), which is caused by infection with Mycobacterium tuberculosis (MTB) complex, remains a major public health concern worldwide. The emergence of drug-resistant TB, especially rifampicin (RIF)- and multidrug-resistant TB, has greatly impeded progress toward controlling this disease.Citation2 An estimated 10.0 million people developed active TB in 2018, of which 484,000 cases were caused by MTB with resistance to RIF. Although RIF is currently the most effective first-line anti-TB drug, initial RIF resistance has been responsible for most treatment failures experienced by TB patients receiving the standard first-line treatment regimen.Citation3 Therefore, early and accurate diagnosis of RIF resistance is essential in order to administer appropriate treatment as early as possible to prevent further community transmission of this refractory disease.
Conventional diagnosis of drug-resistant TB relies on phenotypic drug susceptibility testing (DST) that is based on mycobacterial growth.Citation4 Unfortunately, due to the slow growth rate of MTB, testing requires months to complete after initiation of isolate cultures.Citation5 Recently, Xpert MTB/RIF (Xpert, Cepheid, USA), a fully automated real-time PCR assay, can detect MTB, including RIF-resistant MTB, directly from clinical samples within 2 hours.Citation6 Based on promising Xpert MTB/RIF clinical performance, the World Health Organization has recently endorsed implementation of this assay for diagnosis of TB and MTB drug resistance based on testing of numerous specimen types.Citation7 To detect RIF resistance, the 81-bp genetic region commonly associated with MTB rifampin resistance, the rifampicin resistance-determining region (RRDR) of the rpoB gene, is probed with molecular beacons.Citation6 The accuracy of the Xpert MTB/RIF assay for detecting RIF resistance thus depends on the prevalence of mutations within the RRDR. A recent observational study from South Africa demonstrated that a substantial number of RIF-resistant TB cases harbor a rpoB mutation that induces an amino acid substitution, Ile491Phe. Of concern, this mutation is found outside the RRDR and is not detected by current WHO-endorsed commercial tests.Citation8 More importantly, this type of resistance is also undetectable using phenotypic DST methods and therefore testing does not exist that can be used to prevent transmission of RIF-resistant TB in the community.Citation8 Ultimately, the frequency of this specific mutation among RIF-resistant isolates will have important implications for clinical diagnostic algorithms used in different settings. Although this mutation has been found in DNA of clinical isolates in several countries,Citation8–Citation10 little is known about its contribution to RIF resistance in China, a global drug resistance hot spot. To address this concern, we conducted a retrospective study to investigate the prevalence of mutations located outside the rpoB RRDR in genomes of RIF-resistant MTB clinical isolates in Beijing, China.
Materials and Methods
Patient Selection and Ethics Statement
In this retrospective observational study, we accessed the BioBank of Tuberculosis System to obtain standardized data for all MTB isolates from culture-positive cases who had sought care from 2016 to 2019 at Beijing Chest Hospital, a national TB-designated hospital in China. Data included routine microscopy results for sputum samples and test results of patient isolates obtained using the Xpert MTB/RIF assay and mycobacterial culture-based methods. We then selected study subjects after retrospectively reviewing Xpert MTB/RIF assay results in records of consecutive patients to identify patients who met study selection criteria. Patients infected with RIF-susceptible MTB, as confirmed via Xpert MTB/RIF assay, were treated with the WHO-recommended RIF-based regimen 2HRZE/HR. Medical records were further reviewed to identify RIF-susceptible TB patients who experienced poor clinical response based on culture-positive results obtained after 3 months of treatment.
All procedures were carried out in accordance with the principles of the World Medical Association’s Declaration of Helsinki. The study was approved by the Ethics Committee of Beijing Chest Hospital, Capital Medical University. Informed consent requirements were waived since the study involved clinical isolates containing tubercle bacilli and involved secondary analysis of existing clinical data.
Bacteria Subculture and Drug Susceptibility Testing
Frozen isolates were selected and cultured on Löwenstein-Jensen (L-J) medium for use in further analyses. Fresh 4-week-old MTB cultures were harvested and then were used for drug susceptibility testing to determine minimal inhibitory concentrations (MICs) of RIF, rifapentine (RFT), and rifabutin (RFB) based on the broth microdilution method, as previously described.Citation3 Briefly, after a suspension of 0.5 McFarland standard was prepared, 100 μL of diluted inoculum was transferred to each well of a microtiter plate that contained 100 μL/well of drug, with drug concentrations in wells varying as two-fold serial dilutions in 7H9 broth over a concentration range of 0.0075 mg/L to 8.0 mg/L. Tubercle bacilli (100 μL/well) were added to each well from a stock containing approximately 105 CFU/mL. After incubation of bacteria and drugs for 7–10 days, 70 μL of alamarBlue solution was added to each well and the plates were incubated at 37°C for another 24 h. MIC breakpoint concentrations for RIF, RFT and RFB were defined as 1 mg/L, 0.50 mg/L and 0.50 mg/L, respectively. All experiments were performed in triplicate.
DNA Extraction and Sequencing
For DNA extraction, one loop of bacteria was resuspended in 500 µL of Tris-EDTA (TE) buffer and incubated for 30 min at 95°C. After centrifugation at 13000 rpm for 5 min, the supernatant was used as the source of template DNA for PCR amplification. The entire rpoB gene was amplified following the method used in our previous work.Citation11 DNA sequencing was carried out by RuiBiotech Company (Beijing, China). Amino acid polymorphisms were identified based on comparison with rpoB of the H37Rv reference strain (GenBank accession no. NC_000962) using the Constraint-based Multiple Alignment Tool (https://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?LINK_LOC=BlastHomeLink).
Results
Collection of Isolates and Mutation Detection
Of 15,630 Xpert-positive isolates, 11,009 (70.4%) were identified as RIF-susceptible and the other 4621 (29.6%) were RIF-resistant. Clinical responses of RIF-susceptible cases were determined based on laboratory testing conducted after patient completion of 3 months of treatment with the standard first-line regimen. Testing demonstrated that 201 patients were still culture-positive after 3 months of treatment, while 81 isolates were not stored in our Biobank, subculturing of 15 isolates failed due to lack of growth, and one isolate was confirmed to be a coculture of MTB and NTM. Ultimately, 104 cases were included for complementary MIC testing and DNA sequencing analysis. As shown in , of these 104 isolates, 101 (97.1%) possessed wild-type allelic rpoB profiles, while Sanger sequencing confirmed the presence of a mutation encoding an amino acid substitution (Ile491Phe) in rpoB of one isolate (1.0%) and another substitution (Ser450Leu) in two isolates (1.9%) ().
MICs of MTB Isolates
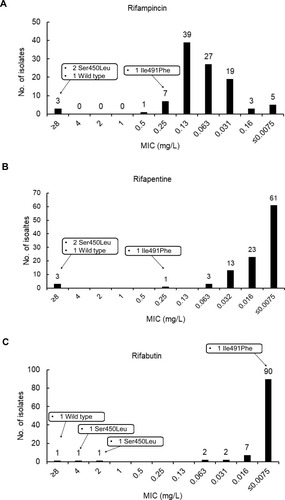
MICs of MTB isolates to three rifamycins are summarized in . Overall, RFB exhibited the highest level of in vitro activity against MTB (with an MIC90 value of 0.016 mg/L) as compared to MIC90 values for RFT (0.032 mg/L) and RIF (0.13 mg/L), the latter of which was only slightly greater than that of reference strain H37Rv. Ultimately, only three isolates were resistant to RIF, RFT, and RFB, including two isolates harboring a rpoB Ser450Leu substitution and one isolate possessing only the wild-type rpoB allele. Notably, one isolate carrying the Ile491Phe mutation had a RIF MIC of 0.25 mg/L, which was below the critical concentration of 1.0 mg/L, while RFT and RFB MICs for this isolate were 0.12 mg/L and 0.25 mg/L, respectively.
Discussion
In order to monitor the efficacy of the Xpert MTB/RIF roll-out worldwide, observational studies tracking the prevalence of a specific rpoB mutation (Ile491Phe) found outside the RRDR are essential for formulating optimal diagnostic algorithms based on Xpert MTB/RIF assay results obtained for RIF-resistant TB cases.Citation8 In this study, our data demonstrated that the rpoB Ile491Phe mutation is rare in China. By contrast, a national survey conducted in Eswatini revealed that 30% of MDR-TB strains carried this mutation.Citation12 Similar results were obtained in South Africa that confirmed that 15% of isoniazid-monoresistant MTB isolates harbored the Ile491Phe rpoB mutation and thus these isolates should be classified as MDR isolates.Citation8 Meanwhile, several other studies have reported occasional detection of this mutation in isolates from European and Asian countries,Citation9,Citation10,Citation13 triggering speculation regarding underlying reasons for differences in Ile491Phe mutation prevalence across countries. On the one hand, this specific mutation has been mainly reported only in Africa where it is prevalent, as determined using whole genome sequencing, and is associated with spoligotypes ST34 and ST92 that are predominant MTB sublineages circulating on that continent.Citation8 Therefore, we speculate that this association is the major reason for the geographic prevalence of this mutation in Africa. On the other hand, different methods used to characterize MTB isolates across studies may have biased the results of these analytical epidemiological studies.
Another interesting finding is that two isolates with high-level RIF resistance due to the Ser450Leu RpoB mutation were not detected using the Xpert MTB/RIF assay. Early studies revealed striking frequencies of genetically heterogeneous bacterial populations in lesions of pulmonary TB patients.Citation14 Although the exact reason for this phenomenon remains unclear, the heterogeneity of bacterial populations in specimens tested via Xpert MTB/RIF assay and mycobacterial cultures may lead to the failure of the Xpert MTB/RIF assay to detect RIF resistant MTB.Citation15 As consistently shown in previous reports,Citation8 one MTB isolate harboring the Ile491Phe mutation was phenotypically susceptible to RIF, while the patient providing this isolate experienced a poor clinical response to treatment with a first-line anti-TB regimen, an indicator of RIF resistance. Nevertheless, despite resistance of the patient MTB infection to RIF, the RFB MIC value (<0.0075 mg/L) of the MTB patient isolate was significantly lower than the corresponding critical concentration. Thus, properly designed trials are urgently needed to assess the clinical efficacy of the standard RFB-containing regimen against MTB harboring the rpoB gene Ile491Phe mutation.
Of note, over 97% of cases infected with RIF-susceptible MTB experience adverse treatment responses, underscoring the complexity associated with tuberculosis treatment. Insufficient blood drug concentration, drug-resistant mutations after long-term anti-TB treatment, potential interactions between anti-TB drugs and other drugs, impaired patient immunity, and poor treatment adherence may contribute to treatment failure.Citation16 Thus, further studies are needed to determine risk factors associated with poor treatment responses so that effective interventions can be implemented to improve clinical responses of this population.
We acknowledge several obvious limitations of the present study. First, enrollment of only patients of one pilot project limited the significance of our conclusions. Further studies are required to validate our findings that evaluate more MTB isolates from China. Second, in this retrospective study we set selection criteria for poor responses based on results obtained after only 3 months of treatment in order to enroll more TB patients with potential RIF resistance. However, this decision may have led to underestimation of the frequency of mutations found outside the rpoB gene RRDR. Third, some isolates may harbor mutations conferring resistance rather than rpoB gene, which were not detected in our study.
In conclusion, our data demonstrate that MTB rpoB mutations outside the RRDR were rare in our Chinese cohort, while also demonstrating the potential of the WHO-approved molecular diagnostics assay to effectively diagnose RIF resistance. In addition, a high proportion of cases infected with RIF-susceptible MTB experienced adverse clinical outcomes after 3 months of treatment, highlighting the urgent need to identify risk factors associated with poor treatment responses toward improving treatment outcomes of this population.
Acknowledgments
This work was supported by the National Key Research Program of China (2018ZX10103-001), and the Beijing Hospitals Authority’ Ascent Plan (DFL20191601). We are grateful to members of the National Clinical Laboratory on Tuberculosis at the Beijing Chest Hospital for their cooperation and technical help.
Disclosure
The authors report no conflicts of interest in this work.
References
- World Health Organization.Global tuberculosis report 2019. Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO.
- Dheda K, Gumbo T, Maartens G, et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir Med. 2017. doi:10.1016/S2213-2600(17)30079-6.
- Pang Y, Lu J, Wang Y, Song Y, Wang S, Zhao Y. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:893–900. doi:10.1128/AAC.01024-1223208715
- Kim SJ. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J. 2005;25:564–569. doi:10.1183/09031936.05.0011130415738303
- Ryu YJ. Diagnosis of pulmonary tuberculosis: recent advances and diagnostic algorithms. Tuberc Respir Dis (Seoul). 2015;78:64–71. doi:10.4046/trd.2015.78.2.6425861338
- Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–237. doi:10.1128/JCM.01463-0919864480
- World Health Organization.Using the Xpert MTB/RIF assay to detect pulmonary and extrapulmonary tuberculosis and rifampicin resistance in adults and children: expert group meeting report. Geneva: World Health Organization; 2013.
- Makhado NA, Matabane E, Faccin M, et al. Outbreak of multidrug-resistant tuberculosis in South Africa undetected by WHO-endorsed commercial tests: an observational study. Lancet Infect Dis. 2018;18:1350–1359. doi:10.1016/S1473-3099(18)30496-130342828
- Aktas E, Durmaz R, Yang D, Yang Z. Molecular characterization of isoniazid and rifampin resistance of Mycobacterium tuberculosis clinical isolates from Malatya, Turkey. Microb Drug Resist. 2005;11:94–99. doi:10.1089/mdr.2005.11.9415910221
- Siu GK, Zhang Y, Lau TC, et al. Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. 2011;66:730–733. doi:10.1093/jac/dkq51921393153
- Huo F, Luo J, Shi J, et al. A 10-year comparative analysis shows that increasing prevalence of rifampin-resistant Mycobacterium tuberculosis in China is associated with the transmission of strains harboring compensatory mutations. Antimicrob Agents Chemother. 2018;62. doi:10.1128/AAC.02303-17.
- Sanchez-Padilla E, Merker M, Beckert P, et al. Detection of drug-resistant tuberculosis by Xpert MTB/RIF in Swaziland. N Engl J Med. 2015;372:1181–1182. doi:10.1056/NEJMc141393025785984
- Heep M, Brandstatter B, Rieger U, et al. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2001;39:107–110. doi:10.1128/JCM.39.1.107-110.200111136757
- Fonseca JD, Knight GM, McHugh TD. The complex evolution of antibiotic resistance in Mycobacterium tuberculosis. Int J Infect Dis. 2015;32:94–100. doi:10.1016/j.ijid.2015.01.01425809763
- Zhang Z, Wang Y, Pang Y, Liu C. Comparison of different drug susceptibility test methods to detect rifampin heteroresistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58:5632–5635. doi:10.1128/AAC.02778-1425022589
- Seid MA, Ayalew MB, Muche EA, Gebreyohannes EA, Abegaz TM. Drug-susceptible tuberculosis treatment success and associated factors in Ethiopia from 2005 to 2017: a systematic review and meta-analysis. BMJ Open. 2018;8:e022111. doi:10.1136/bmjopen-2018-022111