Abstract
Burkholderia pseudomallei, a bacterium that causes the disease melioidosis, is intrinsically resistant to many antibiotics. First-line antibiotic therapy for treating melioidosis is usually the synthetic β-lactam, ceftazidime (CAZ), as almost all B. pseudomallei strains are susceptible to this drug. However, acquired CAZ resistance can develop in vivo during treatment with CAZ, which can lead to mortality if therapy is not switched to a different drug in a timely manner. Serial B. pseudomallei isolates obtained from an acute Thai melioidosis patient infected by a CAZ susceptible strain, who ultimately succumbed to infection despite being on CAZ therapy for the duration of their infection, were analyzed. Isolates that developed CAZ resistance due to a proline to serine change at position 167 in the β-lactamase PenA were identified. Importantly, these CAZ resistant isolates remained sensitive to the alternative melioidosis treatments; namely, amoxicillin-clavulanate, imipenem, and meropenem. Lastly, real-time polymerase chain reaction-based assays capable of rapidly identifying CAZ resistance in B. pseudomallei isolates at the position 167 mutation site were developed. The ability to rapidly identify the emergence of CAZ resistant B. pseudomallei populations in melioidosis patients will allow timely alterations in treatment strategies, thereby improving patient outcomes for this serious disease.
Introduction
Burkholderia pseudomallei is the etiologic agent of melioidosis, a disease endemic to Southeast Asia and northern Australia that is becoming increasingly identified in other tropical regions throughout the world.Citation1 B. pseudomallei is intrinsically resistant to several antimicrobial agents, limiting treatment options for melioidosis.Citation2,Citation3 In Thailand, confirmed melioidosis cases are typically treated with intravenous ceftazidime (CAZ), a third-generation cephalosporin that is routinely administered for a minimum of 10 days, or considerably longer for complex, deep-seated infections.Citation4 CAZ is also routinely administered for suspected melioidosis cases, and for confirmed cases prior to the availability of antimicrobial susceptibility data, on the basis that primary CAZ resistance is rare.Citation2,Citation5 However, recent reports have demonstrated that CAZ resistance can arise in vivo following chemotherapy,Citation3,Citation5–Citation9 especially where CAZ has been administered on multiple occasions, such as in relapse melioidosis cases.Citation5 Early detection of B. pseudomallei resistance towards clinically relevant antibiotics such as CAZ is thus an important factor in reducing the morbidity and mortality of melioidosis.
Several molecular mechanisms for CAZ resistance have been reported in the B. pseudomallei literature, all of which have been shown to arise in vivo following CAZ administration. A single-nucleotide polymorphism (SNP) in the promoter region of the β-lactamase gene, penA (−21) has been shown to cause upregulation of PenA elevating the CAZ minimum inhibitory concentration (MIC) from 1.5 μg/mL in wild-type B. pseudomallei to 6–16 μg/mL in PenA −21A mutants depending on the strain.Citation5,Citation10 Within the penA coding sequence, amino acid substitutions at positions 69Citation5,Citation7 and 167Citation11 lead to markedly increased CAZ hydrolysis compared with wild-type B. pseudomallei isolates (MIC values of ≥256 μg/mL and 64 μg/mL for cysteine to tyrosine at position 69 [C69Y] and proline to serine at position 167 [P167S] mutants, respectively). Lastly, a large chromosomal deletion encompassing penicillin-binding protein-3 causes the loss of a CAZ-binding cell-surface molecule, resulting in high-level CAZ resistance (MIC ≥ 256 μg/mL).Citation6
Case study
Forty B. pseudomallei isolates obtained from a single Thai patient (P45) with acute, nonresolving melioidosis were tested for CAZ resistance. P45, a 51-year-old male rice farmer, presented with a leg abscess and high fever and was treated on admission with intravenous ceftriaxone and clindamycin for 1 day, then switched to intravenous CAZ after confirmation of B. pseudomallei by an immunofluorescence assay.Citation12 P45 initially responded well to therapy, but after 2 weeks of treatment he had a marked deterioration associated with the development of hypotension, dyspnea, recurrence of fever, and pleural effusion on chest radiography. Despite continued CAZ therapy, P45 subsequently died. It was demonstrated previously that the B. pseudomallei isolates obtained from P45 were clonal.Citation13 Thus, having ruled out coinfection or reinfection, it was suspected that in vivo development of CAZ resistance may have accounted for treatment failure in P45.
Materials and methods
Ethics statement
Informed written consent for genetic studies of these isolates was provided by the patient and approved by the Human Research Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (Bangkok, Thailand).Citation14
Culture conditions and antibiotic sensitivity testing
B. pseudomallei isolates were grown as described elsewhere.Citation5 MIC values for CAZ, amoxicillin-clavulanate, imipenem, and meropenem were determined using Etest® (bioMerieux SA, Marcy l’Etoile, France) according to manufacturer’s instructions. CAZ MIC values for susceptible, intermediate resistant, and resistant cutoffs were defined as ≤8 μg/mL, 16 μg/mL, and ≥32 μg/mL, respectively.Citation2
SYBR Green-based mismatch amplification mutation assay (SYBR-MAMA)
SYBR-MAMA polymerase chain reaction setups and thermocycling conditions were carried out as described elsewhere.Citation5 The primers were as follows: penAP167S_ForM (5′-TTTTTTTTTTTTTTTAGCGCCGTGTTCAGCTCAtA) and penAP167S_RevM (5′-GCGACGTTCCGGCTCGAT) to amplify the mutant allele; penAP167S_ForWT (5′-GCCCGCCCGCCCGCCCGCGCCGTGTTCAGCTCAtG) and penAP167S_RevWT (5′-CGACGTTCCGGCTCGATC) to interrogate the wild-type SNP state. Lowercase nucleotides indicate deliberately incorporated antepenultimate mismatches;Citation15 underlined nucleotides represent the SNP. All samples were run in duplicate, and no-template controls were included in every run.
Results
The initial 30 B. pseudomallei isolates collected upon P45’s admission to hospital were sensitive to CAZ (2 μg/mL) as determined by Etest. However, ten isolates collected 18 days later showed a predominately CAZ-resistant collection of nine strains (64 μg/mL), with a single susceptible strain. Given that the CAZ-resistant strains possessed MIC values similar to those from PenAP167S-mutated B. pseudomallei strains,Citation11 it was expected that the latter P45 B. pseudomallei isolates would possess the same penA mutation. Sanger sequencing of penA from representative sensitive and resistant isolates confirmed a P167S substitution (Ambler’s numbering scheme)Citation16 ().
Figure 1 DNA alignments of penA from patient 45 wild-type and mutant alleles against three other wild-type Burkholderia pseudomallei strains – K96243, 1710b, and 668 – showing a cytosine/thymine nucleotide transition at position 517 (according to penA annotation in B. pseudomallei K96243).Citation22
Note: This transition results in an amino acid substitution from proline to serine at position 167 in PenA.
Abbreviation: P45, patient 45.
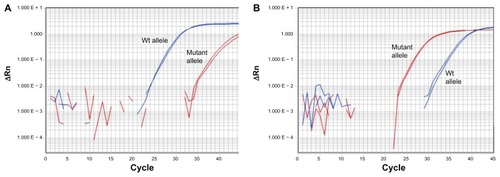
A SYBR-MAMACitation17 was developed to rapidly interrogate the mutant and wild-type variants at PenA 167 in the remaining P45 isolates. The PenA 167 SYBR-MAMA readily discriminated between mutant and wild-type alleles (). In all cases, the PenA 167 genotype corresponded with the CAZ MIC; CAZ-susceptible strains (2 μg/mL) exhibited preferential amplification of the wild-type allele (, left panel) and CAZ-resistant isolates preferentially amplified the mutant PenA 167 allele (, right panel).
Figure 2 SYBR Green mismatch amplification mutation assay interrogation of the proline to serine at position 167 (P167S) PenA mutation, conferred by a cytosine/thymine nucleotide transition in penA at position 517 (according to penA annotation in Burkholderia pseudomallei K96243;Citation22 see ) in patient 45 B. pseudomallei isolates. (A) Example amplification of the wild-type P167S penA allele. (B) Example amplification of the mutated P167S penA allele. All mutant P167S B. pseudomallei strains from patient 45 contained an elevated ceftazidime minimum inhibitory concentration of 64 μg/mL compared with the wild-type strains (2 μg/mL).
Abbreviation: Wt, wild-type.
It has been previously shown that upregulatory penA promoter mutations that increase CAZ resistance can also confer cross-resistance to the alternative treatments for melioidosis; namely, amoxicillin-clavulanate and imipenem.Citation5,Citation18 Therefore, the effect of the PenA P167S substitution on other β-lactam and carbapenem antibiotics commonly used to treat melioidosis was examined to determine whether this mutation also caused cross-resistance in this isolate. MIC values for amoxicillin-clavulanate, imipenem, and meropenem were determined for two CAZ-susceptible and CAZ-resistant B. pseudomallei isolates from P45 using Etest. Both the wild-type and mutated isolates remained susceptible to these alternate treatment options (MIC value: meropenem = 1 μg/mL; imipenem = 0.5 μg/mL; amoxicillin-clavulanate = 1.5 μg/mL), demonstrating that the P167S substitution had no effect on these alternate treatment strategies.
Discussion
The in vivo development of antibiotic resistance can have serious consequences on patient outcomes and is especially problematic where treatment options are already limited. For B. pseudomallei, development of resistance to the first- line antibiotic, CAZ, is becoming increasingly recognized as an important cause of treatment failure and infection relapse. Melioidosis cases reporting CAZ-resistant in vivo B. pseudomallei populations have now been described in Southeast Asia, India, Taiwan, and Australia.Citation5,Citation6,Citation8,Citation9 Given the protracted and lengthy regimen required for effective treatment of melioidosis, and the occurrence of relapse/recrudescent cases in ~6% of patients,Citation19,Citation20 the development of in vivo CAZ resistance is potentially more common than first thought.
In the current study, the molecular basis behind treatment failure in an acute case of melioidosis in Thailand that lead to patient mortality was identified. The P167S mutation appeared to only increase CAZ hydrolysis, with this study and others showing that ceftriaxone, imipenem, meropenem, and amoxicillin-clavulanate MIC values are unaffected by this mutation.Citation11,Citation18 Therefore, the development of the P167S mutation among the P45 in vivo B. pseudomallei population was most likely a result of CAZ administration. Nine out of ten colonies obtained from P45 following melioidosis relapse were CAZ resistant after 17 days of treatment with this drug, indicating strong positive selection of the P167S mutation under CAZ treatment.
Elucidation of resistance mechanisms towards clinically relevant antibiotics is crucial in determining effective treatment regimens for melioidosis. A real-time polymerase chain reaction assay that enables the rapid detection of P167S B. pseudomallei CAZ mutants in vivo is herein described. The availability of real-time assays for resistance detection allows clinicians to monitor infection dynamics and thus alter chemotherapy in real time, leading to more effective antimicrobial treatments and better patient outcomes. In addition, this study further challenges the dogma that a single isolate is representative of the in vivo bacterial population dynamics. Citation13,Citation21 It was demonstrated that such practices can lead to erroneous assumptions regarding antibiotic resistance, as one out of ten isolates collected from P45 18 days after admission still possessed a CAZ-sensitive phenotype.
It is recommended that in acute nonresolving melioidosis cases, good clinical practice should entail collection and characterization of multiple clinical specimens from multiple time points, where possible, to detect for emerging antimicrobial resistance. The introduction of real-time polymerase chain reaction platforms into clinics will lead to improved therapeutic outcomes by enabling minor component resistance to be detected well before conventional culture-based techniques (eg, Etest) can identify resistance.
Acknowledgment
This work was supported by the US Department of Homeland Security via award HSHQDC-10-C-00139.
Disclosure
The authors report no conflicts of interest in this work.
References
- CurrieBJDanceDAChengACThe global distribution of Burkholderia pseudomallei and melioidosis: an updateTrans R Soc Trop Med Hyg2008102Suppl 1S1S419121666
- WuthiekanunVAmornchaiPSaipromNSurvey of antimicrobial resistance in clinical Burkholderia pseudomallei isolates over two decades in Northeast ThailandAntimicrob Agents Chemother201155115388539121876049
- JenneyAWLumGFisherDACurrieBJAntibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosisInt J Antimicrob Agents200117210911311165114
- ChengACCurrieBJMelioidosis: epidemiology, pathophysiology, and managementClin Microbiol Rev200518238341615831829
- SarovichDSPriceEPVon SchulzeATCharacterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from AustraliaPLoS One201272e3078922363490
- ChantratitaNRhollDASimBAntimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomalleiProc Natl Acad Sci U S A201110841171651717021969582
- SamICSeeKHPuthuchearySDVariations in ceftazidime and amoxicillin-clavulanate susceptibilities within a clonal infection of Burkholderia pseudomalleiJ Clin Microbiol20094751556155819297597
- KungCTLeeCHLiCJLuHIKoSFLiuJWDevelopment of ceftazidime resistance in Burkholderia pseudomallei in a patient experiencing melioidosis with mediastinal lymphadenitisAnn Acad Med Singapore2010391294594721274496
- BeheraBPrasad BabuTKamaleshAReddyGCeftazidime resistance in Burkholderia pseudomallei: first report from IndiaAsian Pac J Trop Med20125432933022449529
- HaydenHSLimRBrittnacherMJEvolution of Burkholderia pseudomallei in recurrent melioidosisPLoS One201275e3650722615773
- TribuddharatCMooreRABakerPWoodsDEBurkholderia pseudomallei class A β-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibitionAntimicrob Agents Chemother20034772082208712821450
- WalshALSmithMDWuthiekanunVImmunofluorescence microscopy for the rapid diagnosis of melioidosisJ Clin Pathol19944743773798027383
- PriceEPHornstraHMLimmathurotsakulDWithin-host evolution of Burkholderia pseudomallei in four cases of acute melioidosisPLoS Pathog201061e100072520090837
- LimmathurotsakulDWuthiekanunVChantratitaNSimultaneous infection with more than one strain of Burkholderia pseudomallei is uncommon in human melioidosisJ Clin Microbiol200745113830383217804651
- HezardNCornilletPDroulleCGillotLPotronGNguyenPFactor V Leiden: detection in whole blood by ASA PCR using an additional mismatch in antepenultimate positionThromb Res199788159669336874
- AmblerRPCoulsonAFFrereJMA standard numbering scheme for the class A β-lactamasesBiochem J1991276Pt 12692702039479
- GermerSHiguchiRSingle-tube genotyping without oligonucleotide probesGenome Res19999172789927486
- RhollDAPapp-WallaceKMTomarasAPVasilMLBonomoRASchweizerHPMolecular investigations of PenA-mediated β-lactam resistance in Burkholderia pseudomalleiFront Microbiol2011213921747814
- CurrieBJWardLChengACThe epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective studyPLoS Negl Trop Dis2010411e90021152057
- LimmathurotsakulDChaowagulWChantratitaNA simple scoring system to differentiate between relapse and re-infection in patients with recurrent melioidosisPLoS Negl Trop Dis2008210e32718958279
- PearsonTU’RenJMSchuppJMVNTR analysis of selected outbreaks of Burkholderia pseudomallei in AustraliaInfect Genet Evol20077441642317258514
- HoldenMTTitballRWPeacockSJGenomic plasticity of the causative agent of melioidosis, Burkholderia pseudomalleiProc Natl Acad Sci U S A200410139142401424515377794