Abstract
Purpose
To detect and differentiate co-infection with influenza and respiratory syncytial virus during the COVID pandemic, a rapid method that can detect multiple pathogens in a single test is a significant diagnostic advance to analyze the outcomes and clinical implications of co-infection. Therefore, we validated and evaluated the performance characteristics of TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay for the detection of SARS-CoV-2, Flu A/B, and RSV using nasopharyngeal and saliva samples.
Materials and Methods
The method validation was performed by using culture fluids of Influenza A virus (H3N2) (A/Wisconsin/67/2005), Influenza B virus (B/Virginia/ATCC4/2009), RSV A2 cpts-248, SARS-CoV-2 (USA-WA1/2020) and quantitative RNA controls of Influenza A virus (H1N1) strain A/PR/8/34 (VR-95DQ), RSV A2 (VR-1540DQ) and SARS-CoV-2 (MN908947.3 Wuhan-Hu-1) from ATCC and Zeptometrix, NY, USA. A total of 110 nasopharyngeal specimens and 70 saliva samples were used for the SARS-CoV-2 detection, and a total of 70 nasopharyngeal specimens were used for Influenza and RSV detection. Total RNA was extracted from all the samples and multiplex PCR was performed using TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay. The assay was used for SARS-CoV-2 variant (B.1.1.7_601443, B.1.617.1_1662307, P.1_792683, B.1.351_678597, B.1.1.529/BA.1).
Results
Validation controls showed accurate and precise results. The correlation study found the accuracy of 96.38 to 100% (95% CI) in nasopharyngeal and 94.87 to 100% (95% CI) in saliva for SARS-CoV-2 and 91.1 to 100% (95% CI) for both Influenza A/B and RSV. The diagnostic efficiency of this assay was not affected by SARS-CoV-2 variant, including Omicron.
Conclusion
The TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay is a rapid method to detect and differentiate SAR-CoV-2, Flu A and B, and RSV in nasopharyngeal and saliva samples. It has a significant role in the diagnosis and management of respiratory illnesses and the clinical implications of co-infection.
Introduction
Respiratory tract infections remain a significant public threat with high morbidity and mortality worldwide.Citation1 Viruses are responsible for about 90% of upper respiratory tract infections and, about 30% of lower respiratory tract infections.Citation2 The respiratory syndrome can be caused by several viruses, including rhinoviruses, respiratory syncytial viruses, adenoviruses, influenza viruses, and parainfluenza viruses.Citation1 Most of the viruses have characteristic seasonal patterns, including Influenza virus and respiratory syncytial virus (RSV), which rise mainly in the winter. In addition, the coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) added the potential threats of respiratory viruses. During this pandemic, co-infection with influenza and RSV could pose a challenge to healthcare providers due to their overlapping clinical presentations. Influenza and COVID-19 share very similar symptoms. However, the incubation period of COVID-19 is longer (2–14 days), and people stay infectious longer than with the flu. These viruses can cause fever, chills, headaches, cough, muscle soreness, fatigue, vomiting, diarrhea, shortness of breath, runny nose, and sore throat. The only symptom that is unique to COVID-19 is loss of taste or smell.Citation3 In the case of RSV, symptoms appear four to six days after infection and are usually self-limiting. However, in infants and the elderly, symptoms tend to be more severe and can include fever and wheezing.Citation4
The respiratory illness caused by the influenza virus, SARS-CoV-2 virus, and RSV are all highly contagious. In addition, there is a chance of co-infection with all multiple viruses at the same time complicating the situation.Citation5
The SARS-CoV-2 virus is a newly discovered novel coronavirus, causing a global pandemic of COVID-19. In severe conditions, it can cause severe lower respiratory disease including pneumonia and respiratory failure and, in some cases, death. The SARS-CoV-2 virus is the seventh known coronavirus belonging to the 2B group of the Betacoronavirus family, which is the same family as SARS-CoV and MERS-CoV, and has a 70% similarity in genetic sequence to SARS. Each SARS-CoV-2 virion is 50–200 nm in diameter and composed of several proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N): the N protein holds the RNA genome, and S, E, and M proteins together create the viral envelope. The spike protein is a key player in binding angiotensin-converting enzyme 2 (ACE2) receptors and mediating membrane fusion and virus entry into host cells.Citation6
Influenza, or the flu, is a contagious viral infection caused primarily by the influenza virus A, B, or C and transmitted through respiratory droplets or airborne. It affects mainly the upper respiratory tracts (ie, the nose, throat, bronchi, and infrequently, lungs) but other organs such as the heart, brain, and muscles can be involved. Influenza infection can increase morbidity and mortality in pediatric, elderly, and immunocompromised populations by serious complications of pneumonia. Influenza viruses are segmented negative-strand RNA viruses belonging to the Orthomyxoviridae family with diverse antigenic characteristics.Citation7 Influenza A (Flu A) is the most common type of influenza virus in humans and is generally responsible for seasonal flu epidemics and potential pandemics. Influenza A virus can be further classified into subtypes based on two surface proteins: hemagglutinin (H) and neuraminidase (N). Seasonal flu is normally caused by influenza A subtypes H1, H2, H3, N1, and N2. The influenza B virus has a similar viral structure to type A, but no subtypes due to the fixed antigenic characteristics of HA and NA.Citation8
RSV is a non-segmented negative-strand RNA virus and a member of the Pneumoviridae family (formerly Paramyxoviridae), consisting of two strains (subgroups A and B).Citation9 It is also a contagious disease that affects primarily infants with a mortality rate of 0.3% in hospitalized children and immunocompromised elderly, making them at the highest risk for death.Citation10
Due to the similarity in symptoms, laboratory investigation is the best way to diagnose these viral diseases. Multiplex RT-PCR assays are fast with increased sensitivity for the detection of a wider range of respiratory viruses than immunofluorescence (DFA) and viral culture. The ability to detect multiple pathogens in a single test is an important diagnostic advance as it provides an approach to better understanding the outcomes and clinical significance of co-infection. There are some commercially available kits and CDC’s developed assay (FDA-emergency use authorization) that detect either SARS-CoV-2 only, SARS-CoV-2 and Influenza A/B, SARS-CoV-2, Flu & RSV, or Multiplex respiratory panel.Citation11 They can detect a limited number of samples and take a long time if needed to test a larger volume of samples. Therefore, the rapid, accurate, and economical diagnostic assay is highly desirable in diagnostic laboratories that can detect and differentiate SARS-CoV-2, influenza, and RSV even in a larger volume of samples. The TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay is a newly developed molecular diagnostic assay that enables the detection and differentiation of SARS-CoV-2, influenza A and B, and RSV in respiratory specimens. This is a multiplex real-time PCR assay containing primer and probe sets specific to SARS-CoV-2 (N gene and S gene), Flu A/B, RSV, and MS2 for the detection of RNA from the SARS-CoV-2 virus, influenza A and B viruses, and RSV subtypes A and B, respectively.
In this study, we evaluated the performance characteristics of TaqMan SARS-CoV-2, Flu A/B, and RSV RT-PCR multiplex assay for the detection of SARS-CoV-2, RSV, and Influenza viruses using nasopharyngeal and saliva samples.
Materials and Methods
Validation Controls
A total of four different culture fluids with known controls of Influenza A virus (H3N2) (A/Wisconsin/67/2005), Influenza B virus (B/Virginia/ATCC4/2009), and RSV A2 cpts-248 from ATCC and SARS-CoV-2 (USA-WA1/2020) from Zeptometrix, NY, USA, were used for determining the accuracy, linearity, efficiency, and reproducibility of the TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay. The quantitative genomic RNA, namely Influenza A virus (H1N1) strain A/PR/8/34 (VR-95DQ) and RSV A2 (VR-1540DQ) from ATCC and SARS-CoV-2 (MN908947.3 Wuhan-Hu-1) from Twist bioscience were used for the accuracy, precision, and limit of detection of the assay.
Extraction of RNA from Controls
Total RNA was extracted from the controls using Omega Bio-Tek Mag-Bind Viral RNA Xpress Extraction Kit on the automated liquid handling Hamilton STAR according to the manufacturer’s instruction.Citation12
Multiplex Reverse Transcriptase PCR
Extracted RNA (17.5 μL) was mixed with a 7.5 μL reaction mixer containing TaqPath 1-Step Multiplex Master Mix (No ROX™) and TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay per reaction according to the manufacturer’s instruction.Citation13 This multiplexed assay includes primer and probe set specific to SARS-CoV-2 (N gene and S gene), Flu A/B, RSV, and MS2 for the detection and differentiation of SARS-CoV-2, Flu A/B, and RSV viruses. The PCR reaction was incubated at 50 ℃ for 10 min, followed by 95 ℃ for 2 min and 46 cycles of 95 ℃ for 3s and 60 ℃ for 30s and run in 96 well plates by standard curve mode in QuantStudio 12K flex. The threshold level was determined for each target based on the exponential phase of the amplification curve above the background signal of the highest dilution series.
Accuracy Using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay
Validation controls using culture fluid of Influenza A virus (H3N2) (A/Wisconsin/67/2005), Influenza B virus (B/Virginia/ATCC4/2009), RSV A2 cpts-248 and SARS-CoV-2 (USA-WA1/2020), and quantitative RNA of Influenza A virus (H1N1) strain A/PR/8/34 (VR-95DQ), RSV A2 (VR-1540DQ), and SARS-CoV-2 (MN908947.3 Wuhan-Hu-1) were used for multiplex PCR using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay on QuantStudio 12K Flex to detect SARS-CoV-2, Flu A and B and RSV to check the accuracy of the controls.
Precision or Reproducibility Using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay
The intra-assay and inter-assay reproducibility or variability of the TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay was assessed by using quantitative genomic RNA from Influenza A virus (H1N1) strain A/PR/8/34 (VR-95DQ), RSV A2 (VR-1540DQ), and SARS-CoV-2 (MN908947.3 Wuhan-Hu-1) in triplicates on three runs.
Standard Curve Using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay
To determine the dynamic range of the one-step multiplex RT-PCR assay, four different controls of Influenza A virus (H3N2) (A/Wisconsin/67/2005), Influenza B virus (B/Virginia/ATCC4/2009), and RSV A2 cpts-248 from ATCC and SARS-CoV-2 (USA-WA1/2020) from Zeptometrix were serially diluted in 10-fold with nuclease-free water (108 to 1 copies/reaction) for each target to obtain standard curves and amplification plots of the multiplex RT-PCR assays. The total RNA was extracted from diluted controls, and RT-PCR was run using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay. The standard curve was generated, and cutoff values were established based on the lowest detected copy numbers or the highest Ct. Cycle threshold (Ct) values were plotted against quantities of RNA to establish a standard curve, and linear regression analysis was performed for the four targets, allowing the determination of the correlation coefficient (R2). The amplification efficiencies (E) of the reactions were calculated from the curves using the equation E = −1+10(−1/slope).
Analytical Specificity Using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay
The species-specific sample that includes human coronavirus 229E, MERS, rhinovirus, and negative specimens were used to verify the specificity of TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay.
Validation Using Patients Samples
The TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay were assessed by using two different types of specimens – nasopharyngeal and saliva samples. The nasopharyngeal swab was collected in a disposable virus sampling tube containing 3 mL of virus transport medium. Saliva samples were collected into Omni tubes containing ceramic beads for homogenization in the Omni Bead Ruptor Elite (OMNI International, Perkin Elmer company). All the clinical specimens were assessed in accordance with the Declaration of Helsinki in Patients Choice Laboratories, IN, USA. The study was approved by the Sterling Institutional Review Board (IRB ID-9190) and includes de-identified samples.
A total of 110 de-identified nasopharyngeal specimens, of which 50 were positive and 60 were negative for SARS-CoV-2 by reference method (Taqpath Combo kit, Applied Biosystems), were used for SARS-CoV-2 detection using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay. A total of 70 de-identified saliva samples were included in the study. Of them, five samples were previously positive for SARS-CoV2 (as determined by the reference method). Due to the low number of saliva samples (n = 5) positive for SARS-CoV2, we spiked saliva with positive nasopharyngeal specimens (n = 30) to evaluate the performance of the TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay. In brief, the nasopharyngeal specimens previously tested positive for SARS-COV-2 (reference method) were centrifuged at 3000 rpm for 5 minutes, the supernatant was discarded, and the collected pellet was reconstituted with 30 negative saliva samples. Further, an additional 35 negative saliva samples were used as negative specimens to verify the specificity of assays.
A total of 60 de-identified nasopharyngeal specimens, of which 20 were positive for Flu A/B and 20 were positive for RSV, and 20 were negative for both by reference method (multiplex RT-PCR open array technology, Applied Biosystems) was used for detecting Influenza A/B and RSV.
A total of 15 nasopharyngeal samples, of which 6 were previously positive for SARS-CoV-2, Influenza, and RSV, 3 were positive for SARS-CoV-2 and RSV and 6 were positive for SARS-CoV-2 and Influenza by the reference method were included for co-infection detection by TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay.
Furthermore, a total of 20 nasopharyngeal specimens and 25 saliva samples that were previously tested negative for SARS-CoV-2, Flu A/B, and RSV were selected and spiked with culture fluids of Influenza A virus (H3N2) (A/Wisconsin/67/2005), Influenza B virus (B/Virginia/ATCC4/2009), RSV A2 cpts-248, and SARS-CoV-2 (USA-WA1/2020) controls to check the accuracy of the assay in the co-infection.
Extraction of RNA from the Patient’s Samples
Total RNA was extracted from 200 μL samples in the viral transport medium and 200 μL homogenized saliva samples using Omega Bio-Tek Mag-Bind Viral RNA Xpress Extraction Kit on the automated liquid handling Hamilton STAR according to the manufacturer’s instruction.Citation12
Reference Methods
Taqpath Combo Kit Assay
After total RNA was extracted from nasopharyngeal and saliva specimens, SARS-CoV-2 was detected by the FDA-EUA-approved method validated at Patients Choice Laboratories using the Taqpath combo kit (Applied Biosystems, Thermo Fisher Scientific). This multiplex PCR method includes primer and probe sets specific to the N gene, S gene, and Orf ab gene for detecting a single SARS-CoV-2. The master mix was prepared from TaqPath 1-Step Multiplex Master Mix (No ROX™), COVID-19 Real-Time PCR Assay Multiplex, and Nuclease-free water. Ten microliters of Master mix was dispensed into the 384 well plates followed by the addition of 2.5 μL of the eluted specimen to the appropriate well. Each run includes a SARS-CoV-2 positive control and negative control. The PCR reaction includes an incubation at 50 ℃ for 10 min, followed by 95 ℃ for 2 min and 40 cycles of 95 ℃ for 3 s and 60 ℃ for 30s and run by standard mode in QuantStudio 12K flex.
Multiplex RT-PCR Open Array Technology
After total RNA was extracted from respiratory samples, RNA was reverse transcribed to cDNA in the thermocycler. The Influenza A/B and RSV were determined by multiplex RT-PCR using open array technology method and validated at Patients Choice Laboratories using TaqMan open array master mix and customized respiratory panel open array chips (Applied Biosystems, Thermo Fisher Scientific). The open array chip is a microscope slide–sized plate with 48 sub arrays, each with 64 through-holes. It has multiple targets of respiratory viruses and bacteria including influenza and RSV. The readymade open array master mix is mixed with the sample in the 1:1 ratio in the 384 well plates, and samples are transferred into open array chips with the help of Accufill. Each through hole receives 33 nl sample. The open array is loaded into QuantStudio 12K flex instrument for RT-PCR.
Analytical Sensitivity (Limit of Detection)
The lowest RNA concentration of Influenza A and B, RSV, and SARS-CoV-2 that allows the multiplex RT-PCR to determine the correct target is considered as LOD for TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay. The quantitative genomic RNA from Influenza A virus (H1N1) strain A/PR/8/34 (VR-95DQ), RSV A2 (VR-1540DQ), and SARS-CoV-2 (MN908947.3 Wuhan-Hu-1) were serially diluted in 10-fold with pooled negative nasopharyngeal specimens from 1 x 107copies/μL to 1copy/μL for LOD. The standard curve was generated, and cutoff values were established based on the lowest detected copy numbers or the highest Ct. LOD is considered after >95% positive results from 19/20 replicates.
For LOD in saliva, a ten-fold dilution series of Influenza A virus (H3N2) (A/Wisconsin/67/2005), RSV A2 cpts-248, and SARS-CoV-2 (USA-WA1/2020) was spiked into pooled negative saliva samples.
Data Analysis
Data were analyzed using QuantStudioTM Design and Analysis Software v2.5 for TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay method and SDS software v1.4.1 for reference method. Basic statistical analysis, including mean, standard deviation, and coefficient of variation of the mean Ct value, were calculated using Excel (Microsoft Corp., Redmond, WA, USA). The standard curve was generated and efficiency, R2, and slope were calculated using QuantStudioTM Design and Analysis Software v2.5. Correlation analysis was performed using the MedCalc statistical software, version 16.4.3 (Ostend, Belgium). The cycle threshold (Ct) values of a positive sample between reference method assays and TaqMan multiplex RT-PCR assay by simple regression were assessed using the Pearson correlation coefficient for comparative analysis.
Results
Validation from Controls
Accuracy
Validation controls of Influenza A virus (H3N2) (A/Wisconsin/67/2005), Influenza B virus (B/Virginia/ATCC4/2009) and Influenza A virus (H1N1) strain A/PR/8/34 (VR-95DQ) showed positive for Flu A and B, RSV A2 cpts-248 and RSV A2 (VR-1540DQ) showed positive for RSV and SARS-CoV-2 (USA-WA1/2020) and SARS-CoV-2 (MN908947.3 Wuhan-Hu-1) showed positive for SARS-CoV-2 as expected for known controls when tested in triplicates (Supplementary Table 1).
Precision
The inter-assay and intra-assay precision study using RNA controls of Influenza A virus (H1N1) strain A/PR/8/34 (VR-95DQ), RSV A2 (VR-1540DQ), and SARS-CoV-2 (MN908947.3 Wuhan-Hu-1) was performed in triplicates. Precision was determined by calculating the mean, SD, and CV using cycle threshold (Ct) or Cq. Results from precision showed acceptable reproducibility and repeatability with a CV of less than 5% when Ct was used (Supplementary Table 2).
Standard Curve Using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay
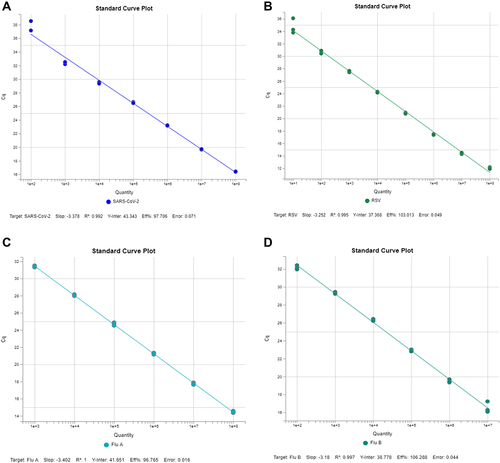
The standard curve using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay showed a linear efficiency between 90 and 110% and R2 values with 0.99 for SARS-CoV-2, Flu A/B, and RSV ().
Figure 1 Standard curve of SARS-CoV-2, RSV and Flu A and B using control organisms. Standard curve of SARS-CoV-2 showing linear efficiency of 97.7% and R2 is 0.99 (A); Standard curve of RSV showing linear efficiency of 103% and R2 is 0.99 (B); Standard curve of Influenza A showing linear efficiency of 96.7% and R2 is 1 (C); and Standard curve of Influenza B showing linear efficiency of 106.2% and R2 is 0.99 (D).
Correlation of Two Methods
For SARS-CoV-2
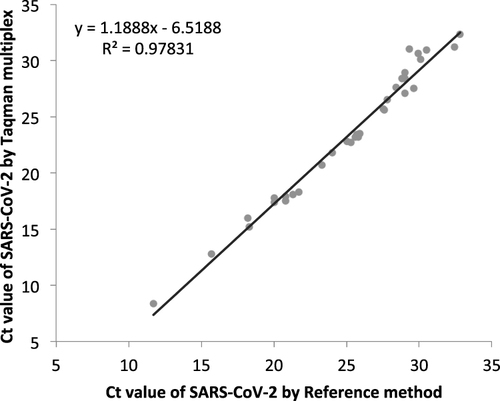
The correlation of the reference method (Taqpath combo kit) and TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay method for Sars-CoV-2 was determined in nasopharyngeal and saliva samples. Considering the reference method as standard, the sensitivity, specificity, and accuracy using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay method in NP specimens are 92.89% to 100%, 92.89% to 100%, and 96.38–100% (95% CI), respectively (). Similarly, the sensitivity, specificity, and accuracy using TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay methods in saliva samples are 90% to 100%, 90% to 100%, and 94.87% to 100% (95% CI), respectively, when compared with the reference method ().
Table 1 Correlation of SARS-CoV-2 in Nasopharyngeal Specimens
Table 2 Correlation of SARS-CoV-2 in Saliva Specimens
The correlation between the cycle threshold (Ct) value of positive SARS-CoV-2 samples by reference method and TaqMan multiplex assays (n = 33) is shown in . There was a strong positive correlation between CT values of SARS-CoV-2 positive samples by two assay methods with Pearson correlation (r) of 0.98.
For Influenza A/B and RSV
The correlation of the reference method (Multiplex RT-PCR-open array technology) and TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay method for influenza and RSV was determined in nasopharyngeal specimens. Considering the reference method as standard, the sensitivity, specificity, and accuracy with TaqMan SARS-CoV-2, Flu A/B, and RSV Multiplex Assay method in NP specimens are 83.1% to 100%, 83.1% to 100%, and 91 to 100% (95% CI), respectively, for both Influenza A/B and RSV ( and ). Due to the low number of saliva samples, only four saliva samples positive for influenza were run and found positive results by TaqMan multiplex assay method (data not shown).
Table 3 Correlation of Influenza A/B in Nasopharyngeal Specimens
Table 4 Correlation of RSV in Nasopharyngeal Specimens
Determination of Co-Infection
Among the total 15 patients’ samples previously detected co-infection with, either SARS-CoV-2 and Influenza or SARS-CoV-2 and RSV or SARS-CoV-2, Influenza and RSV showed concordant results with the reference method. The spiked NP showed 100% agreement with the expected result of co-infection presenting 20 out of 20 NP samples positive for SARS-CoV-2, RSV, and Influenza. Further, spiked saliva samples also showed 100% agreement with the expected result of co-infection in which 16 out of 16 saliva samples are positive for SARS-CoV-2, Influenza, and RSV (Supplementary 3).
Limit of Detection (LOD)
The LOD in nasopharyngeal specimens for SARS-CoV-2 was 100 copies/reaction with a cutoff of 34, 100 copies/reaction for Flu A and B with a cutoff value of 32, and 10 copies/reaction for RSV with a cutoff of 34. LOD was confirmed after >95% positive results from 19/20 replicates ().
Table 5 LOD Determination in Nasopharyngeal Specimens Using Quantitative RNA (RNA Copies/Reaction)
The LOD in saliva for SARS-CoV-2 was 500 copies/reaction at a cutoff of 32, RSV was 100 copies/reaction at a cutoff of 32, and Flu was 500 copies/reaction at a cutoff of 32 ().
Table 6 LOD Determination in Saliva Using Quantitative RNA (RNA Copies/Reaction)
Discussion
Multiplex PCR is considered as a rapid and sensitive technique for the detection of respiratory viruses. Therefore, we evaluated the performance of the TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay for the detection and differentiation of SARS-CoV-2, RSV, and Flu A and B from the nasopharyngeal swabs and saliva. Due to the minimum risk of exposure and ease of collection, saliva is considered as an alternative sample for COVID-19 screening and diagnosis.Citation14 Moreover, the detection of respiratory viruses by multiplex RT-PCR was comparable between nasopharyngeal swabs and saliva.Citation15 The TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay validated using various controls in the present study was accurate and precise with an efficiency of 90–110%. This multiplex PCR assay can detect multiple organisms in a single test. The clinical symptoms of COVID-19 and Flu are very similar. Therefore, rapid, specific, and sensitive assays are required for the effective differentiation of SARS-CoV-2, RSV, and influenza viruses to enable precise treatment, prevention, and control. Multiplex PCR has been in use for rapid detection and precise identification of many respiratory viruses by incorporating several primers within one reaction tube to amplify genomic fragments of many pathogens.Citation16 Previous studies demonstrated a 30% to 50% increase in the diagnostic yield of respiratory viruses compared to direct fluorescent antibody and culture.Citation17 The meta-analysis data showed that multiplex PCR provides highly accurate results in the relevant time frame.Citation16 The Centre for Disease Control and Prevention (CDC) also developed CDC Influenza SARS-CoV-2 Multiplex Assay to help public health laboratories to continue influenza surveillance, while they are also testing for SARS-CoV-2.Citation18 BioFire FilmArray RP, Nanosphere Verigene RV+ Test, and Hologic Gen-Probe Prodesse® assays were found to be highly accurate for the detection of respiratory viruses.Citation16 However, they can detect limited samples and take a long time if needed to test a larger volume of samples. TaqMan SARS-CoV-2, Flu A/B, and RSV RT-PCR multiplex assays are single-well assays that allow large-scale screening in the context of a pandemic and can detect a larger volume of samples with the same rate of sensitivity in one and half hours after RNA is isolated. It is cost-effective when a larger volume of samples needs to be detected for multiple respiratory viruses in less turnaround time. Although many rapid diagnostic kits are available to detect respiratory viruses in less than 15 minutes, their sensitivity is not consistent.Citation19 Apart from influenza viral infection, RSV is one of the leading causes of bronchiolitis and pneumonia in children and immunocompromised patients, and it may cause severe complications.Citation20 Therefore, it requires a definitive diagnosis. Previous studies have revealed that multiplex PCRs are highly sensitive and specific for RSV and flu, and they offer a more rapid and accurate alternative to traditional methods.Citation21
Due to numerous limitations in conventional methods such as antigen tests, direct fluorescent antibody assays, and viral culture, rapid and sensitive PCR-based molecular assays are considered better options to diagnose respiratory viral infections.Citation22 Furthermore, the analytical sensitivity of multiplex was highly comparable with singleplex real-time PCR.Citation23 Therefore, TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay can be one of the rapid and reliable methods for detecting multiple respiratory viruses including co-infection. During the SARS-CoV-2 pandemic, co-circulation of influenza and RSV could pose a serious problem as these viral infections have overlapping clinical presentations. In the present study, we evaluated the co-infection using TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay by spiking with culture fluid of Influenza, SARS-CoV-2, and RSV controls and found 100% accuracy with the assays. The present method was concordant with the reference method to detect co-infection. Reference methods (Taqpath combo kit and multiplex RT-PCR open array technology) are two separate methods. Therefore, TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay can be a cost-effective and rapid method to determine co-infection. A high prevalence of coinfection was elucidated with influenza A (22.3%) virus and RSV (9.7%) in SARS-CoV-2 positive dead cases.Citation5 Other studies also indicated the co-occurrence of respiratory viruses including Influenza virus, Human metapneumovirus, Rhinovirus, and RSV in COVID-19 patients.Citation24 It was indicated that the multiplex PCR format can detect co-infections which is not able to be done using monoplex PCR or culture.Citation25 These data highlight the importance of the multiplex test. When clinical labs focused only on SARS-CoV-2 during this COVID pandemic, they may miss the diagnosis of co-infection with influenza or RSV, which are other important respiratory viruses leading to serious respiratory problems. To minimize the requirement of testing the same sample multiple times, TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay can be a rapid and accurate alternative multiplex test to detect coinfection with a rapid turnaround time.
Further, the current study found accuracy >90% (95% CI) and sensitivity and specificity >80% (95% CI) for the detection of SARS-CoV-2, Flu A/B, and RSV in nasopharyngeal and saliva samples using TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay. The major limitation of this study is the small sample size. There was a low prevalence of Flu A/B and RSV at the time the study was conducted. However, we have evaluated samples recently collected that include new circulating respiratory viral strains. Despite the limitation of sample size, our study provides a fundamental basis to conduct a similar study on a large scale to validate the clinical utility of TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay.
The strong positive correlation between TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay and reference method (TaqPath combo kit, Applied Biosciences, ThermoFisher) shows that the two methods are comparable. We also evaluated the performance of TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay to detect SARS-CoV-2 positivity using SARS-CoV-2 variant controls (B.1.1.7_601443, B.1.617.1_1662307, P.1_792683, B.1.351_678597, B.1.1.529/BA.1) and SARS-CoV-2 delta variant isolated from patients’ sample previously confirmed by sequencing. These controls and samples were found to be positive for SARS-CoV-2 indicating that TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay is not affected by these published variants. Most of the nasopharyngeal and saliva samples of the present study were found to be delta variants with a mutation in L452R, T478K, D614G, and P681R by using TaqMan SARS-CoV-2 mutation panel assay, validated in Patients Choice Laboratories.Citation26 Our data were concordant with the CDC data that shows more than 95% Delta variant from August to November.Citation27 The SARS-CoV-2 positive samples collected after December 24 were found to be Omicron (B.1.1.529/BA.1) indicating that TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay is not impacted by currently circulating variant including Omicron variant which has at least 50 mutations.
Conclusions
The TaqMan SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay performed on the QuantStudio 12k flex is a rapid method to detect and differentiate SAR-CoV-2, Flu A and B, and RSV in nasopharyngeal and saliva samples. It has a significant role in the diagnosis and management of respiratory illnesses and clinical implications of co-infection. In addition, it may help to provide information for epidemiological and surveillance purposes.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgment
We are grateful to the laboratory staff and management of Patients Choice Laboratories for their immense support during the study. We acknowledge Mr Tyler McKee for checking the English in this manuscript.
References
- Avendaño Carvajal L, Perret Pérez C. Epidemiology of respiratory infections. Paediatr Respir Rev. 2020;(1):263–272. doi:10.1007/978-3-030-26961-6_28
- Korsman SNJ, van Zyl GU, Nutt L, Andersson MI, Preiser W. Respiratory viruses, virology. Churchill Livingstone; 2012:108–109.
- Centers for Disease Control and Prevention [homepage on the Internet]. Similarities and differences between Flu and COVID-19. Available from: https://www.cdc.gov/flu/symptoms/flu-vs-covid19.htm. Accessed December 3, 2021.
- Centers for Disease Control and Prevention [homepage on the Internet]. People at high risk for severe RSV infection. Available from: https://www.cdc.gov/rsv/high-risk/index.html. Accessed December 3, 2021.
- Hashemi SA, Safamanesh S, Ghasemzadeh-Moghaddam H, Ghafouri M, Azimian A. High prevalence of SARS-CoV-2 and influenza A virus (H1N1) coinfection in dead patients in Northeastern Iran. J Med Virol. 2021;93(2):1008–1012. doi:10.1002/jmv.26364
- Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi:10.1016/j.ijantimicag.2020.105924
- Poon LL, Song T, Rosenfeld R, et al. Quantifying influenza virus diversity and transmission in humans. Nat Genet. 2016;48(2):195–200. doi:10.1038/ng.3479
- Kanegae Y, Sugita S, Endo A, et al. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J Virol. 1990;64(6):2860–2865. doi:10.1128/jvi.64.6.2860-2865.1990
- Crowe JE Jr, Williams JV. Paramyxoviruses: respiratory syncytial virus and human metapneumovirus. Viral Infect Human. 2014;601–627. doi:10.1007/978-1-4899-7448-8_26
- Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi:10.1001/jama.289.2.179
- Centers for Disease Control and Prevention [homepage on the Internet]. Available from: https://www.cdc.gov/flu/professionals/diagnosis/table-flu-covid19-detection.html. Accessed August 11, 2022.
- Omega biotek. Mag-Bind® Viral RNA XPress Kit. Available from: https://www.omegabiotek.com/product/viral-rna-extraction-kit-mag-bind-viral-rna-xpress/. Accessed August 31, 2022.
- TaqMan™ SARS-CoV-2, Flu A/B, RSV RT-PCR multiplex assay. Pub. No. MAN0019614. Available from: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0019614_TaqManSARS-CoV-2FluAB_RSV_RT-PCRAssay_QR.pdf. Accessed September 05, 2022.
- Teo AKJ, Choudhury Y, Tan IB, et al. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Sci Rep. 2021;11:1–8. PMID: 33542443.
- Kim YG, Yun SG, Kim MY, et al. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J Clin Microbiol. 2016;55(1):226–233. PMID: 27807150. doi:10.1128/JCM.01704-16
- Huang HS, Tsai CL, Chang J, Hsu TC, Lin S, Lee CC. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin Microbiol Infect. 2018;24(10):1055–1063. PMID: 29208560. doi:10.1016/j.cmi.2017.11.018
- Babady NE. The FilmArray® respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev Mol Diagn. 2013;13:779–788. doi:10.1586/14737159.2013.848794
- Centers for Disease Control and Prevention [homepage on the Internet]. CDC influenza SARS-CoV-2 multiplex assay. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/testing.html. Accessed December 4, 2021.
- Centers for Disease Control and Prevention [homepage on the Internet]. Rapid influenza diagnostic tests. Available from: https://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm. Accessed December 4, 2021.
- Piedimonte G, Perez MK. Respiratory syncytial virus infection and bronchiolitis. Pediatr Rev. 2014;35(12):519–530. PMID: 25452661. doi:10.1542/pir.35-12-519
- Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20:49–78. doi:10.1128/CMR.00002-06
- Parker J, Fowler N, Walmsley ML, et al. Analytical sensitivity comparison between singleplex real-time PCR and a multiplex PCR platform for detecting respiratory viruses. PLoS One. 2015;10(11):e0143164. PMID: 26569120. doi:10.1371/journal.pone.0143164
- Zhu X, Ge Y, Wu T, et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi:10.1016/j.virusres.2020.198005
- Bharaj P, Sullender WM, Kabra SK, et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89. PMID: 19558656. doi:10.1186/1743-422X-6-89
- Neopane P, Nypaver J, Shrestha R. Beqaj SS. SARS-CoV-2 variants detection using TaqMan SARS-CoV-2 mutation panel molecular genotyping assays. Infect Drug Resist. 2021;14:4471–4479. PMID: 34737587. doi:10.2147/IDR.S335583
- Centers for Disease Control and Prevention [homepage on the Internet]. Variant proportions. Available from: https://covid.cdc.gov/covid-data-tracker/#variant-proportions. Accessed December 4, 2021.
- ASM article. How ominous is the omicron variant (B.1.1.529)?; 2021. Available from: https://asm.org/Articles/2021/December/How-Ominous-is-The-Omicron-Variant-B-1-1-529. Accessed August 31, 2022.