Abstract
Background
The emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) has garnered international concern due to its significant antibiotic resistance. Notably, children exhibit distinct resistance mechanisms compared to adults, necessitating a differential approach to antibiotic selection. A thorough analysis of CRKP’s epidemiology and drug resistance mechanisms is essential for establishing a robust foundation for clinical anti-infection strategies and precise prevention and control measures.
Methods
This study involved the collection of 31 non-repetitive strains from pediatric and adult patients at a tertiary hospital in China, spanning from July 2016 to July 2022, testing for resistance genes, antimicrobial susceptibility, and homology analysis.
Results
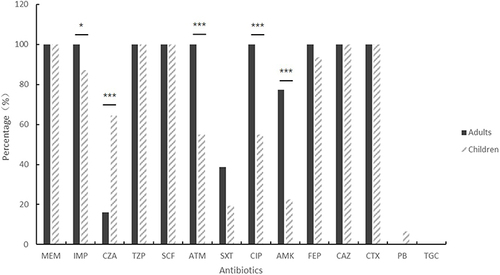
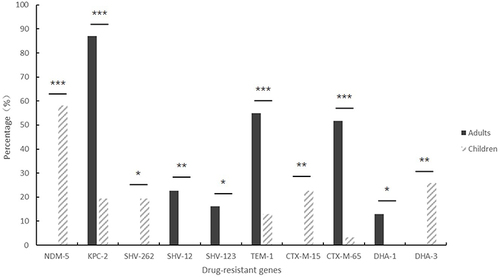
Infants (0–1 year) were the largest pediatric CRKP group, with 61.3% of cases. The neonatal intensive care unit (NICU) and pediatrics were the main departments affected. Adults with CRKP had a mean age of 67 years, with the highest prevalence in neurology and emergency ICU. Antimicrobial susceptibility testing revealed that adult CRKP strains exhibited higher resistance to amikacin, ciprofloxacin, cotrimoxazole, and aztreonam compared to pediatric strains. Conversely, pediatric strains showed a higher rate of resistance to ceftazidime/avibactam. The predominant resistance genes identified were blaNDM-5 in children (58.1%) and blaKPC-2 in adults (87.1%), with over 93% of both groups testing positive for extended-spectrum beta-lactamase (ESBL) genes. Multilocus Sequence Typing (MLST) indicated ST2735 and ST11 as the predominant types in children and adults, respectively. Pulsed-field gel electrophoresis (PFGE) identified clonal transmission patterns of ST11 blaKPC-2 and ST15 blaOXA-232 across both age groups. Notably, this study reports the first instance of ST1114-type CRKP co-producing blaNDM-5 and blaOXA-181 in the NICU.
Conclusion
This study reveals distinct resistance mechanisms and epidemiology in CRKP from children and adults. The identified clonal transmission patterns emphasize the need for improved infection control to prevent the spread of resistant strains.
Introduction
Klebsiella pneumonia (K. pneumoniae), a member of the Enterobacteriaceae family, is an opportunistic pathogen capable of colonizing the human nasopharynx and gastrointestinal tract.Citation1 It is recognized for its potential to cause a spectrum of diseases, including respiratory infections, bloodstream infections, urinary tract infections, wound infections, and meningitis.Citation2 K. pneumoniae is a prevalent pathogen in both healthcare-associated and community-acquired infections. Recent surveillance reports on bacterial resistance in China have indicated a concerning trend: the resistance rate of K. pneumoniae to major carbapenems, such as imipenem and meropenem, has escalated from 2.9% and 3.0% in 2005 to 27.1% and 25.5% in 2021, marking an alarming eight-fold increase.Citation3 This rise underscores the urgency of addressing antimicrobial resistance. According to the 2019 Global Burden of Disease (GBD) study, antimicrobial resistance ranks as the third leading cause of mortality, following closely behind ischemic heart disease and stroke.Citation4 In the context of antibiotic resistance-related mortality, Escherichia coli (E.coli) is the most prevalent pathogen, with K. pneumoniae ranking as the second. It is noteworthy that several low-income countries bear the brunt of deaths related to antibiotic resistance. A study by Cai Yun et al revealed a stark disparity in mortality rates between carbapenem-resistant Klebsiella pneumoniae (CRKP) and carbapenem-susceptible Klebsiella pneumoniae (CSKP), with CRKP exhibiting a significantly higher mortality rate of 42.1% compared to CSKP’s 17.5%. This underscores the pressing need to address CRKP as a critical global public health challenge.Citation5
Carbapenemase production is a primary mechanism behind carbapenem resistance in K. pneumoniae, alongside other factors such as efflux pump overexpression, the presence of extended-spectrum beta-lactamase (ESBL), or deficiencies in membrane proteins. In China, KPC-2 is the predominant carbapenemase enzyme, succeeded by NDM and OXA types.Citation6 The predominant genotypes of CRKP in pediatric patients can vary significantly by region,Citation7,Citation8 which contrasts with the more uniform profile observed in adult patients, where KPC-2 is the predominant enzyme.Citation9 In recent years, there has been a growing incidence of CRKP strains co-producing two or even three different carbapenemase enzymes.Citation10,Citation11 These strains, which are increasingly reported in the literature, exhibit an elevated level of resistance to multiple antibiotics. This phenomenon poses a considerable challenge to clinical therapeutics, necessitating innovative approaches to antimicrobial stewardship and treatment strategies.
The ST11 clone has emerged as the predominant strain in numerous regions,Citation12–14 supplanting other typical sequence types (STs) such as ST15 and ST290.Citation15 Furthermore, the dominant ST-type clones in pediatric populations exhibit significant geographical variation,Citation7,Citation16 different from adults, ST11 type is mainly detected.Citation17,Citation18 Given the documented transmission of various molecular types, including ST15, ST48, ST716, and ST147.Citation13,Citation19–21 It is imperative that we maintain a vigilant surveillance of each ST type. The heterogeneity of ST types underscores the complexity of CRKP epidemiology and the necessity for a nuanced approach to infection control and management strategies.
In summary, child’s compromised immune systems, coupled with the significant prevalence of drug resistance, mortality, and transmission rates associated with carbapenem-resistant Klebsiella pneumoniae (CRKP), highlight the urgency of addressing this pathogen. The distinct resistance mechanisms and molecular epidemiological patterns observed in pediatric populations, as opposed to those in adults, further complicate the clinical management of infections. It is noteworthy that the body of research on the epidemiology and resistance mechanisms of CRKP in children remains less extensive compared to that in adults. This study aims to bridge this gap by conducting a comprehensive analysis of the molecular epidemiology, resistance mechanisms, and genetic homology of CRKP strains isolated from both pediatric and adult patients at the Affiliated Hospital of Xuzhou Medical University. The findings from this research will offer valuable insights and a robust evidence base to inform clinical practices, enabling more targeted and effective treatment strategies for diverse patient populations.
Materials and Methods
Strain Collection and Identification
The study period spanned from July 2016 to July 2022, during which a total of 31 non-repetitive strains were collected from each group of pediatric and adult patients at the Affiliated Hospital of Xuzhou Medical University. To ensure comparability, adult CRKP strains corresponding to the same months were selected using a random number generator, aligned with the collection timeline of each strain. Strain identification was conducted using Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS, Bruker Daltonics, Billerica, MA, USA). The VITEK-2 compact automated microbiological analyzer (Biomerieux, Marcy L’Etoile, France) was employed to confirm all isolates as CRKP, given their resistance to at least one carbapenem antibiotic, including imipenem, meropenem, or ertapenem. A comprehensive collection of patients’ clinical data was performed, encompassing variables such as age, gender, infection type, isolation date, sample type, ward, comorbidities, surgical history, invasive procedures, prior antibiotic exposure, antibiotic treatments, patient outcomes, and hospital stay duration. It is important to note that there is no physical interconnection between the pediatric and adult clinics at the hospital, with all staff and equipment operating independently. The study was granted ethical approval by the Affiliated Hospital of Xuzhou Medical University’s Ethics Committee (XYFY2020-KL084).
Antibiotic Susceptibility Testing
The minimal inhibitory concentrations (MIC) of 13 antibiotics were determined by VITEK-2 Compact system, including Imipenem (IMP), Meropenem (MEM), Tigecycline (TGC), Polymyxin B (PB), Cefepime (FEP), Cefotaxime (CTX), Ceftazidime (CAZ), Amikacin (AMK), Ciprofloxacin (CIP), Trimethoprim/Sulfamethoxazole (SXT), Aztreonam (ATM), Cefoperazone/Sulbactam (SCF), and Piperacillin/ Tazobactam (TZP). The ceftazidime/avibactam (CZA) drug sensitivity assay was performed using the E-test method. The drug sensitivity quality control strain was Escherichia coli ATCC 25922. All the above drug sensitivity results were interpreted with reference to the 2021 American Clinical and Laboratory Standards Institute Standards document (CLSI).Citation22
Detection of Resistance Determinant
Carbapenemase resistance genes (blaKPC, blaNDM, blaOXA-48, blaVIM, blaIMP), ESBL resistance genes (blaSHV, blaTEM, blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9), AmpC enzyme genes (blaDHA, blaCMY), and colistin-resistant related gene (pmrA, pmrB, phoP, phoQ, mgrB, mcr-1) were synthesized with reference to relevant literature.Citation23–29 Amplification was performed according to the following reaction system: 2 μL of DNA template, 12.5 μL of Premix Taqpremix, 1 μL each of upstream and downstream primers, and sterile water made up to 25μL. Reaction conditions: 95°C for 5min, 94°Cfor45s, annealing temperature for45s,72°C for 1min, 30 cycles, 72°C for 5 min. PCR condition for colistin: 94°C for 5 min, 94°C for 1 min, annealing temperature for 45s, 72 °C for 1 min, 35 cycles, 72°C for 10 min. See Table S1 for each primer sequence and annealing temperature, and the amplified PCR products were run by 1.5% agarose gel electrophoresis in 1× TAE electrophoresis solution at 100 V for 45 min. Profitable products were sequenced using Sanger sequencing, and the sequencing results were compared online at https://bigsdb.pasteur.fr/klebsiella/.
Molecular Typing
Genotyping was carried out using multilocus sequence typing. The sequence of seven housekeeping genes (rpoB, pgi, tonB, mdh, phoE, gapA, infB) of Klebsiella pneumoniae was amplified by PCR. The gene sequence was synthesized by referring to the website for housekeeping gene primers (https://bigsdb.pasteur.fr/klebsiella/primers-used/). The PCR reaction system is as follows: DNA template 2 μL, Premix Taq Premix 12.5μL, 1 μL upstream and 1 μL downstream primer respectively. Replenish the volume with sterile water to a final concentration of 25 μL. The PCR reaction conditions were: 94°C for 2 min, 94°C 30s, 50°C 1min, 72 °C 30s, 35 cycles in total, 72 °C 5 min.
The pulsed-field gel electrophoresis (PFGE) was used to analyze the clonal relationship of CRKP strains. PFGE is conducted with reference to the Centers for Disease Control and Prevention (Atlanta, GA), and the cluster analysis was carried out by BioNumerics software to draw a dendrogram. A population with a similarity of more than 85% was used to analyze the genetic relationship between different strains.Citation30
Statistical Analysis
The statistical analysis was conducted using IBM SPSS Statistics software (version specified, SPSS Inc., Chicago, IL, United States). Categorical data were analyzed using the Chi-square test (χ2) The threshold for statistical significance was set at a p-value of less than 0.05.
Results
Clinical Characteristics of Adult and Children CRKP Isolates
Children aged 0–1 year made up the majority of the 31 CRKP strains recovered from children, accounting for 61.3% (19/31), followed by children aged 6–15 years (29.0%,9/31). Specimen type was predominantly sputum (54.8%, 17/31) and blood (16.1%,5/31). The departments were mainly concentrated in neonatal ICU and pediatrics, with 54.8% (17/31) and 19.4% (6/31), respectively.
The mean age of patients with 31 strains of CRKP isolated from adults was at 67 years. Specimen type was predominantly sputum (80.6%,25/31) and urine (12.9%,4/31). The department was mainly concentrated in the neurology and emergency ICU, accounting for 29.0% (9/31) and 25.8% (8/31), respectively.
Comorbidities in pediatric CRKP patients were most frequent in preterm infants (38.7%,12/31), followed by neurological disorders (29.0%,9/31) and respiratory infections (19.4%,6/31). The most common comorbidities in 31 adults CRKP patients were neurological disorders (77.4%, 24/31), followed by hypertension (51.6%,16/31) and respiratory disorders (45.2%,14/31). There were 2.4 comorbidities per adult (74/31) compared to 1.2 comorbidities per child (38/31), a twofold difference.
The history of antimicrobial drug use in children and adults was dominated by β-lactamase inhibitor combinations, 54.8% (17/31) and 67.7% (21/31), respectively. Secondly was third-generation cephalosporins (51.6%,16/31) and carbapenems (35.5%,11/31) in children, and 45.2% (14/31) of carbapenems and 38.7% (12/31) of quinolones in adults. The history of 3 or more (inclusive) antimicrobial drug use was 64.5% (20/31) in adults, 1.7 times higher than 38.7% (12/31) in children. Details of the clinical characteristics of the children were shown in , and for adults in .
Table 1 Clinical Characteristics of the Children CRKP Strains
Table 2 Clinical Characteristics of the Adults CRKP Strains
Antibiotic Susceptibility Testing
Thirty-one pediatric and 31 adult CRKP strains were highly resistant to imipenem, meropenem, cefepime, cefotaxime, ceftazidime, cefoperazone/sulbactam, and piperacillin/tazobactam, the drug resistance rate was>85%, and the sensitivity rate to tigecycline and polymyxin was>93%.We also find two polymyxin-resistant strains detected in pediatric CRKP and two intermediately resistant tigecycline strains detected in adult CRKP. There was a significant difference in drug resistance rates between children and adults. CRKP in adults was higher than in children for imipenem, aztreonam, ciprofloxacin and amikacin, and CRKP in children was higher than in adults for ceftazidime/avibactam (P<0.05, ). See for details.
Detection of Resistance Determinants
The blaNDM-5 gene was the predominant resistance gene in pediatric CRKP strains, accounting for 58.1% of the cases (18/31), followed by blaKPC-2 (6/31, 19.4%). Two strains contained both the blaNDM-5 and blaOXA-181 genes. The positive rate for the ESBL resistance gene in pediatric CRKP strains was 93.5%. Among these, 27 strains carried the blaSHV gene, primarily the blaSHV-11 (7/31, 22.6%) and the blaSHV-262 (6/31, 19.4%). Five strains carried the blaTEM, with the blaTEM-1 variant being dominant at 12.9% (4/31). Ten strains carried the blaCTX-M-1 gene cluster, with the blaCTX-M-15 variant accounting for 22.6% (7/31). Three strains carried the blaCTX-M-9 gene cluster, which included the variants blaCTX-M-14, blaCTX-M-65, and blaCTX-M-113. Among the AmpC enzymes, the blaDHA-3 gene was detected in 25.8% (8/31). Two colistin-resistant strains carried the mgrB gene, representing 6.5% (2/31).
Analysis of the 31 adult CRKP strains revealed that all were carbapenemase producers. The predominant resistance gene identified was blaKPC-2, accounting for 87.1% (27/31) of the strains. blaNDM-1 was detected in 16.1% (5/31) of the strains, with three strains co-producing both KPC-2 and NDM-1 enzymes. All strains harbored the extended-spectrum beta-lactamase (ESBL) resistance gene SHV. Notably, blaSHV-12 was found in 22.6% (7/31) of the strains, while blaSHV-31 and blaSHV-123 each represented 16.1% (5/31) of the strains. Among the TEM beta-lactamase genes, blaTEM was present in 27 strains, with blaTEM-1 being the most prevalent, detected in 54.8% (17/31) of the strains. The CTX-M-1 gene cluster was identified in five strains, with blaCTX-M-107 being the most common, found in 12.9% (4/31) of the strains. The CTX-M-9 gene cluster was present in 21 strains, with blaCTX-M-65 being predominant, representing 51.6% (16/31) of the strains. Additionally, the AmpC enzyme gene blaDHA-1 was identified in 12.9% (8/31) of the strains.
In this study, the main carbapenemase of CRKP in children was blaNDM-5, while blaKPC-2 in adults (P<0.001, ). Among the ESBL resistance genes, children carried the blaSHV-262 and blaCTX-M-15 resistance genes more than adults (P<0.05, ), while carried the blaSHV-12, blaSHV-123, blaTEM-1, and blaCTX-M-65 genes less than adults (P<0.05, ), and the positive rate of adults carrying ESBL resistance genes was higher than children. AmpC enzymes were taken more frequently in children, with blaDHA-3 being the main enzyme in children and blaDHA-1 being the main enzyme in adults (P<0.05, ). No strains were detected carrying the blaIMP, blaVIM, blaCTX-M-2, blaCTX-M-8, or blaCMY genes. The differences in drug-resistance genes between children and adults are shown in and Table S2.
Molecular Characteristics of Adult and Children CRKP Isolates
The MLST analysis revealed significant differences in the genetic lineages between pediatric and adult populations. A total of thirteen sequence types (STs) were identified among the pediatric strains, whereas only four STs were detected in the adult strains. Notably, ST2735 was the predominant type in children, representing 19.4% (6/31) of the strains. In contrast, ST11 was overwhelmingly the most common type in adults, accounting for 83.9% (26/31) of the strains. The distribution of MLST types in adults was notably more concentrated, with a higher prevalence of a single ST, indicative of less genetic diversity. Conversely, the pediatric population exhibited a more dispersed distribution of STs, suggesting the presence of multiple small clusters. This disparity underscores the distinct epidemiological patterns and potential transmission dynamics between the two age groups.
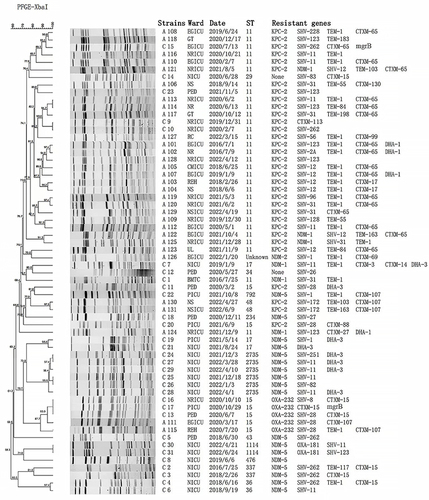
Childhood PFGE was divided into six different clusters, each consisting of different ST types. The main clonal spread of these isolates was mainly from the neonatal ICU (n=11). Six ST2735 isolates indicated high homology (>95%). Adult PFGE was divided into eight different clusters, and 21 ST11-type KPC-2-producing strains were divided into six distinct groups. Major clonal transmission of these isolates was mainly from the neurology ICU (n=7) and emergency ICU (n=6). We also found an outbreak of ST11 CRKP in adults. Moreover, the homology of five ST15-type OXA-232-producing strains was greater than 85%, of which 3 were from children and 2 were from adults. Similarly, the CRKP homology of ST11 producing KPC-2 was greater than 85% in 2 children and 4 adults. The details are shown in .
Figure 3 PFGE profile analysis of 62 strains of CRKP in children and adults. The UPGMA algorithm was performed to construct a dendrogram based on the dice similarity coefficient. Strains were classified as the same clone cluster when their dice similarity index was>85%.
Discussion
K. pneumoniae is a prevalent pathogen associated with both healthcare-related infections and community-acquired infections, capable of causing infections at various sites within the body. The widespread use of antimicrobial drugs has driven the emergence of CRKP, complicating clinical treatment and increasing the risk of CRKP-related mortality. In clinical practice, numerous medications have been explored for the treatment of CRKP. For instance, ceftazidime/avibactam (CZA) is utilized for the treatment of CRKP strains producing class A and D β-lactamase enzymes. In contrast, CRKP strains producing class B metallo-β-lactamase enzymes necessitate a combination therapy involving CZA and aztreonam for effective treatment. This study observed that pediatric CRKP strains exhibited higher resistance rates to CZA compared to adult strains, primarily attributed to distinct resistance mechanisms. The predominant resistance genes in children were associated with metalloenzymes that can hydrolyze CZA. The presence of carbapenemase resistance genes, specifically MBL production, point mutations in blaKPC-2, and elevated expression levels of KPC enzymes, are critical factors contributing to CZA resistance.Citation31,Citation32
Traditionally reserved for their considerable side effects, colistin and tigecycline have been re-evaluated, with dosage adjustments facilitating their use in CRKP treatment. However, the emergence of resistance to these drugs is a growing concern. The study identified two strains exhibiting colistin resistance and two strains showing intermediate resistance to tigecycline, highlighting the ongoing challenge of antibiotic resistance in both pediatric and adult CRKP cases. In Escherichia coli, polymyxin resistance is often due to the plasmid-mediated mcr-1 gene,Citation33 while in Klebsiella pneumoniae strains, it is often due to mutations in mgrB gene.Citation34 In our study, two colistin-resistant CRKP strains were detected to carry the mgrB gene. One exists in the ST11CRKP, while another exists in the ST15CRKP. Tigecycline resistance is mainly due to the overexpression of the AdeABC efflux pump, which is associated with genetic mutations, but this resistance could be reversed into susceptibility by additional mutations in antibiotic-free environments.Citation35 Moreover, a phenomenon of hetero-resistance may develop during infections treated with tigecycline, and hetero-resistance can be an intermediate phase between susceptibility and resistance after exposure to antibiotics.Citation36 Based on the history of antibiotic use, one case of tigecycline intermediate resistance may be hetero-resistance caused by the patient’s history of tigecycline use, and the other case cannot be ruled out as having taken this type of drug before. Further exploration is needed. So, we need to strengthen the monitoring of CRKP to provide a reliable basis for the rational clinical use of antimicrobial drugs and to provide timely data to enable timely responses to the problem of bacterial resistance.
The analysis of clinical data from this study indicated that infants under one year of age are particularly vulnerable to pediatric CRKP infections, with the neonatal intensive care unit (ICU) emerging as a high-risk area for such cases. Similarly, older adults were found to be at an increased risk for adult CRKP infections, with the neurology ICU identified as a high-risk unit for this demographic. The majority of both children and adults in the study presented with comorbidities and were likely to have undergone invasive procedures. Additionally, a significant history of antimicrobial drug use was observed, with the severity of these conditions varying markedly between adults and children. A meta-analysis of CRKP risk factors identified several key risk factors for CRKP infection, including immunosuppressive therapy, ICU admission, exposure to a broad range of antimicrobial drugs (encompassing carbapenems, quinolones, glycopeptides, and beta-lactam/beta-lactamase inhibitors or BL/BLIs), surgical interventions, mechanical ventilation, central venous catheterization, and the presence of indwelling devices such as nasogastric tubes.Citation37 To enhance the provision of a more rational treatment plan, it is crucial to minimize the indiscriminate use of antimicrobial medications and reduce the frequency of unnecessary invasive procedures. This approach is vital for mitigating the risk of CRKP infections and promoting more effective clinical management strategies.
The molecular typing of CRKP strains in children revealed a diverse genetic landscape without a particularly dominant sequence type (ST) phenotype. The distribution of STs was predominantly small and concentrated or scattered. Notably, the ST2735 strain was identified in six cases from December 2021 to April 2022, all originating from the neonatal ICU. These strains harbored the blaNDM-5 gene with a high degree of homozygosity (>85%). This clustering suggests a potential localized neonatal epidemic of ST2735-producing NDM-5 CRKP during the specified period. Further analysis of multilocus sequence typing (MLST) data in children indicated that the ST2735 type is the second most frequently isolated ST in pediatric hospitals, as reported in the literature.Citation38,Citation39 Interestingly, this ST type was not detected in adults at this hospital over the years. The emergence of ST2735 CRKP as a concerning clonal type in children warrants clinical vigilance.
In contrast, the molecular typing in adults showed a more concentrated ST distribution, with ST11 being the predominant group, aligning with our primary prevalent ST type. A retrospective analysis spanning a decade of ST11 type CRKP cases revealed that this strain is nearly universally resistant to clinical antimicrobials. Moreover, it has the potential to acquire pLVPK-like virulence plasmids, which could result in the development of highly virulent carbapenemase-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP).Citation40 Our findings underscore the importance of closely monitoring the prevalent ST11 type within our nation, with a particular emphasis on the spread and evolution of ST11-type CRKP. Seven adult cases of ST11-type CRKP, characterized by the KPC-2 gene and a high degree of homozygosity (>85%), were identified between July 2016 and April 2022. This observation confirms the clonal transmission of ST11-type CRKP within our institution and highlights the highly transmissible and challenging nature of CRKP infections. Additionally, clonal transmission events were observed for ST11 and ST15 types in both pediatric and adult patients at our institution. Despite the absence of a physical link between pediatric and adult clinics, the movement of medical staff between wards for consultations may contribute to the inter-age group clonal spread.
Among these, the clonal transmission of ST15-type CRKP in both children and adults was attributed to the OXA-232 enzyme, with an associated mortality rate of 40%. OXA-232, a variant of OXA-48, has been implicated in recent outbreaks of ST15-type CRKP in regions such as Shanghai and Yancheng.Citation41,Citation42 A study conducted in Zhejiang indicated that these outbreaks of ST15-type OXA-232 enzyme-producing CRKP share similar resistance and virulence genes, as well as plasmid types, suggesting a propensity for environmental colonization and widespread transmission between the environment and patients due to clonal spread.Citation43 The presence of OXA-232-producing CRKP in both pediatric and adult populations at our hospital mandates heightened attention to disinfection and isolation practices between pediatric and adult units. It also calls for robust nosocomial infection prevention and control measures, including environmental decontamination and strategies to reduce colonization transmission.
In this study, the NDM-5 gene emerged as the most prevalent drug-resistance gene in pediatric CRKP, followed by the KPC-2 gene. This contrasts with the adult CRKP population, where the KPC-2 gene was predominantly detected, succeeded by the NDM-1 gene. It is noteworthy that the NDM-5 gene, while most commonly found in E. coli, has become increasingly prevalent in Klebsiella pneumoniae as well.Citation44,Citation45 The NDM-5 gene is known to carry multiple drug-resistance determinants and is associated with a plethora of virulence genes. It has been shown that the NDM-5 gene can be harbored by the IncX3 plasmid in K. pneumoniae and can be efficiently transferred into E. coli J53.Citation46 Importantly, this plasmid exhibits stable inheritance and is resistant to loss, which underscores the potential for the dissemination of resistance genes within clinical settings. Given the implications of such plasmid-mediated gene transfer for the emergence of multi-drug resistant strains, it is crucial to maintain a high index of suspicion for the presence of NDM-5 in CRKP strains. Vigilant surveillance and appropriate infection control measures are essential to prevent potential outbreaks associated with nosocomial plasmid transfer.
This research presents the first report of ST1114-type CRKP co-producing NDM-5 and OXA-181 enzymes identified in neonates. Additionally, in the adult CRKP strains, three instances of KPC-2 and NDM-1 co-producing CRKP were detected. A study conducted by Hui Wang et alCitation47 highlighted that ST11 has emerged as the dominant clone, likely due to its co-evolution with certain plasmids, which has facilitated its predominance in clonal transmission. The coexistence of KPC-2 and NDM-1 genes, which are stably evolved, presents a significant challenge to clinical management. KPC-2 is predominantly found in IncFII plasmids, while NDM-1 is located in various plasmids, such as IncN, IncX, and IncHIB. This genetic diversity complicates clinical diagnosis and treatment strategies. A global multicenter studyCitation48 has underscored the significant regional differences in the epidemiology of CRKP, encompassing baseline patient characteristics, clinical outcomes, and bacterial features. Findings from one region may not be directly applicable to others, emphasizing the heterogeneity of CRKP dynamics on a global scale. To ensure accurate guidance for the therapeutic use of antimicrobial medications and to inform prevention and control measures for hospital-acquired infections, it is imperative to conduct continuous evaluations of bacterial resistance patterns within our local area and hospital. This approach is essential for tailoring clinical practices to the specific epidemiological context and for preserving the effectiveness of antimicrobial agents.
Conclusion
The study conducted at the Affiliated Hospital of Xuzhou Medical University revealed significant differences in the predominant carbapenemase enzymes among pediatric and adult CRKP strains. The blaNDM-5 gene was identified as the primary carbapenemase in children, whereas the blaKPC-2 gene was the main enzyme in adult CRKP strains. Notably, there were substantial differences in resistance rates to key antimicrobial agents such as imipenem, ceftazidime/avibactam, aztreonam, ciprofloxacin, and amikacin between the pediatric and adult populations. Our findings highlight the presence of several clonal transmissions of blaNDM-5-producing CRKP within the pediatric cohort and blaKPC-2-producing CRKP within the adult cohort. Additionally, we observed co-occurring clonal transmissions of ST11 type blaKPC-2 and ST15 type blaOXA-232 resistance genes in both children and adults. Importantly, this research marks the first report of clonal prevalence of ST1114-type CRKP co-producing blaNDM-5 and blaOXA-181 in the neonatal ICU. These findings underscore the fact that clonal transmission of CRKP is not confined to a single unit but can occur across different units, affecting both children and adults. Therefore, it is imperative to strengthen infection prevention and control measures to mitigate the spread of CRKP clones. Continuous surveillance, coupled with the implementation of targeted strategies, is essential to curb the dissemination of these resistant strains and to ensure the delivery of effective clinical care.
Ethical Approval and Consent to Participate
The Medical Ethics Committee of The Affiliated Hospital of Xuzhou Medical University approved this study and waived the requirement for informed written consent, as it was a retrospective study. This study kept patient data confidential and complied with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
Additional information
Funding
References
- Martin RM, Bachman MA. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4.
- Chang D, Sharma L, Dela Cruz CS, et al. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front Microbiol. 2021;12:750662.
- Shen S, Shi Q, Han R, et al. Isolation of a Ceftazidime-Avibactam-Resistant blaKPC-71-Positive Klebsiella pneumoniae Clinical Isolate. Microbiol Spectr. 2022;10(1):e0184021.
- Antimicrobial Resistance C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655.
- Zhang H, Wang J, Zhou W, et al. Risk Factors and Prognosis of Carbapenem-Resistant Klebsiella pneumoniae Infections in Respiratory Intensive Care Unit: a Retrospective Study. Infect Drug Resist. 2021;14:3297–3305.
- Chen R, Liu Z, Xu P, et al. Deciphering the Epidemiological Characteristics and Molecular Features of bla (KPC-2)- or bla (NDM-1)-Positive Klebsiella pneumoniae Isolates in a Newly Established Hospital. Front Microbiol. 2021;12:741093.
- Tian D, Pan F, Wang C, et al. Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infect Drug Resist. 2018;11:1935–1943.
- Yang Y, Liu J, Muhammad M, et al. Factors behind the prevalence of carbapenem-resistant Klebsiella pneumoniae in pediatric wards. Medicine (Baltimore). 2021;100(36):e27186.
- Shao C, Wang W, Liu S, et al. Molecular Epidemiology and Drug Resistant Mechanism of Carbapenem-Resistant Klebsiella pneumoniae in Elderly Patients With Lower Respiratory Tract Infection. Front Public Health. 2021;9:669173.
- Zhao Y, Liao Y, Zhang N, et al. Four Types of ST11 Novel Mutations From Increasing Carbapenem-Resistant Klebsiella pneumoniae in Guangdong, 2016-2020. Front Microbiol. 2021;12:702941.
- Li Z, Ding Z, Yang J, et al. Carbapenem-Resistant Klebsiella pneumoniae in Southwest China: molecular Characteristics and Risk Factors Caused by KPC and NDM Producers. Infect Drug Resist. 2021;14:3145–3158.
- Rocha VFD, Barbosa MS, Leal HF, et al. Prolonged Outbreak of Carbapenem and Colistin-Resistant Klebsiella pneumoniae at a Large Tertiary Hospital in Brazil. Front Microbiol. 2022;831770.
- Kong Z, Liu X, Li C, et al. Clinical Molecular Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae Among Pediatric Patients in Jiangsu Province, China. Infect Drug Resist. 2020;13:4627–4635.
- Zhao D, Shi Q, Hu D, et al. The Emergence of Novel Sequence Type Strains Reveals an Evolutionary Process of Intraspecies Clone Shifting in ICU-Spreading Carbapenem-Resistant Klebsiella pneumoniae. Front Microbiol. 2021;12:691406.
- Yang Y, Yang Y, Chen G, et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect. 2021;10(1):700–709.
- Fu B, Yin D, Sun C, et al. Clonal and Horizontal Transmission of blaNDM among Klebsiella pneumoniae in Children’s Intensive Care Units. Microbiol Spectr. 2022;10(4):e0157421.
- Cheng J, Zhao D, Ma X, et al. Molecular epidemiology, risk factors, and outcomes of carbapenem-resistant Klebsiella pneumoniae infection in a tertiary hospital in eastern China: for a retrospective study conducted over 4 years. Front Microbiol. 2023;14:1223138.
- Peng C, Feng DH, Zhan Y, et al. Molecular Epidemiology, Microbial Virulence, and Resistance of Carbapenem-Resistant Enterobacterales Isolates in a Teaching Hospital in Guangzhou, China. Microb Drug Resist. 2022;28(6):698–709.
- Jia H, Zhang Y, Ye J, et al. Outbreak of Multidrug-Resistant OXA-232-Producing ST15 Klebsiella pneumoniae in a Teaching Hospital in Wenzhou, China. Infect Drug Resist. 2021;14:4395–4407.
- Gu B, Bi R, Cao X, et al. Clonal dissemination of KPC-2-producing Klebsiella pneumoniae ST11 and ST48 clone among multiple departments in a tertiary teaching hospital in Jiangsu Province, China. Ann Transl Med. 2019;7(23):716.
- Ochonska D, Klaminska-Cebula H, Dobrut A, et al. Clonal Dissemination of KPC-2, VIM-1, OXA-48-Producing Klebsiella pneumoniae ST147 in Katowice, Poland. Pol J Microbiol. 2021;70(1):107–116.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing[S]. Wayne, PA: CLSI; 2021.
- Yu Y, Ji S, Chen Y, et al. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect. 2007;54(1):53–57.
- Yang J, Chen Y, Jia X, et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect. 2012;18(12):E506–13.
- Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162.
- Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123.
- Poirel L, Heritier C, Tolun V, et al. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22.
- Choi MJ. Pathways Regulating the pbgP Operon and Colistin Resistance in Klebsiella pneumoniae Strains. J Microbiol Biotechnol. 2016;26(9):1620–1628.
- Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6).
- Song S, Zhao S, Wang W, et al. Characterization of ST11 and ST15 Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae from Patients with Ventilator-Associated Pneumonia. Infect Drug Resist. 2023;16:6017–6028.
- Wang Y, Wang J, Wang R, et al. Resistance to ceftazidime-avibactam and underlying mechanisms. J Glob Antimicrob Resist. 2020;22:18–27.
- Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):124 e1–124 e4.
- Li Q, Qian C, Zhang X, et al. Colistin Resistance and Molecular Characterization of the Genomes of mcr-1-Positive Escherichia coli Clinical Isolates. Front Cell Infect Microbiol. 2022;12:854534.
- Khoshbayan A, Shariati A, Razavi S, et al. Mutation in mgrB is the major colistin resistance mechanism in Klebsiella pneumoniae clinical isolates in Tehran, Iran. Acta Microbiol Immunol Hung. 2022.
- Jo J, Ko KS. Tigecycline Heteroresistance and Resistance Mechanism in Clinical Isolates of Acinetobacter baumannii. Microbiol Spectr. 2021;9(2):e0101021.
- Korczak L, Majewski P, Iwaniuk D, et al. Molecular mechanisms of tigecycline-resistance among Enterobacterales. Front Cell Infect Microbiol. 2024;14:1289396.
- Liu P, Li X, Luo M, et al. Risk Factors for Carbapenem-Resistant Klebsiella pneumoniae Infection: a Meta-Analysis. Microb Drug Resist. 2018;24(2):190–198.
- Zhou Y, Zhao Z, Zeng L, et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae in a paediatric hospital in China revealed the dynamics of carbapenemase and the prevalence of ST2735 K. pneumoniae. J Med Microbiol. 2022;71(1).
- Yin D, Zhang L, Wang A, et al. Clinical and molecular epidemiologic characteristics of carbapenem-resistant Klebsiella pneumoniae infection/colonization among neonates in China. J Hosp Infect. 2018;100(1):21–28.
- Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180.
- Li X, Ma W, Qin Q, et al. Nosocomial spread of OXA-232-producing Klebsiella pneumoniae ST15 in a teaching hospital, Shanghai, China. BMC Microbiol. 2019;19(1):235.
- Zhu Z, Huang H, Xu Y, et al. Emergence and genomics of OXA-232-producing Klebsiella pneumoniae in a hospital in Yancheng, China. J Glob Antimicrob Resist. 2021;26:194–198.
- Han X, Chen Y, Zhou J, et al. Epidemiological Characteristics of OXA-232-Producing Carbapenem-Resistant Klebsiella pneumoniae Strains Isolated during Nosocomial Clonal Spread Associated with Environmental Colonization. Microbiol Spectr. 2022;10(4):e0257221.
- Sun P, Xia W, Liu G, et al. Characterization Of bla NDM-5-Positive Escherichia coli Prevalent In A University Hospital In Eastern China. Infect Drug Resist. 2019;12:3029–3038.
- Li Y, Tang M, Dai X, et al. Whole-Genomic Analysis of NDM-5-Producing Enterobacteriaceae Recovered from an Urban River in China. Infect Drug Resist. 2021;14:4427–4440.
- Tian D, Wang B, Zhang H, et al. Dissemination of the bla NDM-5 Gene via IncX3-Type Plasmid among Enterobacteriaceae in Children. mSphere. 2020;5(1):e00699–19.
- Gao H, Liu Y, Wang R, et al. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599.
- Wang MEM, Chen L, et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis. 2022;22(3):401–412.