Abstract
Background
To explore the plasmid characteristics and transfer mechanisms of an extensive drug resistant (XDR) clinical isolate, Citrobacter portucalensis L2724hy, co-producing blaSFO-1, blaNDM-1, and blaKPC-2.
Methods
Species confirmation of L2724hy was achieved through 16S rRNA sequencing and Average Nucleotide Identity (ANI) analysis. Antimicrobial susceptibility testing (AST) employed the agar dilution and micro broth dilution methods. Identification of resistance genes was carried out by PCR and whole-genome sequencing (WGS). Essential resistance gene locations were verified by S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and southern hybridization experiments. Subsequent WGS data analysis delved into drug resistance genes and plasmids.
Results
The confirmation of the strain L2724hy as an extensive drug-resistant Citrobacter portucalensis, resistant to almost all antibiotics tested except polymyxin B and tigecycline, was achieved through 16S rRNA sequencing, ANI analysis and AST results. WGS and subsequent analysis revealed L2724hy carrying blaSFO-1, blaNDM-1, and blaKPC-2 on plasmids of various sizes. The uncommon ESBL gene blaSFO-1 coexists with the fosA3 gene on an IncFII plasmid, featuring the genetic environment IS26-fosA3-IS26-ampR-blaSFO-1-IS26. The blaNDM-1 was found on an IncX3 plasmid, coexisting with blaSHV-12, displaying the sequence IS5-IS3000-IS3000-Tn2-blaNDM-1-ble-trpF-dsbD-cutA-gros-groL, lacking ISAa125. The blaKPC-2 is located on an unclassified plasmid, exhibiting the sequence Tn2-tnpR-ISKpn27-blaKPC-2-ISKpn6-korC. Conjugation assays confirmed the transferability of both blaNDM-1 and blaKPC-2.
Conclusion
We discovered the coexistence of blaSFO-1, blaNDM-1, and blaKPC-2 in C. portucalensis for the first time, delving into plasmid characteristics and transfer mechanisms. Our finding highlights the importance of vigilant monitoring of drug-resistance genes and insertion elements in uncommon strains.
Introduction
In the vast microbial landscape, the genus Citrobacter stands as a diverse group of bacteria known for its adaptability and ecological ubiquity. Within this genus, the relatively lesser-explored C. portucalensis has garnered attention for its potential contributions to various fields, especially in human health. Since its first report in 2017, diverse occurrences of C. portucalensis in wastewater, sputum, stool, sludge, poultry and, sea turtles across multiple countries have been discovered.Citation1–6 In 2021, Chinese researcher Cao et al isolated C. portucalensis carrying blaNDM-1 from clinical specimens for the first time.Citation2 Subsequently, in 2022, the coexistence of blaKPC-2 and blaNDM-1 was reported in an extensively drug-resistant C. portucalensis obtained from a hospital in China.Citation7 Additionally, in 2023, researchers isolated C. portucalensis carrying blaNDM-1 from an endangered marine animal that died from sepsis.Citation8 This growing body of evidence underscores the increasing clinical and extra-clinical dissemination of drug-resistant C. portucalensis, warranting heightened attention to address the associated public health concerns.
In 2017, the World Health Organization (WHO) designated carbapenem-resistant and extended spectrum beta-lactamases (ESBLs)-producing Enterobacteriaceae as “extremely important” on its list of antimicrobial resistant “priority pathogens”. Numerous antimicrobial resistance genes (ARGs) encoding carbapenemases and ESBLs were discovered. Among these, the blaSFO-1, an uncommon ESBLs gene exhibiting the ability to hydrolyze beta-lactams excluding cephamycins and carbapenems, was first identified on a self-transferring plasmid from Enterobacter cloacae isolated from Japan in 1999.Citation9 The blaSFO-1 later manifested in an outbreak of Enterobacter cloacae in Spain.Citation10 Recently, its emergence has been documented in Enterobacter hormaechei, Escherichia coli and Klebsiella pneumoniae isolated from China.Citation11–13 Further investigations are crucial to the understanding of the epidemiology and clinical impact of blaSFO-1. Among the carbapenemase encoding genes, blaKPC stands out as the predominant in Enterobacteriaceae bacteria globally.Citation14 In China, the epidemic of blaKPC-2-carrying bacteria strains is of current concern.Citation15 Besides blaKPC, the rapid global spread of blaNDM since its discovery in 2008 has posed significant public health challenges as well.
In our study, we identified an extensively drug-resistant clinical isolate of C. portucalensis featuring a rare coexistence of blaSFO-1, blaKPC-2, and blaNDM-1 genes. This report makes the first documented instance of blaSFO-1 in C. portucalensis. Additionally, we investigated the plasmid characteristics and transfer mechanisms associated with drug-resistant genes in this particular strain.
Materials and Methods
Strain Isolation and Identification
In September 2020, we obtained a clinical isolate (L2724hy) from the faeces of a 56-year-old outpatient male patient with acute diarrhea at the First Affiliated Hospital of Zhejiang University. Initially, this isolate was mistakenly identified as C. freundii using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS).Citation16 To confirm its accurate species, we uploaded the 16s rRNA sequence of this strain to BioEZCloud (https://www.ezbiocloud.net/) and analyzed ANI, and the result indicated it to be C. portucalensis.Citation17,Citation18
Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) of L2724hy, transconjugant L2724hy-KPC-EC600, transconjugant L2724hy-NDM-EC600 and E. coli EC600 were determined by agar dilution method and micro-broth dilution method.Citation19 AST results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) 2023 (https://clsi.org), while the breakpoints for tigecycline and colistin were interpreted based on EUCAST (https://www.eucast.org/).
S1-PFGE-Southern Hybridization and Conjugation Assays
S1-PFGE was used to identify the size and number of plasmids. The location of the blaSFO-1, blaNDM-1, and blaKPC-2 were determined by S1-PFGE and hybridization experiments. Conjugation assays were conducted using E. coli EC600 as recipient and L2724hy as donor.Citation20 Subsequently, individual colonies of donor and recipient bacteria were carefully selected and suspended LB liquid culture medium at 37 °C for 6 hours. Afterwards, the two suspensions were mixed in a 1:1 ratio (donor: recipient), introduced into 3 mL LB liquid medium, and incubated at 37 °C for 20 hours. An appropriate amount of bacterial solution was spread onto Mueller–Hinton agar (MHA) plates containing rifampicin (200 ug/mL) and meropenem (2 ug/mL). After the incubation at 37°C overnight, a single colony was selected for further cultivation. Moreover, the strain was identified through MALDI-TOF/MS and the presence of drug-resistant genes was confirmed through PCR.
Whole-Genome Sequencing and Data Analysis
We utilized a Qiagen DNA purification kit in the extraction of the whole genomic DNA of strain L2724hy, which was then sequenced via Oxford Nanopore technology to generate raw data SRA. Then, we used Unicyler 0.4.9 to assemble the SRA, resulting in the acquisition of the whole-genome sequence.Citation21 The prokka v1.14.6 annotation tool was used to annotate the whole-genome sequence.Citation22 To predict drug-resistant genes, the data were submitted to ResFinder (https://cge.food.dtu.dk/services/ResFinder/), and the identification of insertion sequence and transposons was performed using ISFinder (https://www-is.biotoul.fr/). Plasmid types were confirmed through PlasmidFinder (https://cge.food.dtu.dk/services/PlasmidFinder/), and the MLST of the strains was determined via Pubmlst (https://pubmlst.org/). The discovery of transfer-related components was conducted using OriTfinder (https://bioinfo-mml.sjtu.edu.cn/oriTfinder/). The genetic environment surrounding blaSFO-1, blaNDM-1, and blaKPC-2 on the plasmid was presented using Easyfig 2.2.3.Citation23 Additionally, the comparison of multiple plasmids was carried out using the BLAST Ring Image Generator (BRIG) software.Citation24
Results
Strain Isolation and Identification
L2724hy was isolated from the fecal sample of a patient with acute diarrhea and initially identified as C. freundii using MALDI-TOF/MS. Subsequent sequencing of the 16S rRNA lead to the reclassification of L2724hy as C. portucalensis (99.93% similarity for C. portucalensis and 99.86% similarity for C. freundii). Further ANI analysis confirmed L2724hy as C. portucalensis, showing a 98.73% ANI to C. portucalensis ATCC (CP044098) and a 94.52% ANI to C. freundii ATCC (CP033744).
Antimicrobial Susceptibility Testing
As presented in , the results of antimicrobial susceptibility testing indicated that C. portucalensis exhibited resistance to almost all antibiotics, with the exception of tigecycline and polymyxin B. For both transconjugant L2724hy-KPC-EC600 and L2724hy-NDM-EC600, we observed a decrease in the MIC values of carbapenems (imipenem, meropenem), levofloxacin, ciprofloxacin, amikacin, gentamicin, fosfomycin, chloramphenicol, and trimethoprim/sulfamethoxazole (). Notably, the transconjugant L2724hy-NDM-EC600 was highly resistant to ceftazidime-avibactam (MIC=64 mg/l), as was L2724hy, emphasizing the hydrolysis of ceftazidime-avibactam by NDM-1.
Table 1 The MIC Values of L2724hy, L2724hy-KPC-EC600, L2724hy-NDM-EC600 and EC600
S1-PFGE-Southern Hybridization and Conjugation Experiment
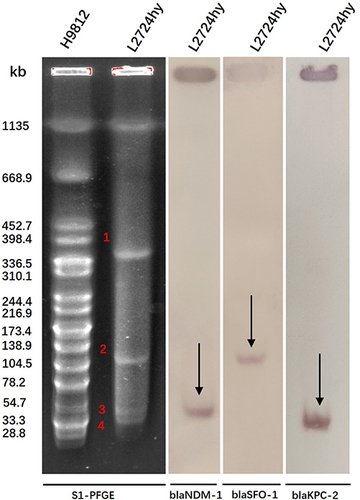
S1-PFGE profiles revealed that L2724hy harbored one chromosome and four plasmids (). However, due to overlap or small size, not all plasmids were discernible on the electrophoresis profiles. The presence of blaSFO-1, blaNDM-1, and blaKPC-2 genes on plasmids of different sizes was verified through Southern blotting hybridization. Specifically, the blaSFO-1 gene was identified on a ~130 kb plasmid, the blaNDM-1 gene was found on a ~54 kb plasmid, and the blaKPC-2 gene was located on a ~41 kb plasmid (). These results are consistent with the data obtained from whole-genome sequencing.
Figure 1 S1-PFGE profiles and southern blotting hybridization for L2724hy. S1-PFGE determines the number and size of plasmids in the strain. Southern blotting hybridization indicates the location of resistance genes blaSFO-1, blaNDM-1, and blaKPC-2.
The transconjugants were identified as E. coli by MALDI-TOF mass spectrometry. PCR confirmed that the transconjugants, namely L2724hy-KPC-EC600 and L2724hy-NDM-EC600, carried the blaKPC-2 and blaNDM-1 genes, respectively. This indicates the transferability of blaNDM-1 and blaKPC-2 to E. coli EC600.
Genomic Characteristics
The MLST analysis showed that strain L2724hy belongs to ST85. Strain L2724hy has a circular chromosome (5,031,620 bp) and eleven plasmids of various sizes (). The chromosome contains 5316 coding sequences and 121 RNAs, including 86 tRNAs, 25 rRNAs, and 10 ncRNAs. Its average G+C content is 51.9%. ResFinder identified ten ARGs in strain L2724hy, with blaCMY-46 and qnrB18 located on the chromosome, while the remaining eight were situated on plasmids. The plasmid type of p2-L2724hy-SFO-1 is IncFII, carrying the blaSFO-1 and fosA3 genes. Meanwhile, p3-L2724hy-NDM-1 carries two ARGs, blaNDM-1 and blaSHV-12, and is classified as IncX3. p4-L2724hy is an IncR plasmid with mph (A), whereas blaKPC-2 is on an unclassified p5-L2724hy-KPC-2. Another unclassified plasmid, p6-L2724hy, contains two resistance genes, including aac(6’)-Ib-cr and aac(6’)-Ib-Hangzhou.
Table 2 The Genomic Characteristics of L2724hy
Our analysis revealed that these distinct drug-resistance genes are consistent with antibiotic phenotypes in L2724hy, which included resistance to carbapenems (blaNDM-1, blaKPC-2), cephalosporins (blaCMY-46, blaSFO-1, blaSHV-12, blaNDM-1, blaKPC-2), 4-quinolones (qnrB18), fosfomycin (fosA3), and aminoglycosides (aac(6’)-Ib-cr, aac(6’)-Ib-Hangzhou).
Plasmid Characteristics of blaKPC-2, blaNDM-1, and blaSFO-1
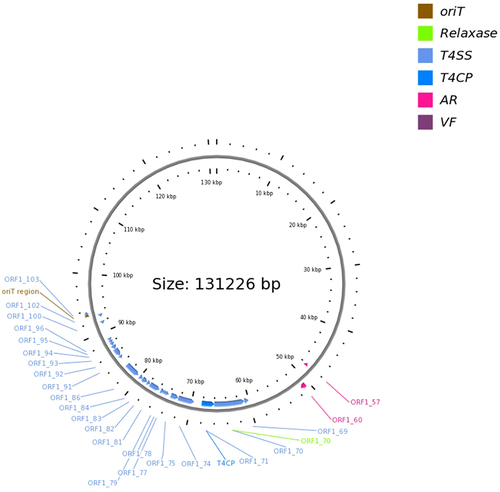
L2724hy carries plasmids of various types, including IncHI2/IncHI2A, IncFII, IncX3, IncR, and unclassified plasmids, as identified by PlasmidFinder. Among these, the IncFII plasmid, designated as p2-L2724hy-SFO-1, has a length of 131,226 bp and carries the resistance genes blaSFO-1 and fosA3. Comparative genomics with three closely related plasmid genomes (CP044029 with 95% coverage and 100% identity, CP042483 with 93% coverage and 100% identity, and CP042519 with 93% coverage and 100% identity) revealed primary variations, most notably the absence of the resistance gene blaSFO-1 and the transcriptional activator ampR (). Additionally, we also identified two strains of K. pneumoniae (Genebank: CP114855, and JQ724541) that closely resembled the genetic environment (IS26-ampR-blaSFO-1-IS26) surrounding blaSFO-1 in L2724hy (). However, these two strains lack the fosA3 gene, which is upstream of the IS26-ampR-blaSFO-1-IS26 sequence in L2724hy. In our study, IS26, which is a critical mobile element, is present on both sides of fosA3. Based on the OriTfinder results, p2-L2724hy-SFO-1 possesses mobile genetic elements associated with transfer, including an oriT region, type IV coupling protein (T4CP), and various type IV secretion system components (T4SS), which includes traI, traD, traQ, traN, traC, traV, traA, traJ, and traM ( and ).
Figure 2 Comparative analysis of plasmids p2-L2724hy-SFO-1, p4-L2724hy-NDM-1, and p5-L2724hy-KPC-2 in L2724hy. The BRIG circle maps show plasmid resistance genes and mobile genetic elements (MGEs). (a) plasmid p2-L2724hy-SFO-1 carrying blaSFO-1 and fosA3. (b) plasmid p4-L2724hy-NDM-1 harboring blaNDM-1 and blaSHV-12. (c) plasmid p5-L2724hy-KPC-2 bearing blaKPC-2.
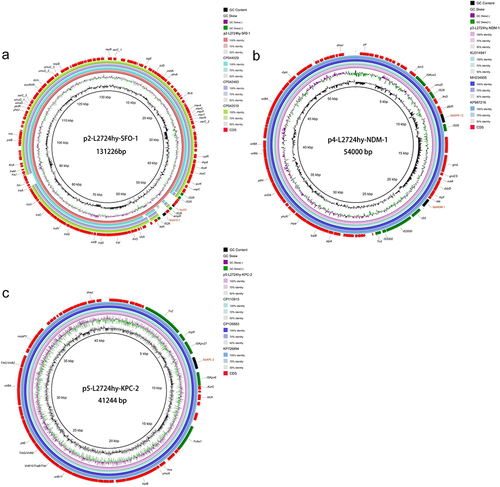
Figure 3 Genetic environment of blaSFO-1, blaNDM-1, and blaKPC-2 in L2724hy. (a) blaSFO-1 (b) blaNDM-1 (c) blaKPC-2. Red arrows denote drug resistance genes, green arrows denote transposons, pink arrows denote other genes, and yellow arrows represent hypothetical proteins.
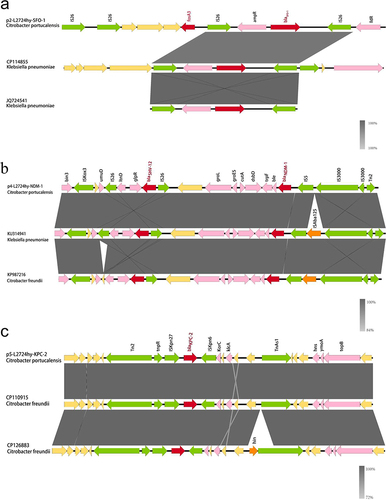
Figure 4 A conjugative plasmid p2-L2724hy-SFO-1. ORF1-57,fosA3; ORF1-60, blaSFO-1.
p4-L2724hy-NDM-1 was classified as an IncX3 plasmid, with a length of 54,000 bp and carried two resistance genes, blaNDM-1 and blaSHV-12 (). The NCBI BLAST search revealed 29 highly similar plasmids to p4-L2724hy-NDM-1, all exhibiting 100% coverage and 100% identity. Specifically, plasmids KU314941, MH234941, and KP987216, originating from distinct strains (K. pneumoniae, E. coli, and C. freundii) were selected for a comparative analysis of plasmid characteristics. The analysis revealed that these plasmids share similar backbones (), underscoring the widespread of blaNDM-1 across different species. Upon analyzing the genetic environment of blaNDM-1 in L2724hy (), a conserved sequence (ble-trpF-dsbD-cutA-gros-groL) downstream of blaNDM-1 on the plasmid was observed. The upstream of blaNDM-1 comprises an IS5-IS3000-IS3000-Tn2 sequence, with the notable deletion of ISAa125 deviating from the typical blaNDM-1 structure. Additionally, an IS26 insertion element surrounding blaSHV-12 was noted suggesting a potential role in facilitating the dissemination of blaSHV-12.
The plasmid p5-L2724hy-KPC-2, carrying blaKPC-2 gene, is currently unclassified and 41,244 bp in length. As shown in , the plasmid contains multiple-insertion elements (ISKpn27, ISKpn6), transposons (Tn2, tnpR, TnAs1), and transfer-related type IV secretion systems (virB2, virB4, virB9, virB10, and virB11). The plasmids CP110915, CP126883, and KP726894, are the most similar to p5-L2724hy-KPC-2, all displaying 100% coverage and 100% identity. Upon examination of these highly similar plasmids, it was observed that CP110915 and CP126883 originated from C. freundii. As illustrated in , a comparison of the genetic context of the two most similar C. freundii plasmids (CP110915, CP126883) with p5-L2724hy-KPC-2 revealed a similar genetic environment for blaKPC-2 (Tn2-tnpR-ISKpn27-blaKPC-2-ISKpn6-KorC) which is entirely consistent with CP110915 from the same region but exhibits a small gap with CP126883 from a different region.
Discussion
In recent years, the coexistence of ESBLs and carbapenemase genes in Enterobacteriaceae, posing challenges to clinical antibiotic treatment.Citation25 To the best of our knowledge, our study is the first to report the coexistence of blaSFO-1 with blaNDM-1 and blaKPC-2. Furthermore, there are no reports of the ESBL gene blaSFO-1 in C. portucalensis.
Since its discovery in 2017, C. portucalensis has been found in humans, animals, and environment. However, the misidentification of C. portucalensis as C. freundii has led to an underestimation of its prevalence in clinical settings.Citation7,Citation26 In this study, L2724hy C. portucalensis was initially misidentified as C. freundii using MALDI-TOF/MS, and was later identified as C. portucalensis by a combination of 16s rRNA sequencing and ANI analysis. Thus, it is essential to enhance the whole-genome sequencing of less common strains.
Data analysis reveals that the plasmids carrying blaSFO-1 include IncA/C, IncHI2, unclassified types.Citation12,Citation13,Citation27,Citation28 In our study, plasmidFinder revealed that stain L2724hy carries the blaSFO-1 gene on an IncFII plasmid that has not been reported before. According to the genetic environment of blaSFO-1 (IS26-ampR-blaSFO-1-IS26), the presence of IS26 on both sides of blaSFO-1 suggests a potential correlation with plasmid-mediated horizontal transmission of blaSFO-1, which is consistent with prior research findings.Citation29 Notably, we also observed that the fosA3, is located upstream of IS26-ampR-blaSFO-1-IS26 and shares the same IS26 with ampR-blaSFO-1. The fosA3 has been reported to be closely associated with the transmission of ESBL gene blaCTX-M by the insertion element IS26.Citation30 Therefore, in our study, we believe that IS26 is a vital insertion element that gradually integrates with fosA3 and blaSFO-1 on the IncFII plasmid, forming a multidrug resistance region, which plays a crucial role in mediating the spread of drug resistance genes. Importantly, this study marks the first instance of blaSFO-1 and fosA3 coexisting on a plasmid.
NDM-1, a carbapenemase, can hydrolyze nearly all beta-lactam antibiotics, including carbapenemase.Citation31 The emergence of carbapenemase-producing strains of NDM-1 represents a significant risk to global public health, necessitating a high level of vigilance. In our study, we observed an absence of ISAba125 from IS5 to IS3000 when comparing it to classical bacteria. According to previous studies, ISAba125 is typically involved in the formation of Tn125, a complex transposon associated with the plasmid-mediated spread of blaNDM-1.Citation32,Citation33 Our findings suggest that the lack of ISAba125 does not impede the horizontal transfer of blaNDM-1. In the conjugation experiment, we successfully transferred blaNDM-1 and blaKPC-2 into EC600 for expression, affirming the transferability of the plasmids in our study. As mentioned above, the genetic environment of blaKPC-2 in this study aligns entirely that of C. freundii (Genebank: CP110915) from the same region, strongly suggesting that blaKPC-2 was transmitted from CP110915.
Conclusion
In conclusion, our study reported for the first time the coexistence of blaSFO-1, blaNDM-1, and blaKPC-2 in a clinical isolated C. portucalensis strain, presenting challenges in multidrug resistance. IS26 is an essential insertion element that facilitates the propagation of drug-resistant genes on IncFII plasmids. These findings emphasize the urgency of continued monitoring and understanding evolving drug resistance dynamics in uncommon strains.
Nucleotide Sequence Accession Numbers
The nucleotide sequences in L2724hy containing a circular chromosome and eleven plasmids have been submitted to GenBank and assigned accession numbers CP136601-136612, respectively.
Ethical Approval
This study was approved by the clinical research ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine [number 2020-IIT-591], and was conducted according to the ethical principles of the Declaration of Helsinki. Parental written consent was obtained.
Disclosure
The authors report no conflicts of interest in this work.
Additional information
Funding
References
- Ribeiro TG, Gonçalves BR, da Silva MS, et al. Citrobacter portucalensis sp. nov. isolated from an aquatic sample. Int J Syst Evol Microbiol. 2017;67(9):3513–3517. doi:10.1099/ijsem.0.002154
- Cao X, Xie H, Huang D, et al. Detection of a clinical carbapenem-resistant Citrobacter portucalensis strain and the dissemination of C. portucalensis in clinical settings. J Glob Antimicrob Resist. 2021;27:79–81. doi:10.1016/j.jgar.2021.04.027
- Chang H, Mishra R, Cen C, et al. Metagenomic analyses expand bacterial and functional profiling biomarkers for colorectal cancer in a Hainan Cohort, China. Curr Microbiol. 2021;78(2):705–712. doi:10.1007/s00284-020-02299-3
- Gupta RK, Singh AK, Bajaj A, Khardenavis AA, Purohit HJ. Phylogenomic analysis of Citrobacter sp. strain AAK_AS5 and its metabolic capabilities to support nitrogen removal behavior. J Basic Microbiol. 2023;63(3–4):359–376. doi:10.1002/jobm.202200323
- Hasan MS, Sultana M, Hossain MA. Complete genome arrangement revealed the emergence of a poultry origin superbug Citrobacter portucalensis strain NR-12. J Glob Antimicrob Resist. 2019;18:126–129. doi:10.1016/j.jgar.2019.05.031
- Thomas SG, Abajorga M, Glover MA, et al. Aeromonas hydrophila RIT668 and Citrobacter portucalensis RIT669-potential zoonotic pathogens isolated from spotted turtles. Microorganisms. 2020;8(11):1805. doi:10.3390/microorganisms8111805
- Luo X, Yu L, Feng J, et al. Emergence of Extensively Drug-Resistant ST170 Citrobacter portucalensis with Plasmids pK218-KPC, pK218-NDM, and pK218-SHV from a Tertiary Hospital, China. Microbiol Spectr. 2022;10(5):e0251022. doi:10.1128/spectrum.02510-22
- Sellera FP, Fuentes-Castillo D, Fuga B, Goldberg DW, Kolesnikovas CKM, Lincopan N. New Delhi metallo-β-lactamase-1-producing Citrobacter portucalensis belonging to the novel ST264 causing fatal sepsis in a vulnerable migratory sea turtle. One Health. 2023;17:100590. doi:10.1016/j.onehlt.2023.100590
- Matsumoto Y, Inoue M. Characterization of SFO-1, a plasmid-mediated inducible class A beta-lactamase from Enterobacter cloacae. Antimicrob Agents Chemother. 1999;43(2):307–313. doi:10.1128/AAC.43.2.307
- Fernández A, Pereira MJ, Suárez JM, et al. Emergence in Spain of a multidrug-resistant Enterobacter cloacae clinical isolate producing SFO-1 extended-spectrum beta-lactamase. J Clin Microbiol. 2011;49(3):822–828. doi:10.1128/JCM.01872-10
- Qiao J, Ge H, Xu H, et al. Detection of IMP-4 and SFO-1 co-producing ST51 Enterobacter hormaechei clinical isolates. Front Cell Infect Microbiol. 2022;12:998578. doi:10.3389/fcimb.2022.998578
- Zhao JY, Zhu YQ, Li YN, et al. Coexistence of SFO-1 and NDM-1 β-lactamase genes and fosfomycin resistance gene fosA3 in an Escherichia coli clinical isolate. FEMS Microbiol Lett. 2015;362(1):1–7. doi:10.1093/femsle/fnu018
- Zhou K, Yu W, Shen P, et al. A novel Tn1696-like composite transposon (Tn6404) harboring bla (IMP-4) in a Klebsiella pneumoniae isolate carrying a rare ESBL gene bla (SFO-1). Sci Rep. 2017;7(1):17321. doi:10.1038/s41598-017-17641-2
- Hobson CA, Pierrat G, Tenaillon O, et al. Klebsiella pneumoniae Carbapenemase variants resistant to ceftazidime-avibactam: an evolutionary overview. Antimicrob Agents Chemother. 2022;66(9):e0044722. doi:10.1128/aac.00447-22
- Han R et al. (2020). Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front. Cell. Infect. Microbiol., 10 10.3389/fcimb.2020.00314
- Chen C, Xu H, Liu R, et al. Emergence of neonatal sepsis caused by MCR-9- and NDM-1-co-producing Enterobacter hormaechei in China. Front Cell Infect Microbiol. 2022;12:879409. doi:10.3389/fcimb.2022.879409
- Sánchez-Pérez S, Comas-Basté O, Duelo A, Veciana-Nogués M Teresa, Berlanga M, Latorre-Moratalla M Luz and Vidal-Carou M Carmen. (2022). Intestinal Dysbiosis in Patients with Histamine Intolerance. Nutrients, 14(9), 1774 10.3390/nu14091774
- Jin Y, Xu H, Yao Q, et al. Confirmation of the need for reclassification of Neisseria mucosa and Neisseria sicca using average nucleotide identity blast and phylogenetic analysis of whole-genome sequencing: hinted by clinical misclassification of a Neisseria mucosa strain. Front Microbiol. 2021;12:780183. doi:10.3389/fmicb.2021.780183
- Liu R, Xu H, Zhao J, et al. Emergence of mcr-8.2-harboring hypervirulent ST412 Klebsiella pneumoniae strain from pediatric sepsis: a comparative genomic survey. Virulence. 2023;14(1):233–245. doi:10.1080/21505594.2022.2158980
- Xu H, Wang X, Yu X, et al. First detection and genomics analysis of KPC-2-producing Citrobacter isolates from river sediments. Environ Pollut. 2018;235:931–937. doi:10.1016/j.envpol.2017.12.084
- Wick RR, Judd LM, Gorrie CL, Holt KE, Phillippy AM. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005595
- Huang J, Zhu J, Gong D, Wu L, Zhu Y, Hu L. Whole genome sequence of EC16, a bla(NDM-5)-, bla(CTX-M-55)-, and fosA3-coproducing Escherichia coli ST167 clinical isolate from China. J Glob Antimicrob Resist. 2022;29:296–298. doi:10.1016/j.jgar.2022.04.001
- Liu S, Xu H, Guo X, et al. Emergence and genetic characterization of plasmid-encoded VIM-2-producing pseudomonas stutzeri with novel integron in1998 isolated from cerebrospinal fluid. Infect Drug Resist. 2021;14:3415–3424. doi:10.2147/IDR.S320294
- Zheng B, Xu H, Lv T, et al. Stool samples of acute diarrhea inpatients as a reservoir of ST11 hypervirulent KPC-2-producing Klebsiella pneumoniae. mSystems. 2020;5(3). doi:10.1128/mSystems.00498-20
- Seman A, Mihret A, Sebre S, Awoke T, Yeshitela B, Yitayew B, Aseffa A, Asrat D and Abebe T. (2022). Prevalence and Molecular Characterization of Extended Spectrum β-Lactamase and Carbapenemase-Producing Enterobacteriaceae Isolates from Bloodstream Infection Suspected Patients in Addis Ababa, Ethiopia. IDR, Volume 15 1367–1382. 10.2147/IDR.S349566
- Wang L, Li Z, Xiao N, et al. Genetic characterization of bla (NDM-1)-carrying Citrobacter portucalensis sequence type 328 and citrobacter freundii sequence type 98. Infect Drug Resist. 2022;15:2235–2242. doi:10.2147/IDR.S361761
- Ai W, Zhou Y, Wang B, et al. Corrigendum: first report of coexistence of bla (SFO-1) and bla (NDM-1) β-lactamase genes as well as colistin resistance gene mcr-9 in a transferrable plasmid of a clinical isolate of Enterobacter hormaechei. Front Microbiol. 2021;12:741628. doi:10.3389/fmicb.2021.741628
- Guo Q, Wang P, Ma Y, Yang Y, Ye X, Wang M. Co-production of SFO-1 and DHA-1 β-lactamases and 16S rRNA methylase ArmA in clinical isolates of Klebsiella pneumoniae. J Antimicrob Chemother. 2012;67(10):2361–2366. doi:10.1093/jac/dks244
- Zhou K, Zhou Y, Zhang C, et al. Dissemination of a ‘rare’ extended-spectrum β-lactamase gene bla(SFO-1) mediated by epidemic clones of carbapenemase-producing Enterobacter hormaechei in China. Int J Antimicrob Agents. 2020;56(3):106079. doi:10.1016/j.ijantimicag.2020.106079
- Lee S, Park Y, Yu J K, Jung S, Kim Y, Jeong S H and Arakawa Y. (2012). Prevalence of acquired fosfomycin resistance among extended-spectrum -lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. Journal of Antimicrobial Chemotherapy, 67(12), 2843–2847. 10.1093/jac/dks319
- Bose P, Rangnekar A, Desikan P. NDM-beta-lactamase-1: where do we stand? Indian J Med Res. 2022;155(2):243–252. doi:10.4103/ijmr.IJMR_685_19
- Alattraqchi AG, Mohd Rani F, A Rahman NI, et al. Complete genome sequencing of Acinetobacter baumannii AC1633 and Acinetobacter nosocomialis AC1530 unveils a large multidrug-resistant plasmid encoding the NDM-1 and OXA-58 carbapenemases. mSphere. 2021;6(1). doi:10.1128/mSphere.01076-20
- Corrêa LL, Kraychete GB, Rezende AM, et al. NDM-1-encoding plasmid in Acinetobacter chengduensis isolated from coastal water. Infect Genet Evol. 2021;93:104926. doi:10.1016/j.meegid.2021.104926
