Abstract
We analyzed 218 strains of methicillin-resistant Staphylococcus aureus (MRSA) isolated from the septicemia patients in a geriatric hospital for 25 years. These strains were classified into 11 major DNA types, A through K, and 27 minor types. The strains belonging to group A and B isolated before 1990 were susceptible to imipenem (IPM), fluoroquinolone, and most other antibiotics tested, except that they were markedly resistant to gentamicin. Strains mostly isolated in 1985 and thereafter were classified into group C through K, and they were mainly resistant to IPM, fluoroquinolones, and clindamycin. Analysis of the MRSA marker gene, staphylococcal cassette chromosome mec (SCCmec), of these strains revealed that the strains in groups A and B had mainly type IV and type I, respectively, and that strains in groups C through J had mainly type II. These results suggested that the strains holding type II SCCmec were resistant to IPM, fluoroquinolone, and clindamycin and they were dominant-resistant type after late 1980s. The antibiotic resistance profiles of MRSA dramatically changed during late 1980s, and these were correlated with the SCCmec types. The lesson from this study would be that consistent execution of surveillance study is needed to update the resistant profiles.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major causative agent of nosocomial and community-acquired infections, which bring about variety of clinical symptoms, including endocarditis, osteomyelitis, pneumonia, toxic shock syndrome, food poisoning, carbuncles, and boils.Citation1 The nosocomial outbreak of MRSA was first reported in England in 1961 and that had been reported all over the world soon after.Citation2–Citation4 The isolation frequency of MRSA strains dramatically increased in ∼1960s thereafter.
Hospitalization of geriatric patients in general hospital due to bloodstream infection (BSI) has been increasing from 1980s through 2000s in the United States.Citation5–Citation7 The major pathogenic organisms causing septicemia are Escherichia coli, Staphylococci, Streptococci, and Pneumococci.Citation6 Overall BSI-associated mortality has been reported to be 30%–60% in elderly patients, and the highest mortality rate is associated with S. aureus infection.Citation7–Citation9 Associated risk factors causing BSI are high incidence of comorbid conditions, such as chronic heart or lung disease, hypertension, and diabetes mellitus.Citation6,Citation7 It is increasingly reported in the health care-associated acquisition of MRSA in elderly patients.Citation8
For the monitoring of antibiotic susceptibilities, it is helpful to determine the minimum growth inhibitory concentration (MIC) of antibiotics. More recently, the gene(s) responsible for the resistance, such as the staphylococcal cassette chromosome mec (SCCmec) element, have been analyzed.Citation10 It is also powerful to use the molecular grouping technique to classify MRSAs into subgroup by the pulsed-field gel electrophoresis (PFGE) of the chromosomal DNA; such study revealed the presence of highly virulent MRSA, such as USA300. The studies contribute both to the control of nosocomial infection and to a better understanding of the evolutionary relationships among MRSA.Citation11–Citation13
The data lacking is a long-term trace of antibiotic-resistant MRSA in a geriatric hospital. To analyze the fate of antibiotic-resistant MRSA, we collected MRSAs from the patients in a geriatric hospital through two decades. And they were subjected to the SCCmec typing, the antibiotic susceptibility test, and the DNA typing. The results revealed that MRSAs with unique DNA type were carried into the hospital probably with incoming patients and causing the nosocomial dissemination and forming the resistant clones.
Material and methods
Bacterial strains used
A total of 218 strains of MRSA were collected from the patients in a single geriatric hospital in Tokyo from 1978 through 2002. Information on patient’s background and clinical symptoms could not be collected due to the law on the protection of privacy regarding the processing of personal data. All the S. aureus strains were isolated from the blood specimens on the mannitol–NaCl agar plate, and the colonies grown on the medium were subjected to the methicillin and oxacillin susceptibility test, the mecA gene amplification by Tsuchizaki et al method.Citation14 The mecA-positive strains with MIC of methicillin ≥16 mg/L or that of oxacillin ≥4 mg/L were classified as MRSA. These strains were kept frozen at −80°C in the brain heart infusion broth (Nippon Becton Dickinson, Tokyo, Japan) supplemented with 40% glycerol.
Antibiotics
Imipenem (IPM) was a gift from Banyu Pharmaceutical Co (Tokyo, Japan). Arbekacin (ABK) was purchased from Meiji Seika Kaisha (Tokyo, Japan). Gentamicin (GM), minocycline (MINO), rifampicin (RFP), and sulfamethoxazole were purchased from Wako Pure Chemical Industries (Osaka, Japan). Ciprofloxacin (CPFX), clindamycin (CLDM), trimethoprim, teicoplanin (TEIC), and linezolid (LZD) were a gift from Bayer Yakuhin (Osaka, Japan), Dainippon Sumitomo Pharma Co (Osaka, Japan), Shionogi and Co (Osaka, Japan), Astellas Pharmaceutical Inc (Tokyo, Japan), and Pfizer Japan Inc (Tokyo, Japan), respectively. Vancomycin (VCM) was purchased from Sigma-Aldrich (Saint Louis, MO).
Determination of the MIC of antibiotic
The MIC of the antibiotics was determined by the agar dilution method according to the Clinical and Laboratory Standards Institute (CLSI) guideline as described previously.Citation15
Typing of the SCCmec gene
Multiplex polymerase chain reaction (PCR) was carried out using a pair of primers according to the method of Oliveira and de Lencastre.Citation16 The DNA polymerase used was Phusion High-Fidelity DNA polymerase (Finnzymes Oy, Espoo, Finland). The reaction mixture contained the formula recommended for the DNA polymerase. The thermal cycler was set to 95°C for 30 sec, a total 30 cycles of 98°C for 10 sec, 57°C for 10 sec, and 72°C for 15 sec, and 72°C for 5 min at the end using a Gene Amp PCR System 9700 thermal cycler (Applied Biosystems Japan, Tokyo, Japan). The products were subjected to electrophoresis in 4% agarose gel in EDTA buffer at 100 V for 80 min. A 100-bp DNA ladder (New England Biolabs, Hitchin, UK) was used as a size maker. Since on time PCR over 25 years is not practical, all the samples were kept frozen at −80°C and subjected to PCR within about half a year in 2007.
PFGE
Chromosomal DNA was extracted from the MRSA cells and was digested with SmaI according to the method described by Bannerman et al.Citation17 The DNA plugs sliced at a thickness of 1–4 mm were placed in 170 μL of a solution containing 10 units of SmaI, 20 μL of T-buffer (0.1 M tris-HCl, pH 8.0, 700 mM MgCl2, 0.2 M KCl, 700 mM 2-mercaptoethanol), and 20 μL of 0.1% bovine serum albumin. The mixture was incubated at 25°C for 4 h. Samples were loaded on 1% agarose gel prepared in 0.5 × TBE buffer containing 44.5 mM tris, 44.5 mM boric acid, and 1 mM EDTA pH 8.0. The wells were sealed with 1% agarose in the same buffer. PFGE was carried out with a CHER-DRIII electrophoresis cell (Bio-Rad, Hercules, CA) at 6 V/cm for 20 h at 14°C with initial and final pulses conducted for 5.3 and 34.9 sec, respectively. The gel was stained with GelRed (Biotium, Hayward, CA) according to the manufacturer’s manual and visualized under a 254-nm UV light.
The digital images were analyzed with PDQuest™ software (version 4.5; Bio-Rad), according to the unweighted pair-group matching analysis clustering algorithm. The strains with over ∼90% identity were classified into the same group. Those groups that consisted of less than four isolates were excluded from further study. Similar to PCR, PFGE of all strains was carried out in a few months.
Results
Grouping of MRSAs
Because all the MRSA strains subjected to this study were collected through two decades, 1978–2002, they might be suitable for the model study of chronological changes of MRSA with regard to antibiotic susceptibility, DNA type, and the MRSA marker gene. All 218 strains were subjected to PFGE fingerprinting, and 11 major groups (more than five strains per group) and 27 minor groups (less than four strains per group) were found (). Group A through K was aligned along chronologic order of sampling (). Strains isolated from 1978 through 1989 were classified into group A and B. Strains isolated ∼1985 through 2002 were classified into the group C through K. Remaining 47 strains in 27 minor groups were not studied any further.
Figure 1 PFGE profiles and a phylogeny of MRSAs grouped into 11 unique DNA types. The PFGE profiles were digitized, and the degree of homology was calculated by the unweighted pair-group method. The DNA profiles that showed over ∼90% identity were classified into the same group. The groups of MRSA that showed unique DNA profiles but consisted of less than four strains were excluded from this figure. A dendrogram was derived from PFGE analysis of SmaI-digested chromosomal DNA.
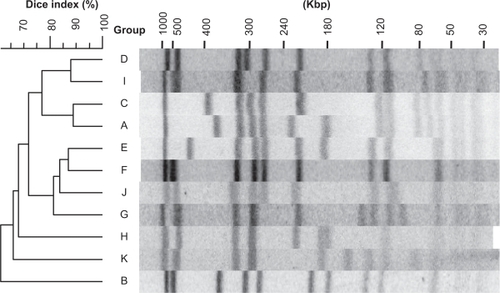
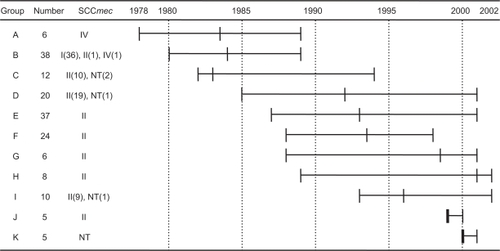
Figure 2 Length and the chronology of the MRSA isolation and their SCCmec type. Length and the chronology of the MRSA isolation and their SCCmec type A through K represents the groups of DNA type and number represents numbers of strain belonging to each type of DNA group. The SCCmec type was given. A horizontal bar shows duration, in which the same types of MRSA were isolated. A vertical cross bar within the horizontal bar is a median value.
Strains isolated from 1978 through 1989, the groups A and B
As seen in , six strains in the group A, which was isolated from 1978 through 1989 had the type IV SCCmec without exception. Strains in the group B, consisting of 38 strains, isolated in 1980–1989, showed predominantly the type I SCCmec with only exception of two strains.
MIC50 values were the antibiotic concentration that inhibits the growth of 50% of strains and the ranges were shown in . The MIC50 of antibiotics in the respective groups was determined and the results were shown in . The strains in the groups A and B showed susceptibility to all the antibiotics tested with an exception against GM. These strains were collected during the period of 1980 through 1989 and were having the type IV or type I SCCmec predominantly. The strains in the group A and B were resistant to GM and that turned to be susceptible in the strain in the groups C, E, F, G, and K. The strains in the groups A and B were susceptible to MINO.
Table 1 MIC50 of antibiotics in MRSA grouped to 11 DNA types by pulsed-field gel electrophoresis
Table 2 The antibiotic susceptibility and SCCmec type of the strains grouped by the DNA type
Strains isolated ∼1985 through 2002, the groups C through K
All the strains in the groups C through K isolated during the period of ∼1985–2002 showed the type II SCCmec excepting nine nontypeable strains. Strains in group K, isolated in 2000–2001, had all nontypeable SCCmec by the present method. This seems to be understandable because the MRSAs in this group were isolated in the latest period during this study. Therefore, the strains in the group K likely had the new type of SCCmec. The MINO-susceptible strains were found in the groups C, D, F, and I and the strains in the rest of groups were MINO resistant.
The strains classified into the group C, which was isolated during the period of 1982 through 1994, suddenly became resistant to the β-lactam antibiotics (IPM) and this trend has been kept for the strains in the groups D through K with only an exception that strains in the group G showed IPM susceptibility. Strains in the groups C through K, which were isolated during the period of 1985 through 2002, showed marked resistance to CPFX, though the strains in the group C isolated in 1982–1994 were intermediately resistant. These results suggested that strong β-lactam antibiotics and fluoroquinolone antibiotics might be introduced for clinical use in this period.
The GM-resistant strains in group A and B turned out to be more susceptible than those strains in group C, E, F, and G. However, reemergence of the GM-resistant MRSAs in the groups C, H, I, and J suggested that GM-resistant clones were selected and spread within the hospital. The strains in the group E, G, H, J, and K were resistant to MINO suggesting that tetracycline derivatives might be used during the period for the groups G through J. The strains in the group C through K except the group I were CLDM susceptible.
Antibiotics effective against MRSA
All the strains tested showed the susceptible range of MIC50 of ABK, RFP, ST, TEIC, VCM, and LZD, whereas they were resistant to IPM, GM, and CLDM according to the CLSI criteria. Thus, the analysis clearly revealed that the susceptibility to these drugs was unrelated with the PFGE fingerprinting and the SCCmec types.
Discussion
PFGE is a useful method for genomic fingerprinting of micro-organisms, and the data could be used for epidemiological study of virulent organisms. SCCmec typing is also used for the epidemiological study of MRSA.Citation18–Citation21 However, it might be difficult to use the SCCmec typing for the clonal identification of MRSA because the SCCmec typing only analyzes a small fraction of the chromosome. Our investigation was conducted to analyze clonal identity of 218 blood-isolated MRSAs from 1978 through 2002, and the results were used for the interpretation of the origins of the strain, possible dissemination, and persistency in the hospital. Attempts have been made to link the MIC of antibiotics with the DNA types of the strain and SCCmec type.
The DNA typing of the strain revealed four important facts: 1) Three quarters of the isolates, which belonged to only 11 DNA types among 39, were inhabited and spread within the hospital. 2) The strains inhabited in the hospital persisted for a long period of time causing the presence of multiple types of MRSA clones of different genotype in the same time. 3) Roughly, one quarter of strains were either successfully treated or phased out. 4) Group A (SCCmec type I) and B (type IV) were highly susceptible to both carbapenem and fluoroquinolone than group C–J (type II) and group K (nontype). In addition, it was generally said in Japan that the strains resistant to less numbers of antibiotics tend to be SCCmec type I and IV.
It was well recognized that the recently isolated MRSAs from septicemia were found to have SCCmec type II.Citation22 However, the result shown in suggested that septicemia in early 1980s was caused by the strain having SCCmec type I (group B) and IV (group A). The strains having SCCmec type I and IV seemed to disappear from hospital probably because they were highly susceptible to multiple antibiotics except against GM. Because the SCCmec typing provides information mainly on β-lactam antibiotic susceptibility, drastic change of the SCCmec types might be related to the use of β-lactam. In fact, the SCCmec type of strains in the group A and B has been changed largely from type IV and I to type II in the group C through J that is consistent with the approval of carbapenem. Close examination of the dendrogram also showed drastic change of the DNA types in groups A, B, and C through K. Moreover, the CPFX susceptibility profile has been markedly changed between the strains in the group A (SCCmec type IV), B (SCCmec type I), and C through K (SCCmec type II and nontype) that is consistent with the approval of the fluoroquinolone antibiotics. One other thing notable in this study would be the GM susceptibility. The groups A and B, those isolated in the early phase of this study, were GM-resistant and the groups C, E, F, G and K turned out to be GM-susceptible. This is probably due to the fact that the isolation frequency of the strain producing aminoglycoside-modifying enzyme decreased markedly.Citation23 The GM-resistant clones reemerged again later, which could be interpreted in two ways: 1) the susceptible strain gained GM resistance, or 2) new GM-resistant clone(s) were carried in from outside. Eventually, the strains resistant to GM, IPM, and CPFX, the multiantibiotic-resistant MRSA survived in the hospital.
Conclusions
The conclusions of this study are as follows: 1) SCCmec type had been changed from type I and IV to type II in the late 1980s. 2) The strains having SCCmec type I or IV were more susceptible to antibiotics than type II. 3) The strains having SCCmec type II were highly antibiotic resistant and therefore they could be persisted in the hospital. To eradicate MRSA from hospitals, a long-term usage of antimicrobial agents must be designed cautiously. Therefore, preventing the dissemination of MRSA in the hospital environments may be the first priority among the infection-control programs. Active surveillance of MRSAs, proper use of antibiotics, and hygienic hand-washing are recommended for prevention of nosocomial spread of MRSAs.
Disclosure
The authors report no conflicts of interest in this work.
References
- ShopsinBKreiswirthBNMolecular epidemiology of methicillin-resistant Staphylococcus aureusEmerg Infect Dis20017232332611294733
- JevonsM“Celbenin”-resistant staphylococciBr Med J19611124125
- StewartGTHoltRJEvolution of natural resistance to the newer penicillinsBr Med J19631532630831113984091
- HaleyRWHightowerAWKhabbazRFThe emergence of methicillin-resistant Staphylococcus aureus infections in United States hospitals. Possible role of the house staff-patient transfer circuitAnn Intern Med19829732973087114626
- PinnerRWTeutschSMSimonsenLTrends in infectious diseases mortality in the United StatesJAMA199627531891938604170
- McBeanMRajamaniSIncreasing rates of hospitalization due to septicemia in the US elderly population, 1986–1997J Infect Dis2001183459660311170985
- MalaniPNRanaMMBanerjeeMBradleySFStaphylococcus aureus bloodstream infections: the association between age and mortality and functional statusJ Am Geriatr Soc20085681485148918662207
- BradleySFStaphylococcus aureus infections and antibiotic resistance in older adultsClin Infect Dis200234221121611740710
- TacconelliEPop-VicasAED’AgataEMIncreased mortality among elderly patients with meticillin-resistant Staphylococcus aureus bacte-raemiaJ Hosp Infect200664325125616978733
- ItoTKatayamaYHiramatsuKCloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315Antimicrob Agents Chemother19994361449145810348769
- CrisóstomoMIWesthHTomaszAChungMOliveiraDCde LencastreHThe evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clonesProc Natl Acad Sci U S A200198179865987011481426
- OliveiraDCTomaszAde LencastreHThe evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elementsMicrob Drug Resist20017434936111822775
- DiepBCarletonHAChangRFSensabaughGFPerdreau-RemingtonFRoles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureusJ Infect Dis2006193111495150316652276
- TsuchizakiNIshikawaJHottaKColony PCR for rapid detection of antibiotic resistance genes in MRSA and enterococciJpn J Antibiot20005342242910955238
- WaynePAPerformance Standards for Antimicrobial Susceptibility Testing 15th Informational Supplement. CLSI/NCCLS Document M100-S15Wayne, PAClinical and Laboratory Standards Institute2005
- OliveiraDCde LencastreHMultiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureusAntimicrob Agents Chemother20024672155216112069968
- BannermanTLHancockGATenoverFCMillerJMPulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureusJ Clin Microbiol19953335515557751356
- MaXXItoTChongtrakoolPHiramatsuKPredominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985J Clin Microbiol200644124515452717050818
- ShibuyaYHaraMHiguchiWTakanoTIwaoYYamamotoTEmergence of the community-acquired methicillin-resistant Staphylococcus aureus USA300 clone in JapanJ Infect Chemother200814643944119089559
- VandeneschFNaimiTEnrightMCCommunity-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergenceEmerg Infect Dis20039897898412967497
- ItoTKuwaharaKHiramatsuKStaphylococcal cassette chromosome mec(SCC mec) analysis of MRSAMethods Mol Biol20073918710218025671
- BaradaKHanakiHYamaguchiYTrends of beta-lactam antibiotic susceptibility in blood-borne methicillin-resistant Staphylococcus aureus (MRSA) and their linkage to the staphylococcal cassette chromosome mec (SCCmec) typeJ Infect Chemother200713421321817721683
- BaradaKHanakiHIkedaSTrends in the gentamicin and arbekacin susceptibility of methicillin-resistant Staphylococcus aureus and the genes encoding aminoglycoside-modifying enzymesJ Infect Chemother2007132747817458673