Abstract
Background
Thyroid cancer is one of the most common cancers with rising incidence worldwide, and papillary thyroid carcinoma (PTC) accounts for 80–85% of thyroid malignancy. Although it has been reported that many genes relate to the carcinogenesis of PTC, the molecular mechanisms remain mostly unclear.
Methods
QRT-PCR assay was performed to detect circRNA_104565, miR-134 and ELF2 expression. CCK8 assay was exercised to examine cell proliferation. Western blot was used to detect ELF2 expression.
Results
We found that circRNA_104565 was highly expressed in PTC tissue and cell and promoted cell proliferation in vitro and in vivo. In addition, circRNA_104565 promoted cell proliferation in PTC by regulating the miR-134/ELF2 axis.
Conclusion
Hence, revealing the function of circRNA_104565 in PTC is important for understanding the molecular mechanism of carcinogenesis and providing new biomarkers or therapeutic targets for PTC.
Introduction
Thyroid cancer is one of the most common cancers with rising incidence worldwide,Citation1 and papillary thyroid carcinoma (PTC) accounts for 80–85% of thyroid malignancy.Citation2–Citation5 Although it has been reported that many genes relate to the carcinogenesis of PTC, the molecular mechanisms remain mostly unclear.
Circular RNAs (circRNAs), non-coding RNAs, are widely expressed in mammals.Citation6,Citation7 Increasing studies indicate circRNA are dysregulated and may display significant roles in tumor pathogenesis. For example, circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271.Citation8 Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via the miR-1275/FOXK1 axis.Citation9 circFBLIM1 acts as a competing endogenous RNA (ceRNA) to promote hepatocellular cancer progression by sponging miR-346.Citation10 Notably, dysregulation of circRNA can also cause formation and progression of PTC. For example, knockdown of circPVT1 inhibits progression of papillary thyroid carcinoma by sponging miR-126.Citation11 Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression.Citation12 CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway.Citation13 Thus, revealing the function of circRNA in PTC is important for understanding the molecular mechanism of carcinogenesis and identifying new biomarkers or therapeutic targets for PTC.
Mechanically, the function of circRNA mainly acts as a ceRNA by sponging miRNA. For example, circRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis.Citation14 Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-8a to regulate oral squamous cell carcinoma cells growth via glucose transporter (GLUT1) and glycolysis.Citation15 Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway.Citation16 Thus, analysis of the circRNA/miRNA network have crucially great significance.
In this study, we firstly found circRNA_104565 were increased in PTC tissue and cell. In addition, we also found upregulation of circRNA_104565 promotes cell proliferation in PTC by sponging miR-134.
Materials and Methods
Human Tissue Specimens
62 pairs of PTC tissues and adjacent normal tissues were obtained from Tinglin Hospital of Jinshan District. This project complied with the principles of the Declaration of Helsinki and was endorsed by the Research Ethics Committee of the Tinglin Hospital of Jinshan District. All patients signed the written informed consent.
Cell Culture
PTC cell lines (BHP5–16, TPC-1, BHP2–7 and BCPAP) and normal human thyroid follicular epithelial cell (Nthy-ori-3–1) were brought from American Type Culture Collection (ATCC, USA). All cells were cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), and were maintained at 37 °C with 5% CO2.
QRT-PCR Assay
TRIzol kits (Invitrogen, Grand Island, NY, USA) were performed to extract total RNAs following the manufacturer's instructions, and ReverTra Ace qPCR RT Kit (TOYOBO) were used to transcribe RNAs into cDNAs. QRT-PCR were performed to examine circRNA_104565 expression by using Light Cycler 480 SYBR Green I Master (Roche) on an ABI7300 cycler (Applied Biosystems) through ABI7300 software. Relative gene expression levels were quantified using the 2−ΔΔCt method. The primer sequences of circRNA_104565 were 5ʹ-CTGCAAGGGGTAAATGAGGGTG-3ʹ (forward primer) and 5ʹ-CCTTTTGGCTGCTTCCACATTG-3ʹ (reverse primer). The primer sequences of GADPH were 5ʹ-GTCAACGGATTTGGTCT-3ʹ (forward primer) and 5ʹ-AGTCTTCTGGGTGGCAGTGAT-3ʹ.
Animal Experiments
The 4-week-old female BALB/c nude mice were purchased from The Model Animal Research Center of Nanjing University, Nanjing, China, and were chosen for xenografts experiments. All procedures were approved by Tinglin Hospital of Jinshan District Animal Care and Use Committee, which was in accordance with Institutional Animal Care and Use Committee and animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. The mice were subcutaneously inoculated with BCPAP/sh-circRNA_104565 or BCPAP/sh-NC cells. The tumor volume was calculated in accordance with the formula (length × width2/2) and measured weekly. After 28 days, the mice were euthanized and the tumors were excised, and tumor weight was determined.
CCK8 Assay
Cell proliferation was detected by CCK8 assay, according to the manufacturer's instructions. Absorbance values at 450 nm were measured.
Luciferase Reporter Assays
We used Lipofectamine 3000 to transfect the luciferase reporter vectors into PTC cells. We then co-transfected the miR-134 mimic or miR-134 inhibitor with the reporter gene into PTC cells. For the last step, we used the Dual Luciferase Reporter System Kit (E1910, Promega, USA) to detect firefly and renilla luciferase activity according to the manufacturer's instructions.
Western Blot
Total proteins were harvested by using RIPA buffer (Sangon Biotech) and separated by 12% SDS-PAGE. Then, the protein in SDS-PAGE were transferred onto a PVDF membrane (Millipore, USA). The membranes were blocked with 4% skimmed milk powder and incubated with rabbit anti-ELF2 (1:1000 dilution; Abcam) primary antibodies. An anti-GAPDH antibody (1:1000, Affinity, USA) was used as control.
Statistical Analysis
Statistical analysis was exercised by using GraphPad Prism 6 software (GraphPad). Student’s t-test was performed to assess statistical differences between two groups, and comparisons among more than two groups were performed using analysis of variance (ANOVA). The correlation between groups was assessed with Pearson correlation coefficient. Data were presented as mean ± SD. Statistical differences were set at P < 0.05.
Results
CircRNA_104565 Was Highly Expressed in PTC Tissue and Cell
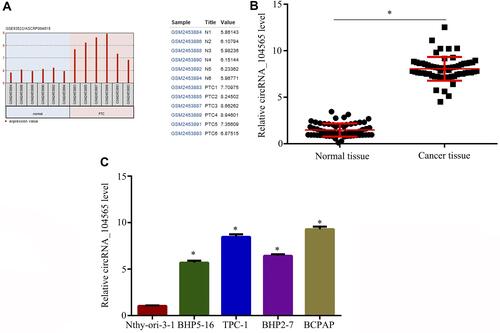
Using the bioinformatics tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE93522), GEO dataset (GSE93522) for circRNA were analyzed from PTC tissue. The results indicated circRNA_104565 expression was increased in PTC tissue (). Subsequently, we detected 62 pairs of PTC tissues and normal tissues, and the result showed circRNA_104565 expression was increased in PTC tissue compared with normal tissue (). In addition, it was also indicated circRNA_104565 expression was increased in PTC cells compared with normal human thyroid follicular epithelial cell (Nthy-ori-3–1) (). Thus, circRNA_104565 was highly expressed in PTC tissues and cells.
Figure 1 CircRNA_104565 was highly expressed in PTC tissue and cell. (A) GEO dataset (GSE93522) showed that circRNA_104565 was significantly increased in PTC tissues compared to normal tissues. (B) CircRNA_104565 expression was significantly increased in PTC tissues compared to normal tissues. (C) CircRNA_104565 was significantly increased in PTC cells compared to Nthy-ori-3–1. *P < 0.05.
Silencing circRNA_104565 Inhibited Cell Proliferation in PTC Cell
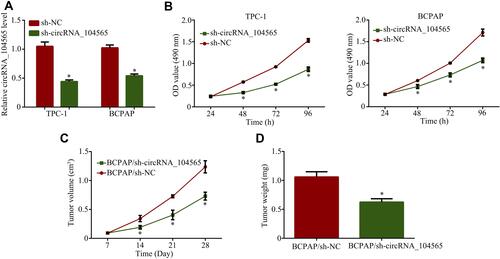
Since circRNA_104565 were increased in PTC, we applied RNA interference to downregulate circRNA_104565 expression to assess its biological functions in PTC. QRT-PCR analysis showed that knockdown of circRNA_104565 was successful (). Subsequently, CCK8 assay indicated knockdown of circRNA_104565 expression remarkably inhibited cell proliferation in the PTC cell (). Moreover, animal experiments further revealed knockdown of circRNA_104565 expression significantly inhibited tumor volume in vivo (). The tumor weight of the sh-circRNA_104565 group were lighter than the sh-NC group (). Thus, silencing circRNA_104565 inhibited cell proliferation in the PTC cell.
Figure 2 Silencing circRNA_104565 inhibited cell proliferation in PTC cell. (A) CircRNA_104565 expressions were detected by qRT-PCR. (B) Cell proliferations were detected by CCK8 assay. (C) The tumor volumes were measured every 5 days. (D) Tumor weight was measured after mouse was surgically dissected. *P < 0.05.
CircRNA_104565 Acts as a ceRNA for miR-134 to Promote Cell Proliferation in PTC
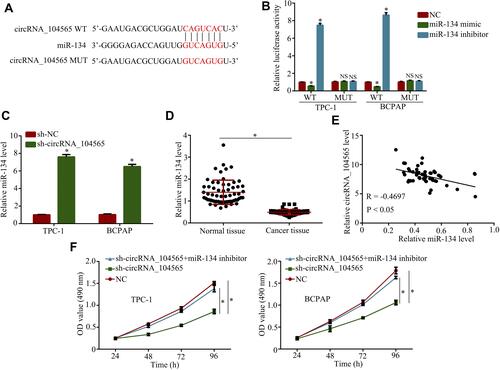
To explore miRNA's interactions with circRNA_104565, we analyzed the overlap from results of miRDB and starbase tool to predict potential binding sites. The results indicated circRNA_104565 may bind to miR-134 (), and luciferase reporter assay had further confirmed their complementary combinations (). Moreover, silencing circRNA_104565 promoted miR-134 expression in PTC cells (). Subsequently, we further examined miR-134 expressions in PTC tissue and normal tissue, and the results showed miR-134 expressions were decreased in PTC tissue compared with normal tissue (). In addition, Pearson correlation analysis revealed circRNA_104565 expression was inversely correlated with miR-134 in PTC tissue (). Furthermore, CCK8 assay showed transfected miR-134 inhibitor restored the cell proliferation of transfected sh-circRNA_104565 PTC cell (). Thus, circRNA_104565 acts as a ceRNA for miR-134 to promote cell proliferation in PTC.
Figure 3 CircRNA_104565 acts as a ceRNA for miR-134 to promote cell proliferation in PTC. (A) The potential binding sites of circRNA_104565 and miR-134 were predicted by miRDB and starbase tool. (B) Luciferase reporter assay had confirmed complementary combinations between circRNA_104565 and miR-134. (C) MiR-134 expressions were detected by qRT-PCR. (D) MiR-134 expression was significantly decreased in PTC tissues compared to normal tissues. (E) Pearson correlation analysis revealed circRNA_104565 expression was inversely correlated with miR-134 in PTC tissue. (F) Cell proliferations were detected by CCK8 assay. *P < 0.05.
CircRNA_104565 Accelerates PTC Cell Growth by Regulating miR-134/ELF2 Axis
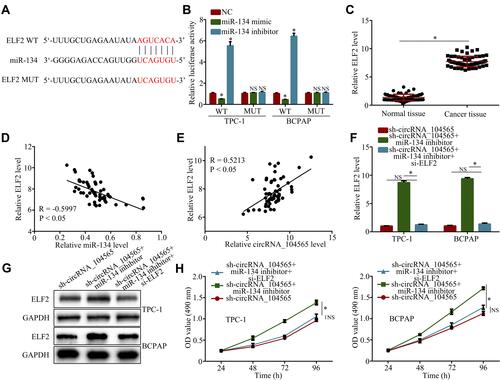
We found that miR-134 had potential interactions with ELF2 via a biological website (). The luciferase reporter assay had further confirmed their complementary combinations (). Subsequently, we further examined ELF2 expressions in PTC tissue and normal tissue, and the results showed ELF2 expressions were increased in PTC tissue compared with normal tissue (). In addition, Pearson correlation analysis revealed ELF2 expression was inversely correlated with miR-134 () and positively correlated with circRNA_104565 (), and silencing circRNA_104565 suppressed ELF2 expression in PTC cells (Figure S1). In addition, transfected si-ELF2 suppressed ELF2 expressions at the mRNA level () and protein level (). Moreover, CCK8 assay showed transfected si-ELF2 suppressed cell proliferation of transfected sh-circRNA_104565 and miR-134 inhibitor in PTC cell (). Hence, circRNA_104565 accelerates PTC cell growth by sponging miR-134, and then up-regulating ELF2.
Figure 4 CircRNA_104565 accelerates PTC cell growth by regulating miR-134/ELF2 axis. (A) MiR-134 had potential interaction with ELF2 via a biological website. (B) Luciferase reporter assay had confirmed complementary combinations between ELF2 and miR-134. (C) ELF2 expression was significantly increased in PTC tissues compared to normal tissues. (D) Pearson correlation analysis revealed ELF2 expression was inversely correlated with miR-134. (E) Pearson correlation analysis revealed ELF2 expression was positively correlated with circRNA_104565. ELF2 expression were detected by qRT-PCR (F) and Western blot (G) (H) Cell proliferations were detected by CCK8 assay. *P < 0.05.
Discussion
Recently, an increasing number of studies indicated circRNAs were discovered to be involved in the progression of tumors. For instance, circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer.Citation17 Circular RNA NSD2 promotes metastasis of colorectal cancer by targeting miR-199b-5p mediated DDR1 and JAG1 signalling.Citation18 Silencing hsa-circ-0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells.Citation19 Notably, circRNAs were also found to be involved in the progression of PTC. For example, circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression.Citation12 CircRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway.Citation13 Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis.Citation20 In this study, we firstly found circRNA_104565 was highly expressed in PTC tissue and cell and promoted PTC cell proliferation in vitro and in vivo.
Mechanically, the function of circRNAs are mainly to act as ceRNAs by sponging miRNA. In this study, we found circRNA_104565 acts as a ceRNA for miR-134. Early study indicated miR-134 functions as a tumor suppressor in cell proliferation and epithelial-to-mesenchymal transition by targeting KRAS in renal cell carcinoma cells.Citation21 MiR-134 regulates the proliferation and invasion of glioblastoma cells by reducing Nanog expression.Citation22 MiR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells.Citation23 However, the function of miR-134 in PTC remains unclear. Hence, we firstly reported downregulation of miR-134 promoted cell proliferation in PTC and confirmed circRNA_104565 promoted cell proliferation in PTC by sponging miR-134. In addition, we found that miR-134 had potential interactions with ELF2 via a biological website, and transfected si-ELF2 suppressed cell proliferation of transfected sh-circRNA_104565 and miR-134 inhibitor in PTC cell, suggesting that circRNA_104565 promotes cell proliferation in papillary thyroid carcinoma by regulating the miR-134/ELF2 axis.
In summary, circRNA_104565 was highly expressed in PTC tissue and cell and promoted cell proliferation in vitro and in vivo. In addition, circRNA_104565 promoted cell proliferation in PTC by regulating the miR-134/ELF2 axis. Hence, revealing the function of circRNA_104565 in PTC is important for understanding the molecular mechanism of carcinogenesis and providing new biomarkers or therapeutic targets for PTC.
Acknowledgments
The present study was supported by The Medical Subject Construction Fund Project of Shanghai Jinshan District (grant no. JSZK2019B03).
Disclosure
The authors have no conflict of interest to declare.
References
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. doi:10.3322/caac.21565
- LiVolsi VA. Papillary thyroid carcinoma: an update. Modern Pathol. 2011;24(Suppl 2):S1–9. doi:10.1038/modpathol.2010.129
- Pillai S, Gopalan V, Smith RA, Lam AK. Diffuse sclerosing variant of papillary thyroid carcinoma–an update of its clinicopathological features and molecular biology. Crit Rev Oncol Hematol. 2015;94(1):64–73. doi:10.1016/j.critrevonc.2014.12.001
- Pusztaszeri M, Auger M. Update on the cytologic features of papillary thyroid carcinoma variants. Diagn Cytopathol. 2017;45(8):714–730. doi:10.1002/dc.23703
- Sowder AM, Witt BL, Hunt JP. An update on the risk of lymph node metastasis for the follicular variant of papillary thyroid carcinoma with the new diagnostic paradigm. Head Neck Pathol. 2018;12(1):105–109. doi:10.1007/s12105-017-0835-9
- Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11(1):21. doi:10.1186/s13045-018-0569-5
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
- Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566–1576.
- Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498(4):1009–1015. doi:10.1016/j.bbrc.2018.03.105
- Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Experimental Clinical Cancer Res. 2018;37(1):172. doi:10.1186/s13046-018-0838-8
- Tao L, Yang L, Tian P, Guo X. Knockdown of circPVT1 inhibits progression of papillary thyroid carcinoma by sponging miR-126. RSC Adv. 2019;23(9):13316–13324. doi:10.1039/C9RA01820D
- Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503(1):56–61. doi:10.1016/j.bbrc.2018.05.174
- Bi W, Huang J, Nie C, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of beta-catenin pathway. J Experimental Clinical Cancer Res. 2018;37(1):275. doi:10.1186/s13046-018-0936-7
- Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–632. doi:10.1016/j.bbrc.2018.02.119
- Chen X, Yu J, Tian H, et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol. 2019;234(11):19130–19140. doi:10.1002/jcp.28692
- Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234(1):500–508. doi:10.1002/jcp.26612
- Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Molecular Ther. 2019;27(9):1638–1652. doi:10.1016/j.ymthe.2019.05.011
- Chen LY, Zhi Z, Wang L, et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol. 2019;248(1):103–115. doi:10.1002/path.5238
- Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(1):118–126. doi:10.26355/eurrev_201801_14108
- Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Experimental Clinical Cancer Res. 2019;38(1):318. doi:10.1186/s13046-019-1321-x
- Liu Y, Zhang M, Qian J, et al. miR-134 functions as a tumor suppressor in cell proliferation and epithelial-to-mesenchymal Transition by targeting KRAS in renal cell carcinoma cells. DNA Cell Biol. 2015;34(6):429–436. doi:10.1089/dna.2014.2629
- Niu CS, Yang Y, Cheng CD. MiR-134 regulates the proliferation and invasion of glioblastoma cells by reducing Nanog expression. Int J Oncol. 2013;42(5):1533–1540. doi:10.3892/ijo.2013.1844
- Li J, Wang Y, Luo J, et al. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett. 2012;586(20):3761–3765. doi:10.1016/j.febslet.2012.09.016
