Abstract
Purpose
This study aimed to explore the expression level and mutation of LRP1B in hepatocellular carcinoma (HCC) and to analyse the relationship between its prognostic value and immune invasion.
Methods
HCC mutant gene sets were obtained from the Cancer Genome Atlas and International Cancer Genome Consortium databases. The Kaplan–Meier method was used to evaluate the prognostic value of LRP1B expression and mutation load in HCC. The relationships between LRP1B expression level and immune cells and immune marker molecules were analysed by using the TIMER database. The association of LRP1B expression with drug sensitivity was obtained by using CellMiner. Gene set enrichment analysis and co-expression by Spearman correlation analysis were used to explore the internal mechanism of LRP1B in HCC.
Results
Seventeen most commonly mutated genes were screened out, and LRP1B was the only gene associated with HCC prognosis. The copy number variations were significantly correlated with T cell CD8+ (P < 0.05). LRP1B expression level was positively correlated with the infiltration degree of macrophage (P < 0.05, R = 0.132), myeloid dendritic cell (P < 0.05, R = 0.093), neutrophil (P < 0.05, R = 0.134) and T cell CD8+ cells (P < 0.05, R = 0.102) and negatively correlated with B cell (P < 0.05, R = −0.014) and T cell CD4+ (P < 0.05, R = −0.075). LRP1B expression level was significantly correlated with immunomarker molecules and drug sensitivity (all P < 0.05). The prediction of lncRNA RUSC1-AS1/hsa-miR-215-5p/LRP1B axis by bioinformatics may be the potential mechanism underlying LRP1B’s effect on HCC prognosis and progression.
Conclusion
LRP1B plays a vital role in HCC prognostic value, which is expected to be a new potential therapeutic target for HCC. LRP1B provides a theoretical basis for the clinical targeted therapy of HCC.
Introduction
Primary liver cancer (PLC), a difficult health problem affecting people all over the world, is one of the most common gastrointestinal malignancies, ranking fifth in the United States and second in China.Citation1,Citation2 Hepatocellular carcinoma (HCC) is the most common pathological type, accounting for more than 80%. It is the leading cause of cancer-related death worldwide. HCC is mostly diagnosed at an advanced stage with a 5-year survival rate of only 12%. Traditional treatments, such as surgical resection, liver transplantation, radiotherapy and chemotherapy, lack effective radical treatment and have a high fatality rate.Citation3,Citation4 Therefore, improving early diagnosis and developing new molecular therapeutic targets are key to improving the efficacy of HCC treatment and survival rate.
Lipoprotein receptor-related protein 1B (LRP1B), which is located on chromosome 2q, can bind to fibrinogen and APOE lipoproteins and regulate blood coagulation and lipoprotein metabolism.Citation5,Citation6 It can be prevalent in a variety of malignant tumours. Wang’s study showed that the LRP1B expression level in colon cancer was significantly lower than that in para-cancer tissues, and the low expression level of LRP1B promoted the proliferation and metastasis of colon cancer cells.Citation7 In prostate cancer, up-regulated miR-301b-3p induced by hypoxia can inhibit the expression level of LRP1B, thereby enhancing the proliferation and migration ability of prostate cancer cells.Citation8 Downregulated expression of LRP1B was involved in malignant proliferation, angiogenesis, differentiation and metastasis in human non-small cell lung cancer, neuroglioma, renal cell cancer and cervical squamous cell carcinoma.Citation9–Citation12 In these types of tumours, LRP1B often acts as a tumour suppressor. The value of LRP1B in tumours is closely related to its expression level, and its mutational significance cannot be ignored, because it is one of the 10 most commonly mutated genes in human cancer.Citation13 Recent studies have shown that LRP1B gene mutation is significantly correlated with the occurrence and prognosis of human tumours, including non-small cell lung cancer, ovarian clear cell carcinoma, urachal carcinoma and renal cell cancer.Citation14–Citation17 Li’s study results showed that LRP1B in HCC serves as a mediator of lipid metabolism in patients with HCC, and its expression level is regulated by HSF1.Citation18 Although Liu’s study pointed out that LRP1B mutation is a new independent prognostic factor in HCC,Citation19,Citation20 it only explains the effect of LRP1B on HCC from the perspective of mutation and does not explain the role of its expression level in HCC and internal mechanism analysis.
Immunotherapy is the most promising new treatment technology for cancer patients. In recent years, with the development of precision medicine, the immunotherapy of HCC has attracted extensive attention. Although CTLA-4 and PD-1, the two most common immune checkpoint inhibitors in tumour immunotherapy, have certain effects on the treatment of HCC, even the efficiency of PD-1 in HCC is only approximately 20% due to the unique immunosuppressive state of the liver.Citation21,Citation22 Therefore, revealing the deeper mechanism of immune checkpoints, developing new immunotherapeutic drugs for HCC and improving the clinical therapeutic effect of HCC are urgent.
The important role of LRP1B in HCC from the perspective of mutation was identified in this study. More important, the mutation types and variable shear forms of LRP1B in HCC were specifically analysed. The association between the expression level and the clinicopathology, immune infiltration, immune checkpoint, drug sensitivity and prediction of underlying mechanisms was comprehensively analysed to identify potential therapeutic targets and provide a theoretical basis for early HCC diagnosis and treatment.
Materials and Methods
Data Collection and Preparation
The mRNA expression data of LRP1B and clinical information, including survival time, survival status, age, sex, tumour grade and pathological stage in 374 patients with HCC and 50 normal cases, were obtained from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The gene mutation and copy-number alterations (CNA) data were obtained from the TCGA database, International Cancer Genome Consortium (ICGC) database (https://dcc.icgc.org/) and the online site cBioportal (http://www.cbioportal.org/). Cases with incomplete information were excluded from the analysis of clinical relevance of the LRP1B gene expression levels. The information of the patients with HCC who were involved in this study is shown in .
Table 1 Clinical Characteristics of HCC Patients in the TCGA Cohort
The Relationship Between Genes Expression and HCC Prognosis
The HCC prognostic difference between the varied expression level groups and betwixt mutation and wild-type patients was evaluated by R package “survival.” The relationship between the two groups of patients with HCC and their survival was assessed through the progression-free interval (PFI), overall survival (OS), disease-specific survival (DSS) and disease-free interval (DFI). Patients were divided into high- and low-expression groups according to the median value of the LRP1B expression level, which served as the cut-off value. The prognostic value of LRP1B expression level and mutation in HCC was analysed by the Kaplan–Meier method.
Immune Correlation Assessment
TIMER is an online comprehensive website that can analyse the expression profile data of tumour samples in TCGA, the association among tumour-infiltrating immune cells with gene expression, gene mutation, somatic cell copy number variation and other data. TIMER 2.0 (http://timer.cistrome.org/) was used to evaluate the correlation between the abundance of immune cell infiltration and the mutation, CNVs, expression level of LRP1B in patients with HCC. The relationship between LRP1B expression and common immune checkpoint inhibitors, such as CD274, PDCD1 and CTLA4, was analysed by GEPIA (http://gepia.cancer-pku.cn/) and TIMER.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was used to assess the distribution trend of genes from a pre-defined Gene Set in a Gene table ordered by correlation with the phenotype to determine their contribution to the phenotype. GSEA software (version 4.1.0) was used to explore the molecular mechanism of LRP1B in HCC. The differences of gene enrichment pathways between the two groups of high and low LRP1B expression in HCC were analysed. The number of gene set permutations was set to 1000 each time. A gene set was regarded as significantly enriched when the P-value was < 0.05.
Construction of Gene Network Regulation Mechanism
MicroRNAs that may have binding sites with LRP1B were obtained from TargetScan and ENCOR websites. miRNAs with significantly negative correlation with LRP1B expression and with differential expression and prognosis in HCC, were screened out by R package “ggpubr.” Screening criteria were Spearman correlation coefficients of > 0.1 and P-value of < 0.05. Similar methods were used to identify lncRNAs that bind the same microRNA competing with LRP1B.
Drug Susceptibility Prediction
The CellMiner database is based on the 60 cancer cells listed by the National Cancer Institute’s Cancer Research Center (NCI). The NCI-60 cell line is the most widely used cancer cell population for cancer drug testing, which involved 22,379 genes and 20,503 compounds.Citation23 The significance of LRP1B gene expression in the prediction of drug sensitivity was analysed by the NCI-60 data through the R package “CellMiner”, “impute” and “limma”.
Cell Culture and Quantitative Real Time Polymerase Chain Reaction (qRT-PCR) Analysis
Human HCC cell lines (HepG2, SMMC-7721 and MHCC97-H) and normal hepatic cell line LO2 were cultured in DMEM medium with 10% foetal bovine serum under the conditions of 37 °C and 5% CO2. Total cell RNA was extracted according to the instructions of RNA-easy isolation reagent (Vazyme). The extracted RNA was used for reverse transcription into cDNA using the HiScript II Q RT SuperMix for qPCR(+gDNA WIper) kit and 04-MR101-02miRNA 1st Strand cDNA Synthesis Kit (by stem-loop) for microRNA (Vazyme). Finally, SYBR Green I chimeric fluorescence method was used for the qPCR reaction, which followed the instructions of ChamQ SYBR qPCR Master Mix kit and miRNA Universal SYBR qPCR Master Mix kit for microRNA (Vazyme). The primer sequences of LRP1B were as follows: forward: TTTCTCCTCGCCTTACTCACT, reverse: CACACAACTGCTGATCTCGGT; RUSC1-AS1 forward: CAGGGTCCCACTATGTTGCT; RUSC1-AS1 reverse: CCATTTTATAGGCGGGGAGT; hsa-miR-215-5p stem-loop: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTCTGTCA; hsa-miR-215-5p forward: ACACTCCAGCTGGGATGACCTATGAATTG, hsa-miR-215-5p reverse: TGGTGTCGTGGAGTCG.
Statistical Analysis
R (version 3.6.3) software was performed for the statistical analysis. Single-factor correlation analysis was performed by Pearson or Spearman correlation analysis. The relationship between LRP1B expression level and OS, DFI, DSS and PFI in patients with HCC was analysed by the Kaplan-Meier survival method. A P-value of < 0.05 was statistically significant.
Results
Mutated Genes in HCC
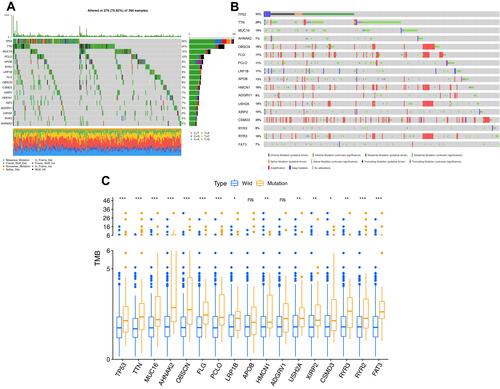
The 17 mutation genes were obtained from the intersection of the top 40 genes with high mutation frequency in HCC obtained from the TCGA and ICGC databases. The intersection genes, such as TP53, TTN, MUC16, AHNAK2, OBSCN, FLG, PCLO, LRP1B, APOB, HMCN1, ADGRV1, USH2A, XIRP2, CSMD3, RYR3, RYR2 and FAT3, were used for the further analysis of the mutation load characteristics. The results ( and ) showed that the top three genes with the highest mutation frequency in HCC were TP53, TTN and MUC16. The average mutation frequency for LRP1B was approximately 10% at a median level and involved depthdeep deletion. The tumour mutation burden (TMB) of mutated LRP1B was significantly higher than that of the wild-type in HCC patients (, P < 0.05).
Figure 1 Mutation results of the 17 intersection genes in HCC. (A) The same frequently mutated genes in both TCGA and ICGC cohorts, (B) Mutations of 17 intersection genes in HCC patients obtained from cBioportal online website, (C) Tumor mutation burden in patients in HCC patients with wild and mutation types. *P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05.
Copy Number Variation Analysis of Mutant Genes
Most genes revealed a higher CNV frequency of gain than that of loss, but LRP1B showed the opposite result with the loss CNV frequency of 10% and the gain CNV frequency of approximately 5% ( and ). The mutation and CNA analysed by cBioportal database () showed consistent results with .
Figure 2 Copy number variation analysis of 17 genes in HCC. (A) Frequency of genes copy number variation in HCC patients, (B) 17-gene mutations and CNA distributions in hepatocellular carcinoma analyzed by cBioportal online website, (C) The location of the mutation genes on the chromosome and the dominant CNVs type. Red indicates that the copy number variation frequency of gain is higher than that of loss with the blue opposite.
Differences in Prognosis Between HCC Patients in Mutation- and Wild-Type
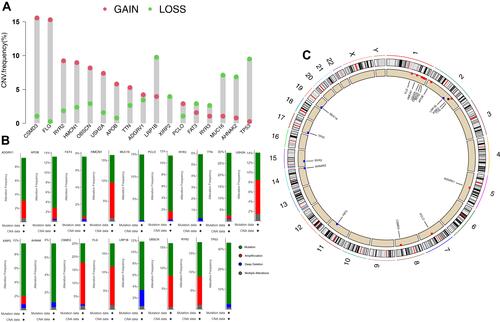
Kaplan-Meier method was further used to analyse the relationship between gene mutation and prognosis of HCC patients. Patients with LRP1B mutation had significantly shorter OS time than those without mutation (, P = 0.027). The relationship between LRP1B expression level and HCC patients’ clinical characteristics was analysed by logistic regression. Results in Supplementary Figure 1 show that no significant difference was found in the expression level of LRP1B among patients with varying ages, genders, grades and stages.
Characteristic Analysis of LRP1B Expression Level in HCC
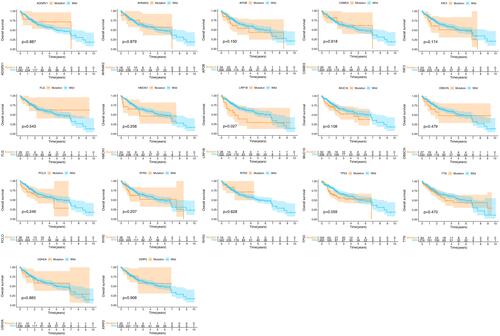
Only the mutation of LRP1B among the 17 genes analysed in the previous analysis is of great value in HCC prognosis, TMB and CNVs. Thus, we continued to analyse the significance of the expression level of LRP1B in HCC. The expression level of LRP1B in HCC tumour patients was significantly higher than that in normal patients in the TCGA database (, P < 0.05). The paired difference analysis of LRP1B expression level in HCC tumour and normal tissue showed that the LRP1B expression level was significantly increased in the tumour (, P < 0.05). HCC patients with down-regulated LRP1B expression had a longer OS time compared with high LRP1B expression patients, but no significant difference was found in DSS, DFI and PFI between the two groups ().
Figure 4 Analysis of the expression level of LRP1B in hepatocellular carcinoma. (A) Expression difference between HCC tumor patients and normal patients, (B) The paired difference analysis of LRP1B expression level in HCC tumor and normal tissue, (C) Effect of LRP1B expression on prognosis of hepatocellular carcinoma. The bottom of the graph showed the patients number in the two groups at different time points.
The Association Between Immune Cell Infiltration and LRP1B Mutation and Copy Number Variation
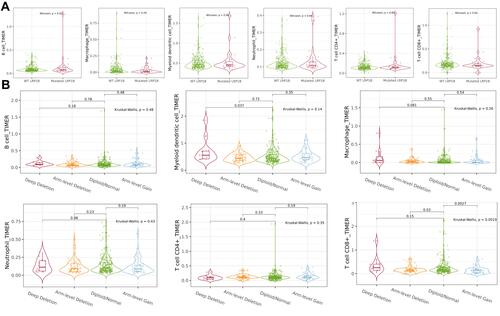
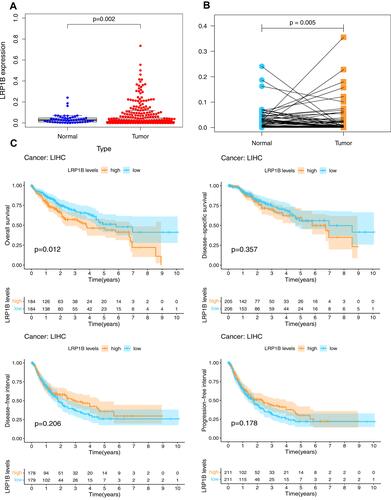
The infiltration of six kinds of immune cells, namely, B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells, was analysed by TIMER online database. The results () showed that no significant correlations existed between LRP1B mutation and immune cell infiltration (all P > 0.05). CNVs were only significantly correlated with CD8+ T cells (P < 0.05), and the content of CD8 + T cells decreased with increasing gene copy number ().
The Association Between LRP1B Expression Level and Immune Cell Infiltration and Immune Checkpoint
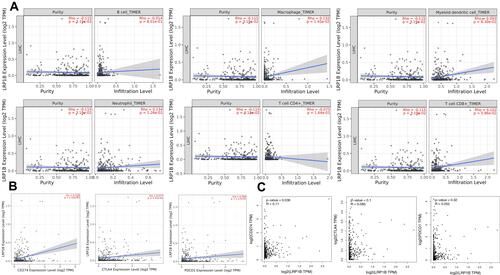
TIMER online database analysis () showed that the LRP1B expression level was positively correlated with the infiltration degree of macrophage (P < 0.05, R = 0.132), myeloid dendritic cell (P < 0.05, R = 0.093), neutrophil (P < 0.05, R = 0.134) and T cell CD8+ (P < 0.05, R = 0.102), whereas it was negatively correlated with B cell (P < 0.05, R = −0.014) and T cell CD4+ (P < 0.05, R = −0.075). The results of TIMER online database analysis showed the expression level of LRP1B and immune cells. The results of TIMER analysis showed that the expressions of CD274, CTLA4 and PDCD1 increased when the expression of LRP1B was significantly up-regulated (, all P < 0.05). However, only the expression level of CD274 was positively correlated with LRP1B according to the results of GEPIA website analysis (, P < 0.05).
Figure 6 Relationship between LRP1B expression level and immunity. (A) The relationship between LRP1B expression level and infiltration degree of 6 kinds of immune cells in HCC. On the left of the graph is a correction of gene expression levels. (B) Correlation analysis between LRP1B expression level and immune checkpoints in HCC by TIMER online database. (C) Correlation analysis between LRP1B expression level and immune checkpoints in HCC by GEPIA online database.
Relationship Between Immune Cell Marker Molecules and the Expression Level of LRP1B
To further clarify the relationship between the expression level of LRP1B and tumour immunity, 21 kinds of common immune molecular markers were further analysed by Spearman correlation analysis. Through the analysis of common immune cell markers (), LRP1B was found to have a significant positive correlation with 10 kinds of immune cell marker genes, namely, ITGAM, VSIG4, ITGAX, MS4A4A, PTGS2, HLA-DPA1, CEACAM8, CD163, HLA-DQB1 and HLA-DRA (all R > 0, P < 0.05). The corresponding immune cell marker genes are shown in below.
Table 2 Correlation Between Immune Cell Marker Molecules and the LRP1B Expression Level
Figure 7 The correlations between LRP1B expression and immune cell marker genes. The strongest correlation with LRP1B expression was found in ITGAM (R = 0.314, P < 0.05). R > 0 and P < 0.05 indicates a significant positive correlation. R < 0 and P < 0.05 indicates a significant negative correlation.
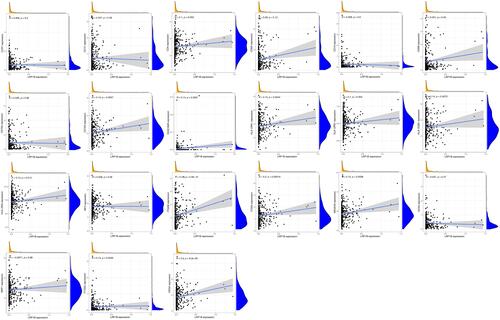
Correlation Between LRP1B Expression Level and Drug Sensitivity
LRP1B expression level was significantly correlated with the sensitivity of 12 kinds of drugs by analysing drug responses in the CellMiner database (). With increasing LRP1B expression, the cell sensitivity to drugs, including Lapatinib (R = −0.26, P = 0.04), Allopurinol (R = −0.37, P = 0.004) and 6-Mercaptopurine (R = −0.29, P = 0.02), decreased.
Figure 8 Correlation analysis of LRP1B expression level with drug sensitivity. The horizontal axis shows gene expression, and the vertical axis shows drug sensitivity. Correlation coefficient R > 0 was considered positive correlation and P < 0.05 was considered significant difference.
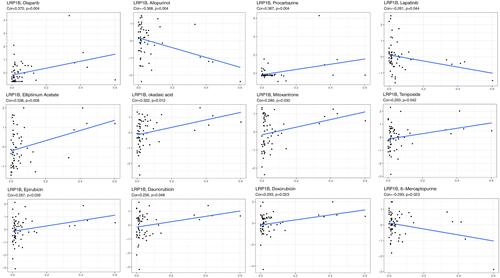
LRP1B expression had significant positive correlations with daunorubicin (R = 0.25, P = 0.047), teniposide (R = 0.26, P = 0.042), epirubicin (R = 0.27, P = 0.038), mitoxantrone (R = 0.28, P = 0.030), doxorubicin (R = 0.29, P = 0.022), okadaic acid (R = 0.32, P = 0.012), elliptinium (R = 0.34, P = 0.008), procarbazine (R = 0.37, P = 0.003) and olaparib (R = 0.37, P = 0.003).
GSEA Enrichment Analysis
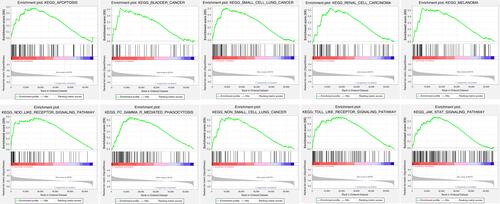
The tumour-related pathways are shown in below through the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the LRP1B single gene. When LRP1B was highly expressed, the following 10 pathways were significantly enriched, including apoptosis, bladder cancer, FcγR mediated phagocytosis, JAK-STAT signalling pathway, melanoma, NOD-like receptor signalling pathway, non-small cell lung cancer, renal cell carcinoma, small cell lung cancer and TOLL-like receptor signalling pathway. All enrichment pathways affected by the different LRP1B expression levels are shown in .
Table 3 All Enrichment Pathways Affected by Different Expression Levels of LRP1B
Construction of Gene Network Regulation Mechanism
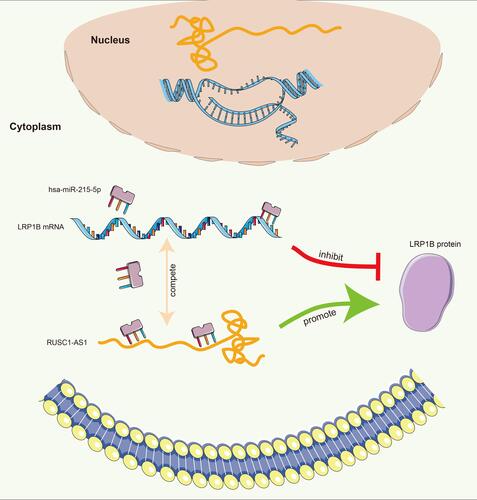
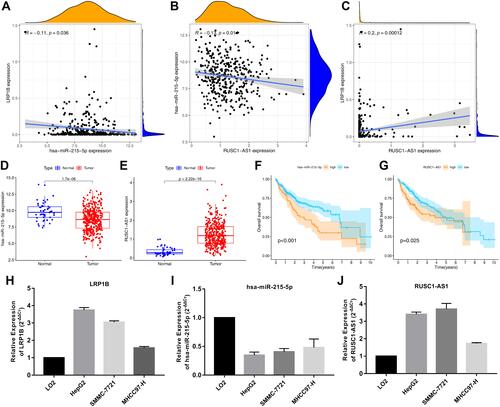
The mechanism of LRP1B promoting tumour progression in HCC was further revealed through bioinformatics methods (). The results of TargetScan and ENCOR online analysis showed that hsa-miR-215-5p had binding sites with LRP1B and was negatively correlated with LRP1B expression level in HCC (). The lncRNA RUSC1-AS1 could combine with hsa-miR-215-5p, which was significantly negatively correlated with hsa-miR-215-5p () and positively correlated with LRP1B (). The hsa-miR-215-5p and RUSC1-AS1 were significantly correlated with the expression and prognosis of patients with HCC (–). The expression levels of LRP1B and RUSC1-AS1 were up-regulated in HCC, whereas the expression levels of hsa-miR-215-5p were significantly reduced in HCC cells, according to the results of the qRT-PCR experiments (–). The molecular interactions are shown in .
Figure 10 Molecular mechanism of LRP1B in hepatocellular carcinoma. (A) Correlation analysis of LRP1B and hsa-miR-215-5p expression in hepatocellular carcinoma, (B) Correlation analysis of hsa-miR-215-5p and RUSC1-AS1 expression, (C) Correlation analysis of LRP1B and RUSC1-AS1 expression in hepatocellular carcinoma, (D and E) Expression difference of hsa-miR-215-5p and RUSC1-AS1 between tumor and normal patients in HCC, (F and G) Prognostic analysis of hsa-miR-215-5p and RUSC1-AS1 in HCC patients. (H–J) The expression levels of LRP1B, hsa-miR-215-5p and RUSC1-AS1 at the cellular level.
Discussion
Tumour-targeted therapy takes the characteristic changes of tumour cells as the target to exert stronger anti-tumour activity while reducing the toxic and side effects on normal cells.Citation24 Targeted therapy for cancer is often used for patients who are too advanced for other treatments; it has sufficient efficacy as a fourth tumour therapy option after surgery and chemoradiotherapy.Citation25,Citation26 Although advances in molecular targeted therapy, immunotherapy and other methods have brought great benefits to clinical patients in recent years, these methods are only effective for tumour patients expressing specific biomarkers; the emergence of resistance to each administered target drug may occur.Citation27,Citation28 This means that more new molecular markers need to be found to improve the diagnostic rate and therapeutic targets of HCC. LRP1B is a tumour suppressor gene in a variety of tumours that is involved in human non-small cell lung cancer, neuroglioma, renal cell cancer and cervical squamous cell carcinoma. These results suggested that LRP1B may be a new target for tumour targeting therapy. In addition to the guiding significance of LRP1B mutation in HCC prognosis,Citation19 some researchers also attempted to screen differentially expressed genes through the LRP1B mutation in HCC and establish a prognostic model to divide patients with HCC into high- and low-risk groups. The two groups showed differences in immune status, including immune cell infiltration rate and checkpoint.Citation20 The specific mechanism of LRP1B on HCC is insufficient, and research works on the immune, prognosis and drug sensitivity of patients with HCC at the level of LRP1B expression are lacking. Therefore, we designed this study to comprehensively analyse the role of LRP1B mutation and expression level in HCC and its potential correlation with tumourigenesis and prognosis of patients with HCC.
Mutations that result in the enhancement of the activity of some genes, thereby affecting cell division and proliferation and leading to out-of-control cell division, occur when DNA is copied as human cells divide and cannot be corrected in time.Citation29,Citation30 Firstly, 17 genes with high mutation frequency in HCC, including LRP1B, were identified by analysing the mutation data of TCGA and ICGC databases. The mutation frequency of LRP1B in HCC was approximately 10%, and the LRP1B mutation was significant associated with poor prognosis in HCC, which indicated the significance of LRP1B gene in the development of HCC. This study showed that LRP1B was based on the gain CNV frequency. Gene copy number variation, a variant form of DNA mutation, is reportedly closely associated with human tumours.Citation31 LRP1B single gene mutation was significantly associated with higher TMB in patients with HCC, thereby adding to the special role of LRP1B mutation in predicting HCC. This comprehensive analysis of the impact of LRP1B gene mutation on HCC prognosis, mutation types, and CNVs supplement and enrich the findings obtained in previous studies. We have enough reason to predict that LRP1B can be used as a new potential therapeutic target for HCC; the growth rate, invasion ability, drug sensitivity and prognosis of HCC cells are affected by tumour heterogeneity caused by somatic mutations, CNVs and epigenetic differences.Citation32,Citation33
Furthermore, the role of LRP1B expression level in HCC was explored in this study. The expression level of LRP1B was significantly different between tumour and normal tissues. The expression level of LRP1B had no significant effect on the length of DSS, DFI and PFI of patients with HCC, possibly due to the fact that patients are treated with certain drugs and surgery to interfere with the natural progression of the disease after HCC diagnosis. However, HCC patients with down-regulated LRP1B expression had a longer OS time compared with patients with high LRP1B expression. The expression level of LRP1B can possibly be used as a prognostic marker for HCC, which is the opposite of the role LRP1B plays in the other tumours.
To better verify whether LRP1B can be used as a new target for HCC targeted therapy, the relationship among LRP1B mutation, expression and tumour immunity was analysed. Although the relationship among LRP1B mutations, CNVs and tumour immune infiltrating cells was not significant, LRP1B expression level was associated with a variety of Immune infiltrating cells, immune cell markers and immune checkpoints. The complexity of the tumourigenesis mechanism shows that the treatment of tumours should be a multi-molecule and multi-mechanism interaction. Previous studies have confirmed that the tumour microenvironment contains a large number of immunosuppressive cells and molecules that can induce T cells to up-regulate the expression of immune checkpoint receptors, leading to the loss of anti-tumour function of T cells and inducing immune escape of tumour cells.Citation34,Citation35 Immune checkpoint, an important effector molecule in the immune microenvironment, plays a key role in the immune regulation of the tumour microenvironment.Citation36 It may be directly involved in HCC occurrence, development and metastasis. Immunosuppressive molecules, including CD274, CTLA4 and PDCD1, have become research hotspots in recent years.Citation37 The results of TIMER analysis showed that the LRP1B expression level was positively correlated with CD274, CTLA4 and PDCD1 expression levels. According to this study, the high expression of LRP1B may up-regulate the expression level of related immunosuppressive molecules, thus promoting the occurrence of the tumour immune escape. This finding indirectly suggested the effectiveness of using LRP1B as a targeted therapeutic molecule for HCC.
LRP1B expression level was also correlated with the sensitivity of many chemotherapeutic drugs. This indicated that LRP1B expression level can also be used to analyse whether patients are suitable for certain chemotherapy drugs in the clinic.
Finally, this study revealed the internal molecular mechanism of LRP1B that is affecting HCC through GSEA analysis and the construction of gene regulatory network. KEGG analysis showed that LRP1B could activate multiple oncogenic pathways and cellular activities, which may explain the poor prognosis in HCC induced by the high expression of LRP1B.
MicroRNAs (miRNAs) are small endogenous non-coding RNA molecules composed of approximately 21–25 nucleotides. These small miRNAs typically target one or more mRNAs and regulate gene expression by inhibiting the target mRNAs at the translation level or by directly degrading the mRNAs.Citation38,Citation39 In recent years, the well-known competitive endogenous RNA (ceRNA) hypothesis holds that endogenous RNA molecules have miRNA action sites and can competitively bind to miRNA to indirectly regulate the expression of miRNA target genes.Citation40–Citation42 This competitive binding miRNA effect is also called miRNA sponges. This study attempted to analyse the regulation mechanism of LPP1B expression level from the perspective of ceRNA. The lncRNA RUSC1-AS1 and LRP1B simultaneously had binding sites with hsa-miR-215-5p. This finding was consistent with the ceRNA regulatory network by analysing the relationships of expression levels and HCC prognosis among these three molecules. The expression levels of LRP1B and RUSC1-AS1 were up-regulated in HCC at the cellular level according to the results of qRT-PCR experiments, whereas the expression levels of hsa-miR-215-5p were significantly reduced, which was in line with the change trend of our ceRNA model. RUSC1-AS1 possibly competitively binds hsa-miR-215-5p with LRP1B to relieve the inhibitory effect of hsa-miR-215-5p on LRP1B mRNA and to up-regulate the expression level of LRP1B in HCC, thereby promoting HCC occurrence and poor prognosis.
There are some deficiencies in our study. Firstly, the subjects of this study were obtained from the TCGA public database. Information on some patients is lacking, such as the specific details of patient’s medication or surgical treatment, which may also affect the prognosis of patients. Secondly, the LRP1B’s expression and its direct role of the progression and metastasis in HCC still need to be verified by experiments. Although this study has some limitations, it provided clues for studying the role of LRP1B in HCC and provided possible therapeutic targets and potential prognostic markers for HCC.
Conclusions
In conclusion, the CNV type of LRP1B is in the form of the loss of the CNVs. The mutation and expression level of LRP1B were correlated with poor prognosis, immune infiltration and sensitivity to chemotherapy drugs in patients with HCC. The possible mechanism underlying LRP1B’s effect on the prognosis and expression of HCC was realised through the RUSC1-AS1/hsa-miR-215-5p/LRP1B axis by bioinformatics prediction. Further experiments are needed to confirm this prediction. This study enriched the understanding on the molecular mechanism of HCC pathogenesis and helped develop new therapeutic strategies and predict prognosis. The results of this study provided theoretical basis and reference ideas for subsequent in vivo, in vitro and clinical studies.
Ethics Approval and Consent to Participate
This article does not contain any studies with human participants or animals performed by any of the authors. These are not applicable.
Acknowledgments
The results of this study are based on the data from TCGA (https://www.cancer.gov/tcga). We thank all the authors who provided the data for this study.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Funding
References
- Zhu RX, Seto WK, Lai CL, Yuen MF. Epidemiology of hepatocellular carcinoma in the Asia-Pacific Region. Gut Liver. 2016;10(3):332–339. doi:10.5009/gnl15257
- Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11(4):317–370.
- Hilmi M, Neuzillet C, Calderaro J, Lafdil F, Pawlotsky J-M, Rousseau B. Angiogenesis and immune checkpoint inhibitors as therapies for hepatocellular carcinoma: current knowledge and future research directions. J Immunother Cancer. 2019;7(1):333. doi:10.1186/s40425-019-0824-5
- Villanueva A, Longo DL. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263
- Haas J, Beer AG, Widschwendter P, et al. LRP1b shows restricted expression in human tissues and binds to several extracellular ligands, including fibrinogen and apoE-carrying lipoproteins. Atherosclerosis. 2011;216(2):342–347. doi:10.1016/j.atherosclerosis.2011.02.030
- Liu CX, Li Y, Obermoeller-Mccormick LM, Schwartz AL, Bu G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J Biol Chem. 2001;276(31):28889–28896. doi:10.1074/jbc.M102727200
- Wang Z, Sun P, Gao C, et al. Down-regulation of LRP1B in colon cancer promoted the growth and migration of cancer cells. Exp Cell Res. 2017;357:1–8.
- Hz A, Lb B. Hypoxia induced microRNA-301b-3p overexpression promotes proliferation, migration and invasion of prostate cancer cells by targeting LRP1B. Exp Mol Pathol. 2019;111:104301. doi:10.1016/j.yexmp.2019.104301
- Beer AG, Zenzmaier C, Schreinlechner M, et al. Expression of a recombinant full-length LRP1B receptor in human non-small cell lung cancer cells confirms the postulated growth-suppressing function of this large LDL receptor family member. Oncotarget. 2016;7(42):68721–68733. doi:10.18632/oncotarget.11897
- Liu CX, Ranganathan S, Robinson S, Strickland DK. Gamma-secretase-mediated release of the low density lipoprotein receptor-related protein 1B intracellular domain suppresses anchorage-independent growth of neuroglioma cells. J Biol Chem. 2007;282(10):7504–7511. doi:10.1074/jbc.M608088200
- Ni S, Hu J, Duan Y, et al. Down expression of LRP1B promotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling in renal cell cancer. Cancer Sci. 2013;104(7):817–825. doi:10.1111/cas.12157
- Liu L, Ren M, Han S, Sun L, Zhu L. Expression level and clinical significance of low-density lipoprotein receptor-related protein 1B gene in cervical squamous cell carcinoma. Int J Clin Exp Pathol. 2018;11(3):1701.
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi:10.1038/nature08822
- Maru Y, Tanaka N, Ohira M, Itami M, Hippo Y, Nagase H. Identification of novel mutations in Japanese ovarian clear cell carcinoma patients using optimized targeted NGS for clinical diagnosis. Gynecol Oncol. 2017;144(2):377. doi:10.1016/j.ygyno.2016.11.045
- Lee S, Lee J, Sim SH, et al. Comprehensive somatic genome alterations of urachal carcinoma. J Med Genet. 2017;54(8):572–578.
- Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol. 2019;10:1113.
- Elgendy M, Fusco JP, Segura V, et al. Identification of mutations associated with acquired resistance to sunitinib in renal cell cancer. Int J Cancer. 2019. doi:10.1002/ijc.32256
- Li M, Hu J, Jin R, et al. Effects of LRP1B regulated by HSF1 on lipid metabolism in hepatocellular carcinoma. J Hepatocell Carcinoma. 2020;7:361–376. doi:10.2147/JHC.S279123
- Liu F, Hou W, Liang J, Zhu L, Luo C. LRP1B mutation: a novel independent prognostic factor and a predictive tumor mutation burden in hepatocellular carcinoma. J Cancer. 2021;12(13):4039–4048. doi:10.7150/jca.53124
- Xu J, Shen X, Zhang B, et al. Development and validation of LRP1B mutation-associated prognostic model for hepatocellular carcinoma. Biosci Rep. 2021;41:9.
- Zhu AX, Finn RS, Edeline J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19(7):940–952. doi:10.1016/S1470-2045(18)30351-6
- El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, Phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492. doi:10.1016/S0140-6736(17)31046-2
- Pommier Y, Reinhold WC, Sunshine M, Varma SH, Kohn KW, Doroshow JH. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Eur J Cancer. 2012;48(Suppl 6):82. doi:10.1016/S0959-8049(12)72066-X
- Ting LY, Tan YJ, Ein OC. Molecular targeted therapy: treating cancer with specificity. Eur J Pharmacol. 2018;834:188–196. doi:10.1016/j.ejphar.2018.07.034
- Qin SK, Ren ZG, Meng ZQ, et al. LBA27A randomized multicentered Phase II study to evaluate SHR-1210 (PD-1 antibody) in subjects with advanced hepatocellular carcinoma (HCC) who failed or intolerable to prior systemic treatment. Ann Oncol. 2018;29(suppl_8):viii719–viii720. doi:10.1093/annonc/mdy424.029
- Contratto M, Wu J. Targeted therapy or immunotherapy? Optimal treatment in hepatocellular carcinoma. World J Gastrointest Oncol. 2018;10(5):108. doi:10.4251/wjgo.v10.i5.108
- Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287.
- Chen DH, Zhang XS. Targeted therapy: resistance and re-sensitization. Chin J Cancer. 2015;11:496–501.
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719. doi:10.1038/nature07943
- Calderaro J, Couchy G, Imbeaud S, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67(4):727–738.
- Hovhannisyan G, Harutyunyan T, Aroutiounian R, Liehr T. DNA copy number variations as markers of mutagenic impact. Int J Mol Sci. 2019;20(19):4723. doi:10.3390/ijms20194723
- Assenov Y, Brocks D, Gerhauser C. Intratumor heterogeneity in epigenetic patterns. Semin Cancer Biol. 2018;51:12–21. doi:10.1016/j.semcancer.2018.01.010
- Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54(5):716–727. doi:10.1016/j.molcel.2014.05.015
- Curdy N, Lanvin O, Laurent C, Fournié J, Franchini DM. Regulatory mechanisms of inhibitory immune checkpoint receptors expression. Trends Cell Biol. 2019;29(10):777–790. doi:10.1016/j.tcb.2019.07.002
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi:10.1038/nrc3239
- Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut J Br Soc Gastroenterol. 2015.;64(5):842–848.
- Xu F, Jin T, Zhu Y, Dai C. Immune checkpoint therapy in liver cancer. J Exp Clin Cancer Res. 2018.;37(1):110.
- Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi:10.1038/nature03315
- Grimson A, Farh KH, Johnston WK, Garrett-Engele P, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi:10.1016/j.molcel.2007.06.017
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
- Qu J, Min L, Wen Z, Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8(10):17110–17116.