Abstract
Background
Lung cancer (LUCA) is one of the most prevalent human malignancies, and the leading cause of cancer-related deaths worldwide. Previous reports have shown that miR-21-5p plays a vital role in development of various tumors. Here, we explored the relationship between miR-21-5p/PIK3R1 axis and prognosis of patients with lung adenocarcinoma (LUAD).
Methods
MiRNAseq data, deposited in The Cancer Genome Atlas (TCGA) database, was downloaded and used to determine patterns of miR-21-5p expression in both LUAD and normal lung tissues. Statistical analyses and data visualization were performed using dbDEMC v3.0 platform, starBase v3.0 database and packages implemented in R software. Next, we employed TargetScan Human, miRDB and DIANA Tools databases to predict miR-21-5p target genes, then analyzed their expression patterns as well as prognostic value in LUAD.
Findings
Most human cancers overexpressed miR-21-5p. Specifically, miR-21-5p was significantly upregulated in LUAD tissues relative to normal lung tissues (P < 0.001), and this high expression was significantly correlated with poor patient prognosis (hazard ratio [HR]=1.45, P = 0.014). PIK3R1 was predicted as a miR-21-5p target gene, and both were negatively correlated (r=−0.218, P < 0.01). Notably, PIK3R1 was significantly downregulated in LUAD, relative to normal lung tissues (P < 0.01), with its overexpression significantly associated with poor prognosis of LUAD patients (HR = 0.62, P = 0.0014).
Conclusion
miR-21-5p is a potential prognostic biomarker for LUAD patients. Moreover, it might be playing a role in LUAD progression by regulating PIK3R1 expression.
Introduction
Lung cancer (LUCA) is one of the most frequently occurring human malignant cancers, and a leading cause of cancer-related deaths worldwide.Citation1 Histologically, LUCA is categorized into two major types, namely small cell lung cancer or non-small cell lung cancer (NSCLC). The latter is the most common histological type, accounting for approximately 85% of all lung carcinoma cases.Citation2 NSCLC is further classified into three specific pathological subtypes, namely LUAD, lung squamous cell carcinoma and large-cell lung carcinoma, of which LUAD accounts for about 40% of all LUCA cases.Citation3–Citation6 Despite significant progresses in development of diagnostic and therapeutic techniques, prognosis of LUAD patients remains poor due to its unapparent symptoms and signs.Citation7,Citation8 Therefore, elucidating the molecular mechanisms underlying LUAD progression is imperative to development of more effective prognostic and treatment therapies for this group of patients.
MicroRNAs (miRNAs), a type of non-coding small RNA molecules encoded by endogenous genes with a length of approximately 19 to 22 nucleotides, have been shown to inhibit gene translation and promote post-transcriptional mRNA degradation by binding to the 3ʹ-untranslated region (UTR) of target mRNAs.Citation9–Citation11 Accumulating evidences have suggested that many miRNAs play a vital role, and serve as oncogenic factors or tumor suppressors, in initiation and progression of cancers.Citation12 For example, miR-21-5p was identified as a distinct oncogenic factor, owing to its over-expression in numerous human cancers, such as prostate cancer, breast cancer (BRCA) and gastric cancer.Citation13–Citation15 Recent reports have demonstrated that miR-21-5p is up-regulated and exerted oncogenic activity in NSCLC.Citation16–Citation18 Furthermore, it has been hypothesized that miR-21-5p may be regulating numerous target genes, thereby causing the unclear biological mechanisms of LUAD. However, the underlying mechanism of this phenomenon remains unclear, necessitating further explorations.
In the present study, we carried used bioinformatics analysis to analyze the correlation between differential expression of miR-21-5p with prognosis of LUAD patients. Next, we predicted miR-21-5p’s target genes and found PIK3R1 to be a key target. Furthermore, we explored the relationship between levels of expression and prognostic value of PIK3R1 in LUAD, relative to adjacent normal lung tissues.
Materials and Methods
Analysis of Differential miR-21-5p Expression in Various Human Cancer
Analysis of differential miR-21-5p expression in various human cancer types was performed using the ggplot2 bioconductor package (version 3.3.3) implemented in R statistical software (version 3.6.3) and dbDEMC v3.0 web platform (https://www.biosino.org/dbDEMC/index).
Determination of Differential miR-21-5p Expression Between LUAD and Normal Tissues
Differential expression of miR-21-5p between LUAD and normal lung tissues was conducted using a MiRNAseq dataset deposited in the TCGA database (https://portal.gdc.cancer.gov/). The data were converted to log2(RPM+1) values and analyzed statistically using the Mann–Whitney U-test. Statistical analysis and data visualization were performed using the starBase v3.0 platform (starbase.sysu.edu.cn/index.php) and the ggplot2 package in R statistical software, respectively.
Correlation Between miR-21-5p Expression and LUAD Prognosis
To investigate the potential relationship between miR-21-5p expression and prognosis of LUAD patients, we employed the starBase v3.0 database and R software. Statistical analyses were conducted the survival package (version 3.2–10) in R and visualization were performed using the survminer package (version 0.4.9) in R. Overall survival (OS) was chosen as the main prognostic parameter. Next, we stratified the patients into high or low expression groups, based on the median value of their levels of miR-21-5p expression, then applied a Cox regression model for comparisons.
Prediction of Target miR-21-5p Genes in LUAD
Target genes of miR-21-5p in LUAD were predicted using TargetScan Human (http://www.targetscan.org/), miRDB (http://mirdb.org/) and DIANA Tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php). To accurately identify and narrow down the range of predicted target genes, data from TargetScan Human, miRDB and DIANA Tools databases were combined, and a Venn diagram constructed using Venny 2.1 online tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html) to visualize the intersection among the three datasets. Several targets, including CREBRF, HGF, LIFR, PIK3R1 and XKR6, were identified in LUAD tissues, and their relationship with miR-21-5p visualized using scatter plots generated using starBase v3.0 project. This allowed selection of target genes with the strongest correlation. Furthermore, we investigated the link between target gene expression levels and survival rates in LUAD.
Analysis of Differential PIK3R1 Expression Between LUAD and Normal Tissues
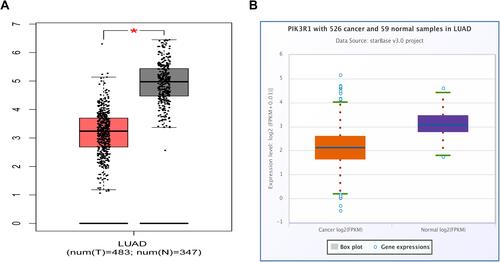
Differential PIK3R1 expression was analyzed in 526 LUAD cases alongside 59 normal samples using the starBase v3.0 project. Data were visualized using plots generated using the GEPIA web platform (http://gepia.cancer-pku.cn/).
Correlation Between Levels of PIK3R1 Expression and Prognosis of LUAD Patients
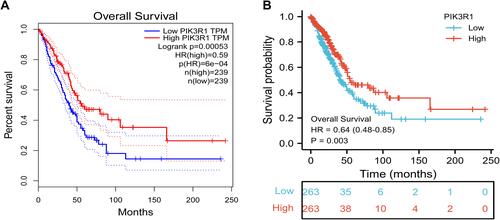
The relationship between levels of PIK3R1 expression and OS rates of LUAD patients was analyzed using Kaplan–Meier (K-M) survival curves, generated using the GEPIA platform and the survminer and survival packages implemented in R software.
Results
MiR-21-5p is Upregulated in LUAD Relative to Normal Lung Tissues
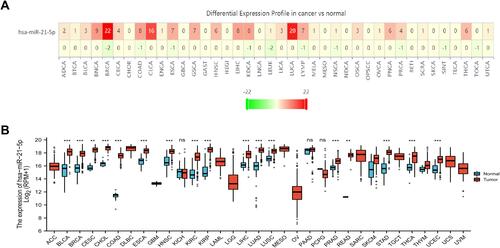
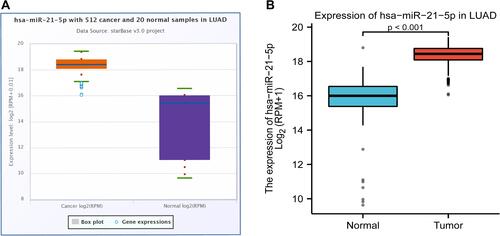
miR-21-5p was significantly up-regulated in most of human malignancies (P < 0.001), including BRCA, colorectal cancer, and LUCA, among others ( and ). Interestingly, miR-21-5p has been confirmed as oncogenes or tumor suppressors in all these tumors, where it plays a regulatory role during initiation and progression of tumors. Analysis of differential expression between LUAD and normal lung tissues revealed that miR-21-5p was significantly upregulated in tumor, relative to adjacent normal tissues (P < 0.001) ( and ).
miR-21-5p Expression is Correlated with Prognosis of LUAD Patients
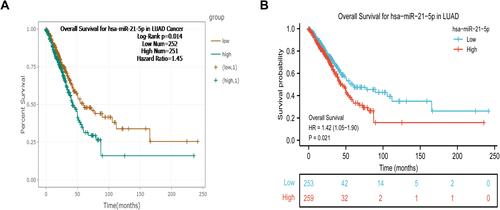
The potential association between levels of miR-21-5p expression and OS rates of 503 LUAD patients was investigated using K-M survival curves ( and ). miR-21-5p was significantly upregulated and downregulated in 251 (49.9%) and 252 (50.1%) cases, respectively. Notably, high miR-21-5p expression was significantly correlated with poor prognosis of LUAD patients (HR = 1.45, P = 0.014), relative to the low miR-21-5p expression group (). Samples from the 512 patients were stratified into high and low expression groups, based on the median value of miR-21-5p expression, with 259 (50.6%) and 253 (49.4%) Cases, respectively (). As expected, we found statistically significant differences in survival rates in patients between both groups, with low miR-21-5p expression significantly correlated with better prognosis (HR = 1.42, P = 0.021, ).
Predicted Target Genes for miR-21-5p in LUAD
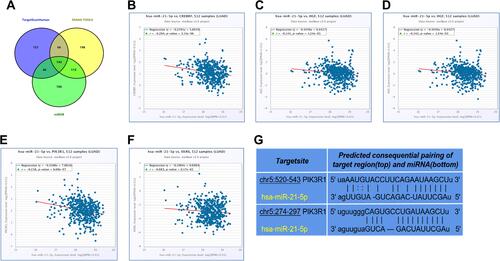
To identify miR-21-5p target genes in LUAD tissues, we extracted hundreds of potential candidates using three different and independent miRNA target prediction platforms, namely TargetScan Human, DIANA Tools and miRDB. A Venn diagram combining datasets from the three platforms revealed 142 overlapping target genes (). A comprehensive analysis of the intersecting target genes consequently revealed five targets, namely CREBRF, HGF, LIFR, PIK3R1 and XKR6. Further analysis revealed that the five targets were negatively correlated with miR-21-5p (–). Among them, PIK3R1 had the strongest correlation to miR-21-5p, as evidenced by a correlation coefficient of −0.218 (P < 0.05) (). In addition, analysis of miRNA-circRNA interactions using Ago CLIP-seq data in ENCORI platform (http://starbase.sysu.edu.cn/agoClipRNA.php?source=circRNA) revealed that the miR-21-5p sequence was complementary to the 3ʹUTR end of PIK3R1 mRNA ().
PIK3R1 is Differentially Expressed in LUAD Relative to Normal Lung Tissues
We generated box-and-whisker plots using GEPIA web platform and starBase v3.0 project to depict differential PIK3R1 expression in LUAD relative to normal lung tissues ( and ). Notably, PIK3R1 was significantly downregulated in LUAD than normal lung tissues (P < 0.01), while the numbers of tumors and normal tissues were 483 and 347, respectively. We performed the same analyses using another database and obtained similar results ().
Correlation Between PIK3R1 Expression and Prognosis of LUAD Patients
Survival curves revealed that differential PIK3R1 expression had a significant effect on OS rates of LUAD patients ( and ). Specifically, high PIK3R1 expression was significantly correlated with high OS rates, while the opposite was true for low expression (HR = 0.59, P = 0.00053), indicating that the high PIK3R1 expression contributes to OS of LUAD patients. Similarly, the results obtained from were consistent with mentioned above information (HR = 0.64, P = 0.003).
Discussion
Until the present time, the incidence and mortality of LUCA dominate a leading position in all human cancers around the world.Citation1,Citation19 Accumulating evidences have demonstrated that many miRNAs play a crucial part in initiation and progression of LUCA. Functionally, some miRNAs have been shown to promote tumorigenesis and progression by repressing expression of tumor-suppressor genes. In fact, some of these miRNAs are considered biomarkers for diagnosis, prognosis and treatment of the disease owing to their significant differential expression in most malignant cancer, relative to adjacent normal tissues. Numerous studies have shown that miR-21-5p, a vital member of miRNA families that is located on human chromosome 17q23.1, is differentially expressed between malignant tumor and adjacent normal tissues. Therefore, miR-21-5p has strongly been associated with proliferation, migration, invasion and apoptosis of tumor cells. For example, Tao et alCitation16 reported that silencing miR-21-5p repressed progression of paclitaxel resistance in BRCA via targeting PDCD4, indicating that it may serve as a therapeutic target in drug-resistant BRCA. In addition, Liu et alCitation20 demonstrated that overexpressing miR-21-5p in gastric cancer cells promoted glycolysis and progression of the disease by downregulating levels of PDHA1 expression. Furthermore, miR-21-5p overexpression in LUAD was associated with disease occurrence and progression through regulation of target genes.Citation21 To date, however, the underlying molecular function of miR-21-5p in LUAD remains unclear.
In the present study, we analyzed patterns of miR-21-5p expression in pan-cancer, by comparing expression levels between cancerous and normal tissues and found that it was significantly upregulated in most human malignancies, and more pronounced in LUAD. Our results further indicated that miR-21-5p expression was significantly correlated with and prognosis of LUAD patients, with its high expression associated with poor survival rates of patients. Furthermore, we predicted downstream targets of miR-21-5p across three different programs and analyzed the resulting intersection. Results revealed five candidate target genes, namely CREBRF, HGF, LIFR, PIK3R1 and XKR6, which were all negatively correlated with miR-21-5p expression. Among them, PIK3R1, a regulatory subunit of the phosphoinositide 3-kinase (PI3K) that participates in biological processes in numerous human malignancies,Citation22 was the most relevant target gene.
Nonetheless, the molecular function and mechanism of PIK3R1 in LUAD has not been fully elucidated. Further analysis revealed that PIK3R1 is downregulated in LUAD patients with this low expression associated with poor OS of patients relative to those in the high expression group.
Conclusion
Taken together, our results suggest that miR-21-5p might lower OS rates of LUAD patients by down-regulating PIK3R1 expression. However, this molecular mechanism needs to be validated using laboratory experiments to ascertain. We next sought to explore the potential biological mechanism of miR-21-5p/PIK3R1 axis in the initiation and development of LUAD.
Abbreviation
BRCA, breast cancer; HR, hazard ratio; K-M, Kaplan–Meier; LUAD, lung adenocarcinoma; LUCA, lung cancer; miRNAs, microRNAs; NSCLC, non-small cell lung cancer; OS, overall survival; PI3K, phosphoinositide 3-kinase; TCGA, The Cancer Genome Atlas; UTR, untranslated region.
Ethical Statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the institutional review board of Fujian Medical University Union Hospital (No.2021KY150), which waived the need for informed consent of the patients.
Author Contributions
All authors contributed to data analysis, drafting and revising the manuscript, gave final approval of the version to be published, agreed to the submitted journal, and agree to be accountable for all aspects of the work.
Acknowledgments
We gratefully acknowledge the support of XianTao platform. We also thank Dr. Jerry for revising the English of this manuscript.
Disclosure
All authors have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics [published correction appears in CA Cancer J Clin. 2011 Mar-Apr;61(2):134]. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
- Chen C, Liu X, Chen C, Chen Q, Dong Y, Hou B. Clinical significance of let-7a-5p and miR-21-5p in patients with breast cancer. Ann Clin Lab Sci. 2019;49(3):302–308.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
- Zarogoulidis K, Zarogoulidis P, Darwiche K, et al. Treatment of non-small cell lung cancer (NSCLC). J Thorac Dis. 2013;5 Suppl 4(Suppl4):S389–S396. doi:10.3978/j.issn.2072-1439.2013.07.10
- Daugaard I, Knudsen A, Kjeldsen TE, Hager H, Hansen LL. The association between miR-34 dysregulation and distant metastases formation in lung adenocarcinoma. Exp Mol Pathol. 2017;102(3):484–491. doi:10.1016/j.yexmp.2017.05.012
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
- Fu HL, Wu DP, Wang XF, et al. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochem Biophys. 2013;67(2):657–668. doi:10.1007/s12013-013-9554-3
- Han Q, Lin X, Zhang X, et al. WWC3 regulates the Wnt and Hippo pathways via Dishevelled proteins and large tumour suppressor 1, to suppress lung cancer invasion and metastasis. J Pathol. 2017;242(4):435–447. doi:10.1002/path.4919
- Yuan B, Yu WY, Dai LS, et al. Expression of microRNA-26b and identification of its target gene EphA2 in pituitary tissues in Yanbian cattle. Mol Med Rep. 2015;12(4):5753–5761. doi:10.3892/mmr.2015.4192
- Hermann A, Wennmann DO, Gromnitza S, et al. WW and C2 domain-containing proteins regulate hepatic cell differentiation and tumorigenesis through the hippo signaling pathway. Hepatology. 2018;67(4):1546–1559. doi:10.1002/hep.29647
- Kao HW, Pan CY, Lai CH, et al. Urine miR-21-5p as a potential non-invasive biomarker for gastric cancer. Oncotarget. 2017;8(34):56389–56397. doi:10.18632/oncotarget.16916
- Tang J, Li X, Cheng T, Wu J. miR-21-5p/SMAD7 axis promotes the progress of lung cancer [published online ahead of print, 2021 Jul 12]. Thorac Cancer. 2021;12(17):2307–2313. doi:10.1111/1759-7714.14060
- Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383(3):280–285. doi:10.1016/j.bbrc.2009.03.077
- Kumar S, Keerthana R, Pazhanimuthu A, Perumal P. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J Biochem Biophys. 2013;50(3):210–214.
- Li BS, Zhao YL, Guo G, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7(7):e41629. doi:10.1371/journal.pone.0041629
- Tao L, Wu YQ, Zhang SP. MiR-21-5p enhances the progression and paclitaxel resistance in drug-resistant breast cancer cell lines by targeting PDCD4. Neoplasma. 2019;66(5):746–755. doi:10.4149/neo_2018_181207N930
- Jin XH, Lu S, Wang AF. Expression and clinical significance of miR-4516 and miR-21-5p in serum of patients with colorectal cancer. BMC Cancer. 2020;20(1):241. doi:10.1186/s12885-020-06715-6
- Wang G, Zhou Y, Chen W, et al. miR-21-5p promotes lung adenocarcinoma cell proliferation, migration and invasion via targeting WWC2. Cancer Biomark. 2020;28(4):549–559. doi:10.3233/CBM-201489
- Merrick DT, Kittelson J, Winterhalder R, et al. Analysis of c-ErbB1/epidermal growth factor receptor and c-ErbB2/HER-2 expression in bronchial dysplasia: evaluation of potential targets for chemoprevention of lung cancer. Clin Cancer Res. 2006;12(7 Pt 1):2281–2288. doi:10.1158/1078-0432.CCR-05-2291
- Liu Z, Yu M, Fei B, Fang X, Ma T, Wang D. miR‑21‑5p targets PDHA1 to regulate glycolysis and cancer progression in gastric cancer. Oncol Rep. 2018;40(5):2955–2963. doi:10.3892/or.2018.6695
- Zhu T, Yu Y, Liu J, Ren K. Identification of a competing endogenous RNA network related to immune signature in lung adenocarcinoma. Front Genet. 2021;12:665555. doi:10.3389/fgene.2021.665555
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi:10.1038/onc.2008.245