Abstract
Background
Lung adenocarcinoma (LUAD) accounts for approximately 40% of all lung cancer cases. The tumour microenvironment (TME) and microRNAs affect the occurrence, metastasis, recurrence and treatment of tumours. However, the role of microRNAs in the TME and LUAD still needs to be further investigated.
Methods
RNA-seq and microRNA-seq data of LUAD and NSCLC samples were downloaded from the TCGA and GEO database. The immune and stromal components in the TME and the abundance of tumour-infiltrating immune cells (TICs) were calculated by the ESTIMATE and CIBERSORT algorithms, respectively. The differentially expressed microRNAs (DEMs) between different StromalScore and ImmuneScore groups were screened out by the edgeR package. Bioinformatics analysis was performed to screen out important DEMs and explore their functional effect.
Results
Our results revealed that a low StromalScore, ImmuneScore and ESTIMATEScore led to poor prognosis of LUAD. Then, 62 DEMs were screened out as downregulated in both the high StromalScore and ImmuneScore groups. Among these DEMs, elevated expression levels of miR-873, miR-105-2 and miR-516a-2 significantly shortened the survival time of LUAD patients. Subsequent analysis revealed that the expression levels of miR-873 and miR-105-2 were increased significantly in tumour tissues. The expression patterns of these 2 microRNAs were confirmed by GSE102286, implying the important roles of these 2 microRNAs in LUAD. Further analysis showed that miR-873 and miR-105-2 were mainly involved in immune-related pathways and that high expression levels of miR-873 and miR-105-2 decreased the abundance of monocytes and resting dendritic cells in the TME.
Conclusion
Although further exploration is still needed, our results revealed that miR-873 and miR-105-2 were closely related to the TME and affected the prognosis of LUAD by altering the abundance of TICs.
Lung cancer, one of the most common malignant tumours, is the leading cause of cancer-related deaths worldwide and causes more than 1.5 million deaths every year.Citation1 Lung adenocarcinoma (LUAD), one of the predominant subtypes of non-small-cell lung cancer (NSCL), accounts for approximately 40% of all lung cancers.Citation2 It usually arises at more distal airways and is now viewed as the most heterogeneous and invasive subtype of lung cancer.Citation3 Although the identification of oncogenic driver mutants and the clinical application of immunotherapy have improved LUAD treatment in recent years, the 5-year survival rate of LUAD is still less than 20%.Citation4 Therefore, the identification of novel biomarkers is extremely important for the early diagnosis and treatment of LUAD.
MicroRNAs are endogenous small noncoding RNAs approximately 20nt long. They are able to regulate the expression of target genes by binding with their 3’ untranslated region (3ʹUTR).Citation5 Although several microRNAs elevate the expression of target genes,Citation6 the vast majority of microRNAs repress the expression of target genes by promoting mRNA degradationCitation7 or inhibiting the translation process.Citation8 By inhibiting the expression of tumour suppressor genes or oncogenes, microRNAs are able to promote or suppress the metastasis and prognosis of lung cancer.Citation9 Moreover, microRNAs play an important role in the TME by regulating the growth and proliferation of immune cells and stromal cells.Citation10
The ESTIMATE algorithm, developed by Yoshihara et al, estimates the relative ratios of stromal and immune components in the tumour microenvironment (TME) by calculating the StromalScore and ImmuneScore.Citation11 The CIBERSORT algorithm is able to calculate the abundance profiles of 22 kinds of immune cells in the TME.Citation12 Mining The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Oncomine and other public databases by these two algorithms, BTK,Citation13 TTC21A,Citation14 JAK1,Citation15 ADAM12 and ERGCitation16 were found to be related to the TME and affected the prognosis of LUAD. These studies revealed that investigating the TME by ESTIMATE and the CIBERSORT algorithm was an effective way to screen out potential prognostic markers and therapeutic targets. However, most of these studies focused on protein-coding genes; there are few reports about the role of microRNAs in the TME of LUAD. Due to the important roles of microRNAs in the TME and LUAD, our study explored microRNAs related to the TME and the prognosis of LUAD by the ESTIMATE and CIBERSORT algorithms.
Materials and Methods
Data Source
Level 3 RNA-seq and microRNA-seq data of 570 LUAD samples, including 57 normal samples and 513 tumour samples, were downloaded from the TCGA database (https://portal.gdc.cancer.gov/, accessed June 30, 2021) along with their corresponding clinical data. GSE102286, containing expression information of microRNAs in 88 pairs of NSCLC and paracancerous tissues, was downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/). This study was approved by the ethics committee of Deyang People’s Hospital.
Calculation of the StromalScore and ImmuneScore
The StromalScore and ImmuneScore were positively correlated with the relative proportions of stromal and immune components in the TME. The ESTIMATEScore, the sum of the StromalScore and ImmuneScore, comprehensively reflects the relative proportion of stromal and immune components in the TME. The StromalScore, ImmuneScore and ESTIMATEScore were calculated by the ESTIMATE package of R language in our study.
Detection of Differentially Expressed microRNAs (DEMs) and Differentially Expressed Genes (DEGs)
LUAD samples were divided into a high-score group and a low-score group according to the median values of the StromalScore and ImmuneScore, respectively. Differential expression analysis between the high- and low-score groups was performed with the edgeR package.Citation17 P < 0.05 and |log2FC| > 1 were used as the criteria to screen out DEMs and DEGs. Heatmaps were generated using the ggplot package of R language.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses
GO and KEGG enrichment analyses of DEGs were performed by the database for annotation, visualization, and integrated discovery (DAVID) (https://david.ncifcrf.gov/home.jsp).Citation18 P < 0.05 was considered statistically significant.
Calculating the Abundance Profile of Tumour-Infiltrating Immune Cells (TICs) in the TME
The CIBERSORT algorithm, which analyses the relative expression levels of genes in each sample according to given gene expression profiles, was applied to estimate the TIC abundance profile in the TME.Citation12 Then, 417 samples with P < 0.05 were selected for subsequent analysis.
Prediction of microRNA Target Genes
Prediction of microRNA targets was performed by miRANDA (http://www.microrna.org/microrna/home.do),Citation19 TargetScan (http://www.targetscan.org/vert_72/)Citation20 and Diana-microT-CDS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index).Citation21 Because miR-873 and miR-105-2 were downregulated in high StromalScore and ImmuneScore patients, upregulated DEGs, which were also predicted to be suppressed by miR-873 and miR-105-2, were screened out as potential targets of miR-873 and miR-105-2.
Statistical Analyses
R language and SPSS 19.0 (IBM, New York, USA) were used for statistical analysis. Differential analysis of transcriptomics data was performed using R language. Qualitative data were analysed by the chi-square test. Difference analyses of normally distributed and nonnormally distributed quantitative data were analysed by t tests and rank-sum tests, respectively. The Pearson and Spearman correlation tests were performed for correlation analyses of normally distributed and nonnormally distributed data, respectively. Kaplan–Meier survival analysis was performed to compare the survival times of different groups. P < 0.05 indicated that the difference was statistically significant.
Results
Relationship Between the StromalScore, ImmuneScore and Clinical Characteristics
A cohort of 513 LUAD patients from TCGA database were included in this study. The StromalScore and ImmuneScore of each patient were calculated by the ESTIMATE algorithm. The relationship between these scores and clinical characteristics was further analysed. Our results revealed that the StromalScore was related to the age (P = 0.001), sex (P = 0.009) and M classification (P = 0.007) of LUAD patients. Meanwhile, the ImmuneScore was associated with the age (P = 0.003), sex (P = 0.007), pathological stage (P = 0.039) and T classification (P = 0.003) of LUAD patients ().
Table 1 The Relationship Between StromalScore, ImmuneScore and Clinical Characteristics of LUAD Patients
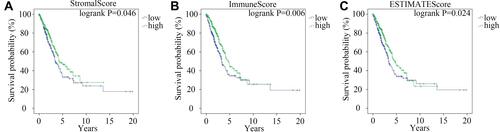
Impact of the StromalScore, ImmuneScore and ESTIMATE Score on the Prognosis of LUAD
Kaplan–Meier survival analysis was performed to explore the impact of the StromalScore, ImmuneScore and ESTIMATEScore on the prognosis of LUAD. Then, a low StromalScore, ImmuneScore and ESTIMATEScore were found to decrease the overall survival time of LUAD patients (–C). These results revealed that the relative proportions of stromal and immune components affected the prognosis of LUAD. Therefore, screening out DEMs and DEGs between the high- and low-score groups was an effective way to identify biomarkers for LUAD.
DEMs and DEGs Between the High- and Low-Score Groups
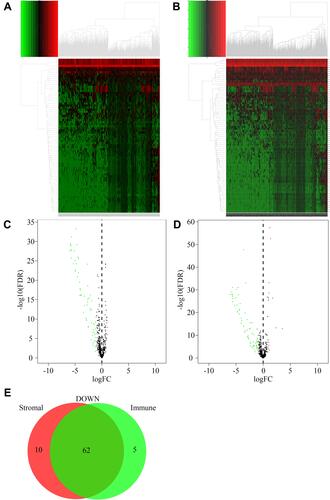
DEMs between the different StromalScore and ImmuneScore groups were screened out. There were 74 microRNAs differentially expressed between the low and high StromalScore groups. Among these microRNAs, 72 and 2 microRNAs were downregulated and upregulated in the high StromalScore group, respectively (). Meanwhile, 78 DEMs, including 67 downregulated and 11 upregulated microRNAs in high ImmuneScore patients, were screened out (). Taken together, 62 microRNAs were downregulated in both high StromalScore and high ImmuneScore patients ().
Figure 2 DEMs between high and low score groups. (A) Heatmap of DEMs between high and low StromalScore groups. Red and green represented upregulated and downregulated microRNAs, respectively. (B) Heatmap of DEMs between high and low ImmuneScore groups. Red and green represented upregulated and downregulated microRNAs, respectively. (C) Volcano plot of DEMs between high and low StromalScore groups. Red and green dots represented up-regulated and down-regulated DEMs, respectively. Black dots represented microRNAs without significantly changed. (D) Volcano plot of DEMs between high and low ImmuneScore groups. Red and green dots represented up-regulated and down-regulated DEMs, respectively. Black dots represented microRNAs without significantly changed. (E) Venn plot of down-regulated DEMs shared by StromalScore and ImmuneScore.
DEGs were also screened out between different StromalScore and ImmuneScore groups. The expression levels of 1947 and 1436 genes were increased and decreased in high StromalScore patients, respectively (). In high ImmuneScore patients, the expression of 1509 and 1684 genes was increased and decreased, respectively (). Taken together, 669 and 946 DEGs were upregulated and downregulated in both high StromalScore and high ImmuneScore patients, respectively (). GO enrichment analysis revealed that these DEGs were mainly involved in biological processes including immune response, regulation of immune response and adaptive immune response (), while KEGG enrichment analysis revealed that these DEGs were mainly involved in signal pathways such as systemic lupus erythematosus, neuroactive ligand–receptor interaction, cytokine–cytokine receptor interaction and alcoholism ().
Figure 3 DEGs between high and low score groups. (A) Heatmap of DEGs between high and low StromalScore groups. Red and green represented genes with high and low expression levels, respectively. (B) Heatmap of DEGs between high and low ImmuneScore groups. Red and green represented genes with high and low expression levels, respectively. (C) Volcano plot of DEGs between high and low StromalScore groups. Red and green dots represented up-regulated and down-regulated DEGs, respectively. Black dots represented genes without significantly changed. (D) Volcano plot of DEGs between high and low ImmuneScore groups. Red and green dots represented up-regulated and down-regulated DEGs, respectively. Black dots represented genes without significantly changed. (E) Venn plot of up-regulated DEGs shared by StromalScore and ImmuneScore. (F) Venn plot of down-regulated DEGs shared by StromalScore and ImmuneScore. (G) GO enrichment result of DEGs shared by StromalScore and ImmuneScore. The size and color of each bubble represented gene number and p value of each term, respectively. (H) KEGG enrichment result of DEGs shared by StromalScore and ImmuneScore. The size and color of each bubble represented gene number and p value of each term, respectively.
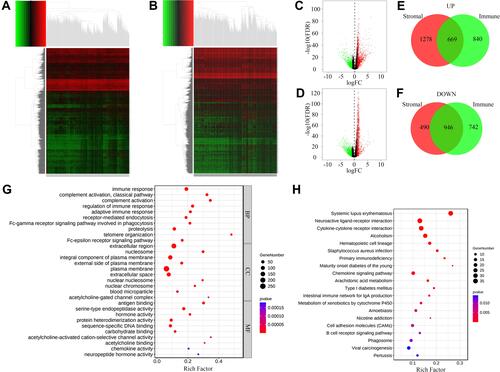
miR-873 and miR-105-2 Were Associated with the Prognosis and Occurrence of LUAD
Survival analysis was performed to explore the effect of DEMs on the prognosis of LUAD. Increased expression levels of miR-873, miR-105-2 and miR-516a-2 significantly shortened the survival time of LUAD patients (). A paired rank-sum test was performed to investigate the expression levels of these microRNAs in paracancerous and LUAD tissues. Among these 3 DEMs, only the expression levels of miR-873 and miR-105-2 increased significantly in tumour tissues (). Therefore, we verified the expression patterns of miR-873 and miR-105-2 using GSE102286. Consistent with the aforementioned results, the expression levels of miR-873 and miR-105 increased significantly in NSCL tissues (). Taken together, these results revealed that miR-873 and miR-105-2 were closely related to the prognosis and occurrence of LUAD.
Figure 4 Survival analysis, difference analysis and functional influence of miR-873 and miR-105-2 on LUAD. Kaplan–Meier survival curve of LUAD patients with high or low expression level of miR-873 (A), miR-105-2 (B) and miR-516a-2 (C). Green and blue lines represent patients with high- and low- expression levels, respectively. Paired rank-sum analysis results of the expression levels of miR-873 (D) and miR-105-2 (E) in LUAD and paracancerous tissues. Red and blue rectangles represented cancerous and paired paracancerous tissues, respectively. Paired rank-sum analysis results of the expression levels of miR-873 (F) and miR-105 (G) in NSCLC and paracancerous tissues. Red and blue rectangles represented cancerous and paired paracancerous tissues, respectively. (H) Predicted targets of miR-873 and miR-105-2. Red rectangles and green triangles represented DEGs upregulated in high score groups and DEMs downregulated in high score groups, respectively. (I) KEGG enrichment result of potential targets of miR-873 and miR-105-2. The size and color of each bubble represented gene number and p value of each term, respectively. (J) GO enrichment result of potential targets of miR-873 and miR-105-2. The size and color of each bubble represented gene number and p value of each term, respectively.
The potential targets of miR-873 and miR-105-2 were predicted to explore their functional effect. Then, 109 and 126 DEGs were predicted to be regulated by miR-873 and miR-105-2, respectively (). KEGG analysis revealed that these target genes were involved in cytokine–cytokine receptor interactions, haematopoietic cell lineages, intestinal immune networks for IgA production, and systemic lupus erythematosus (). Meanwhile, GO enrichment analysis results revealed that these target genes were mainly enriched in immune response, adaptive immune response and inflammatory response (). Interestingly, 17 out of 28 KEGG pathways modulated by miR-873 and miR-105-2, including cytokine–cytokine receptor interaction, haematopoietic cell lineage, intestinal immune network for IgA production and systemic lupus erythematosus, were related to DEGs between different StromalScore and ImmuneScore groups. These results suggested that miR-873 and miR-105 may affect the TME via these pathways.
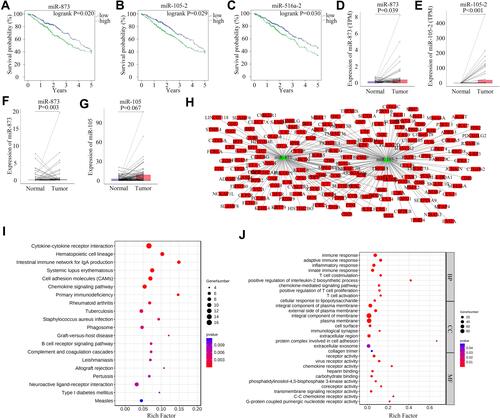
miR-873 and miR-105-2 May Alter the Relative Proportions of TICs
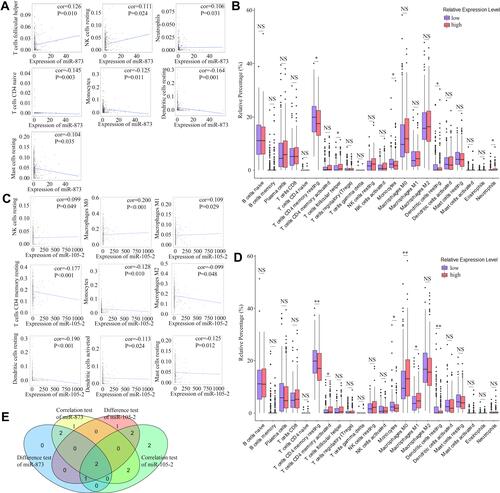
The CIBERSORT algorithm and correlation analysis were used to analyse the relative proportions and the relationships between 22 kinds of TICs in LUAD patients (). Rank-sum tests were performed to compared the relative proportions of 22 kinds of TICs in cancerous and paracancerous tissues. Our results revealed that the relative proportions of most TICs in cancerous and paracancerous tissues were significantly different, implying the important roles of TICs in LUAD (). Then, correlation and difference analyses were performed to explore the effects of miR-873 and miR-105 on TICs. Follicular helper T cells, resting NK cells, neutrophils and CD4 naive T cells, monocytes, resting dendritic cells, and resting mast cells were positively and negatively correlated with the expression of miR-873, respectively (). Meanwhile, high expression of miR-873 elevated and decreased the relative proportions of follicular helper T cells, neutrophils and memory resting CD4 T cells, monocytes, and resting dendritic cells, respectively (). Subsequent analysis revealed that miR-105-2 was positively and negatively correlated with the abundance of resting NK cells, M0 macrophages, M1 macrophages and memory resting CD4 T cells, monocytes, M2 macrophages, resting dendritic cells, activated dendritic cells, and resting mast cells, respectively (). Additionally, a high expression level of miR-105-2 decreased and elevated the relative proportions of resting memory CD4 T cells, monocytes, resting dendritic cells and activated memory CD4 T cells, M0 macrophages, and M1 macrophages, respectively (). Intersection analysis results revealed that elevated expression levels of miR-873 and miR-105-2 decreased the relative proportions of monocytes and resting dendritic cells in the TME (). Taken together, our results implied that miR-873 and miR-105-2 may affect the immune activity of the TME by altering the relative proportions of TICs.
Figure 5 The abundance profile of TICs in LUAD samples and correlation analysis. (A) Bar plot showing the proportion of 22 kinds of TICs in LUAD samples. Each column represented a LUAD sample. Different colors represented different kinds of TICs. (B) Heatmap showing the correlation between 22 kinds of TICs. The color of each box represented the correlation index between two kinds of TICs. (C) Bar plot showing the proportion of 22 kinds of TICs in cancerous and paracancerous tissues. NS, *, ** and *** represented P>0.05, P<0.05, P<0.01 and P<0.001, respectively. Red and blue rectangles represented cancerous and normal tissues, respectively.
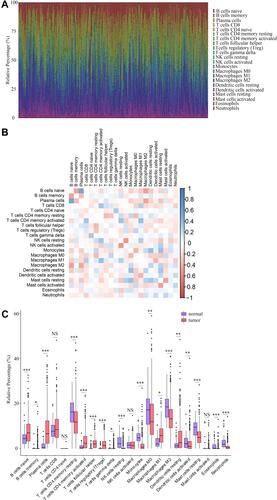
Figure 6 Correlation of proportions of TICs and the expression levels of miR-873 and miR-105-2. (A) Scatter plots indicated the correlation between the expression level of miR-873 and 7 kinds of TICs. The blue line in each plot was fitted linear model indicating the correlation between indicated TIC and the expression level of miR-873. (B) Box plot indicated the influence of high and low expression level of miR-873 on proportions of 22 kinds of TICs. NS and * represented P>0.05 and P<0.05, respectively. (C) Scatter plots indicated the correlation between the expression level of miR-105-2 and 9 kinds of TICs. The blue line in each plot was fitted linear model indicating the correlation between indicated TIC and the expression level of miR-105-2. (D) Box plot indicated the influence of high and low expression level of miR-105-2 on proportions of 22 kinds of TICs. NS, * and ** represented P>0.05, P<0.05 and P<0.01, respectively. (E) Venn plot of TICs correlated with the expression levels of miR-873 and miR-105-2 codetermined by difference and correlation tests.
Discussion
The TME is composed of immune cells, stromal cells, endothelial cells, extracellular matrix, blood vessels and other kinds of cells. Although immunotherapy using immune checkpoint inhibitors such as antibodies against PD-1 and CTLA-4 has improved the prognosis of NSCL in recent years, the efficacy of immunotherapy is affected by the TME.Citation22 Moreover, accumulating evidence has revealed that the TME plays an important role in the occurrence, metastasis, recurrence and treatment of tumours.Citation23 Therefore, investigating TME-related genes will help to further reveal the molecular mechanism of LUAD and identify potential biomarkers.
In our study, the effect of the TME on the prognosis of LUAD was investigated by mining the TCGA database. Consistent with previous reports,Citation13,Citation24,Citation25 our results showed that high values for the StromalScore, ImmuneScore and ESTIMATEScore improved the prognosis of LUAD. However, most of these researches focused on protein-coding genes. Although microRNAs played an important role in the TME, few research explored their function in the TME. Therefore, we explored the effect of microRNAs in the TME of LUAD via bioinformatics analysis. In both the high StromalScore and high ImmuneScore groups, 62 DEMs were screened out to be significantly downregulated. Among these microRNAs, miR-873, miR-105-2 and miR-516a-2 were associated with poor prognosis of LUAD. Moreover, the expression levels of these microRNAs in cancerous and paired paracancerous tissues were verified using a TCGA LUAD dataset and an independent GEO NSCL dataset. Compared with paracancerous tissues, the expression levels of miR-873 and miR-105-2 increased significantly in cancerous tissues. Finally, our bioinformatics analysis results revealed that miR-873 and miR-105-2 may alter the abundance profiles of TICs in the TME and then affect the occurrence and prognosis of LUAD.
MiR-873, located on chromosome 9p21.1, was able to affect the proliferation, invasion and metastasis of many cancers by inhibiting the expression of downstream targets.Citation26 Recent studies revealed that miR-873 was upregulated in LUAD and promoted the proliferation, migration and progression of tumour cells.Citation27,Citation28 Additionally, elevated expression of miR-873 significantly shortened the survival time of LUAD patients.Citation29 MiR-105-2, residing on chromosome Xq28, belongs to the miR-767-105 cluster and leads to poor prognosis of hepatocellular carcinoma.Citation30,Citation31 A high plasma miR-105 level resulted in poor prognosis of NSCL and was a potential biomarker for early diagnosis of NSCL.Citation32 In addition, miR-105 was inhibited by LINC00261 and affected the metastasis and proliferation of NSCLC.Citation33 Consistent with previous reports, our results revealed that miR-873 and miR-105-2 shorten the survival time of LUAD patients by altering the relative proportions of TICs in the TME.
Growing evidence suggests that TICs affect tumour progression and the immunotherapeutic response.Citation34 Therefore, we explored the influence of miR-873 and miR-105-2 on TICs in our study. Our results revealed that miR-873 and miR-105-2 were correlated with the relative proportions of several kinds of TICs. Among these kinds of TICs, we focused on monocytes and resting dendritic cells, which were more closely associated with the expression levels of miR-873 and miR-105-2. Monocytes are a heterogenic population of mononuclear phagocytes that comprise a major population of innate immune cells. By inducing tumour cytotoxicity, suppressing metastasis, engulfing tumour materials and negatively regulating Treg cells, monocytes were able to suppress tumour progression.Citation35 Although the proportions of CD14+ monocytes were similar in tumour and normal tissues, the proportion of CD16+ monocytes decreased significantly in NSCL tumour tissues.Citation36 Dendritic cells are a group of antigen-presenting cells involved in adaptive immune responses. Resting dendritic cells, also known as immature dendritic cells, are primarily localized in peripheral tissues and can capture antigens efficiently. By regulating T-cell activation, resting dendritic cells are mainly involved in maintaining self-tolerance.Citation37 Previous research revealed that immature dendritic cells led to a better prognosis of lung cancer.Citation38 Later studies unveiled that dendritic cells were more likely to remain immature in the TME in NSCLC.Citation39,Citation40 These results implied that monocytes and resting dendritic cells may suppress the progression of lung cancer. Our results revealed that miR-873 and miR-105-2 were negatively correlated with the abundance of monocytes and resting dendritic cells in the TME of LUAD. Taken together, our results implied that miR-873 and miR-105-2 may suppress the progression of NSCLC by decreasing the relative proportions of monocytes and resting dendritic cells.
Conclusion
In conclusion, our results revealed that miR-873 and miR-105-2 were closely related to the TME and may affect the prognosis of LUAD by altering the relative proportions of TICs. Although these findings still need to be further investigated in vitro and in vivo, our results identified 2 potential biomarkers for LUAD.
Data Sharing Statement
The datasets used in our study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Acknowledgments
We are very grateful for TCGA and GEO database.
Disclosure
The authors report no conflicts of interest in this work.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
- Kleczko EK, Kwak JW, Schenk EL, Nemenoff RA. Targeting the complement pathway as a therapeutic strategy in lung cancer. Front Immunol. 2019;10:954. doi:10.3389/fimmu.2019.00954
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi:10.1097/JTO.0b013e318206a221
- Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. doi:10.1016/j.semcancer.2017.11.019
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi:10.1038/nrm3611
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi:10.1126/science.1149460
- Bracken CP, Szubert JM, Mercer TR, et al. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011;39:5658–5668. doi:10.1093/nar/gkr110
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi:10.1016/s0092-8674(04)00045-5
- Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2019;70:3–20. doi:10.1016/j.mam.2018.07.003
- Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–246. doi:10.1158/2159-8290.CD-15-0893
- Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi:10.1038/ncomms3612
- Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi:10.1038/s41587-019-0114-2
- Bi KW, Wei XG, Qin XX, Li B, Has BTK. Potential to be a prognostic factor for lung adenocarcinoma and an indicator for tumor microenvironment remodeling: a study based on TCGA data mining. Front Oncol. 2020;10:424. doi:10.3389/fonc.2020.00424
- Wang W, Ren S, Wang Z, Zhang C, Huang J. Increased expression of TTC21A in lung adenocarcinoma infers favorable prognosis and high immune infiltrating level. Int Immunopharmacol. 2020;78:106077. doi:10.1016/j.intimp.2019.106077
- Cai J, Deng H, Luo L, You L, Liao H, Zheng Y. Decreased expression of JAK1 associated with immune infiltration and poor prognosis in lung adenocarcinoma. Aging. 2020;13:2073–2088. doi:10.18632/aging.202205
- Yue C, Ma H, Zhou Y. Identification of prognostic gene signature associated with microenvironment of lung adenocarcinoma. PeerJ. 2019;7:e8128. doi:10.7717/peerj.8128
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi:10.1093/bioinformatics/btp616
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi:10.1038/nprot.2008.211
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi:10.1371/journal.pbio.0020363
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. doi:10.7554/eLife.05005
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–73. doi:10.1093/nar/gkt393
- He J, Hu Y, Hu M, Li B. Development of PD-1/PD-L1 pathway in tumor immune microenvironment and treatment for non-small cell lung cancer. Sci Rep. 2015;5:13110. doi:10.1038/srep13110
- Lee K, Hwang H, Nam KT. Immune response and the tumor microenvironment: how they communicate to regulate gastric cancer. Gut Liver. 2014;8:131–139. doi:10.5009/gnl.2014.8.2.131
- Wu W, Jia L, Zhang Y, Zhao J, Dong Y, Qiang Y. Exploration of the prognostic signature reflecting tumor microenvironment of lung adenocarcinoma based on immunologically relevant genes. Bioengineered. 2021;12:7417–7431. doi:10.1080/21655979.2021.1974779
- Yang T, Hao L, Cui R, et al. Identification of an immune prognostic 11-gene signature for lung adenocarcinoma. PeerJ. 2021;9:e10749. doi:10.7717/peerj.10749
- Zou Y, Zhong C, Hu Z, Duan S. MiR-873-5p: a potential molecular marker for cancer diagnosis and prognosis. Front Oncol. 2021;11:743701. doi:10.3389/fonc.2021.743701
- Gao Y, Xue Q, Wang D, Du M, Zhang Y, Gao S. miR-873 induces lung adenocarcinoma cell proliferation and migration by targeting SRCIN1. Am J Transl Res. 2015;7:2519–2526.
- Luo J, Zhu H, Jiang H, et al. The effects of aberrant expression of LncRNA DGCR5/miR-873-5p/TUSC3 in lung cancer cell progression. Cancer Med. 2018;7(7):3331–3341. doi:10.1002/cam4.1566
- Zheng R, Mao W, Du Z, Zhang J, Wang M, Hu M. Three differential expression profiles of miRNAs as potential biomarkers for lung adenocarcinoma. Biochem Biophys Res Commun. 2018;507:377–382. doi:10.1016/j.bbrc.2018.11.046
- Liao X, Zhu G, Huang R, et al. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res. 2018;10:787–803. doi:10.2147/CMAR.S161334
- Li W, Kong X, Huang T, Shen L, Wu P, Chen QF. Bioinformatic analysis and in vitro validation of a five-microRNA signature as a prognostic biomarker of hepatocellular carcinoma. Ann Transl Med. 2020;8:1422. doi:10.21037/atm-20-2509
- Dong X, Chang M, Song X, Ding S, Xie L, Song X. Plasma miR-1247-5p, miR-301b-3p and miR-105-5p as potential biomarkers for early diagnosis of non-small cell lung cancer. Thorac Cancer. 2021;12:539–548. doi:10.1111/1759-7714.13800
- Wang Z, Zhang J, Yang B, et al. Long intergenic noncoding RNA 00261 acts as a tumor suppressor in non-small cell lung cancer via regulating miR-105/FHL1 axis. J Cancer. 2019;10:6414–6421. doi:10.7150/jca.32251
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi:10.1038/s41591-018-0014-x
- Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. J Leukoc Biol. 2019;106:309–322. doi:10.1002/JLB.4RI0818-311R
- Lavin Y, Kobayashi S, Leader A, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765 e17. doi:10.1016/j.cell.2017.04.014
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi:10.1038/32588
- Zeid NA, Muller HK. S100 positive dendritic cells in human lung tumors associated with cell differentiation and enhanced survival. Pathology. 1993;25:338–343. doi:10.3109/00313029309090853
- Baleeiro RB, Anselmo LB, Soares FA, et al. High frequency of immature dendritic cells and altered in situ production of interleukin-4 and tumor necrosis factor-alpha in lung cancer. Cancer Immunol Immunother. 2008;57:1335–1345. doi:10.1007/s00262-008-0468-7
- Perrot I, Blanchard D, Freymond N, et al. Dendritic cells infiltrating human non-small cell lung cancer are blocked at immature stage. J Immunol. 2007;178:2763–2769. doi:10.4049/jimmunol.178.5.2763