Abstract
Purpose
Prolyl 4-hydroxylase subunit alpha 3 (P4HA3) has been proven to participate in the occurrence and development of multiple cancers. However, the functional role of P4HA3 in the tumor immune microenvironment (TIME) of colon adenocarcinoma (COAD) and the prognosis of COAD patients has not been clarified. This study aimed to elucidate the immunological role and prognostic value of P4HA3 in COAD.
Methods
P4HA3 expression in COAD tissues was analyzed via experiments and a bioinformatics algorithm. Based on the COAD patients in The Cancer Genome Atlas database, we comprehensively evaluated whether the expression levels of P4HA3 affected clinical prognosis, TIME, and immunotherapy of COAD using the R platforms and several public databases, including GEPIA, TIMER, TISIDB, and TCIA.
Results
The results of the pan-cancer analysis indicated that P4HA3 expression was significantly different in most tumor tissues compared with normal tissues. P4HA3 was overexpressed in COAD tissues, and overexpression of P4HA3 was associated with a worse overall survival and a shorted progression-free interval in COAD patients. The expression of P4HA3 was positively correlated with pathological stage, T stage, N stage, perineural infiltration, and lymphatic infiltration. There were significant correlations of P4HA3 expression levels with immune cell infiltration and their makers, as well as immunomodulators, chemokines, and microsatellite status. Moreover, overexpression of P4HA3 was associated with a lower response rate to immunotherapy in the IMvigor210 cohort.
Conclusion
Overexpression of P4HA3 is closely related to the poor prognosis of COAD patients, and P4HA3 is a potential target for immunotherapy in COAD patients.
Introduction
Colorectal cancer (CRC) is a prevalent malignant tumor of the digestive system and is associated with high degrees of malignancy and mortality. With more than 1.8 million estimated new CRC cases diagnosed and nearly 9 hundred thousand CRC-associated deaths in 2018,Citation1 it remains a huge health challenge and represents a heavy global economic burden. Colon adenocarcinoma (COAD) is the dominant histological subtype of CRC, accounting for more than 90% of cases.Citation2 Surgery, radiotherapy, neoadjuvant therapy, and palliative chemotherapy are the main treatment modalities for CRC.Citation3 Early-stage CRC has a good prognosis, with a 5-year survival rate of more than 90%.Citation4 However, owing to the insidious nature of early symptoms, most CRC patients diagnosed with clear symptoms have already advanced to the middle or late stage of the disease and have missed the chance to undergo surgery. A lack of biomarkers for early diagnosis and prognostic evaluation of COAD patients also contributes to poor prognosis. Moreover, effective treatment options for advanced COAD remain limited. In recent years, immunotherapy for malignant tumors using immune checkpoint blockers has developed rapidly and achieved encouraging results.Citation5–7 Nevertheless, a significant proportion of individuals cannot benefit from immunotherapy owing to a low response rate. Consequently, there is an urgent need for novel practical biomarkers for prognostic evaluation and therapy.
The development of immune checkpoint inhibitors (ICIs) has led to striking clinical improvements in immunotherapy. Programmed death receptor 1 (PD-1) and PD-ligand 1 (PD-L1) inhibitors have achieved remarkable long-term efficacy in the late stages of advanced malignant tumors.Citation8–10 For example, the 5-year overall survival (OS) of advanced lung cancer is generally 20%, whereas it is as high as 40% in patients with high PDL1 expression.Citation11 In COAD, mismatch repair deficient-microsatellite instability-high (dMMR-MSI-H) patients appear to be the group that derive the most advantage from ICIs.Citation7 However, they account for a small proportion compared with mismatch repair proficient-microsatellite instability-low (pMMR-MSI-L) type patients. Therefore, it is necessary to identify more robust and reproducible biomarkers. In addition, as with other treatments, most patients who initially respond to ICIs eventually face resistance. Various mechanisms related to immunotherapy resistance have been studied, and there has been increasing recognition that the tumor immune microenvironment (TIME) has a profound impact on the efficacy of immunotherapy, involving exogenous and endogenous drug resistance pathways. Further understanding of TIME heterogeneity will lay the foundation for further optimization of treatment strategies and stratified immunotherapy.
Proline 4-hydroxylase (P4H) is an enzyme necessary for collagen stability, ensuring that the newly synthesized procollagen strand folds correctly into a triple helix in three dimensions.Citation12 P4H is a tetramer structure composed of 2α and 2β, where the α subunit is the enzyme substrate-binding domain and has P4HA1, P4HA2, and P4HA3 subtypes.Citation12,Citation13 The expression patterns of these subtypes vary in humans. P4HA1 is commonly expressed in most cell types, and P4HA2 is mainly expressed in chondrocytes, osteoblasts, and capillary endothelial cells. By contrast, P4HA3 expression levels are low in human tissues.Citation12,Citation14
As a key enzyme in post-translational processing of collagen, P4HAs is closely related to invasion and metastasis of various solid tumors.Citation13,Citation15 Upregulation of P4HA1 and P4HA2 has been confirmed to be associated with poor prognosis in breast cancer.Citation16,Citation17 Moreover, high expression of P4HA1 promotes invasion and metastasis of lung adenocarcinoma via promoting epithelial–mesenchymal transformation (EMT) and increasing the expression of matrix metalloproteinases.Citation18 Increased expression of P4HA1 in colon cancer can upregulate the expression of HIF1α and activate the Wnt signaling pathway to promote invasion and metastasis.Citation19 In addition, upregulation of P4HA2 has been shown to be associated with aggressive behavior in cervical and prostate cancers.Citation20,Citation21 Notably, overexpression of P4HA3 may also promote tumorigenesis; in recent years, it has gradually emerged that P4HA3 is highly upregulated in gastric cancer and COAD.Citation22,Citation23 For example, Zhou et al demonstrated that P4HA3 promoted the proliferation, migration, invasion and EMT of colon cancer cells by activating the TGF-β/Smad signaling pathway.Citation23 However, there have been relatively few studies on the clinical characteristics of P4HA3 and COAD patients, and the role of P4HA3 in the TIME of COAD remains to be clarified.
Hence, the primary purpose of the present study was to comprehensively evaluate whether expression levels of P4HA3 influence clinical prognosis and the TIME in COAD using multiple publicly available databases including The Cancer Genome Atlas (TCGA), Gene Expression Profiling Interactive Analysis (GEPIA), TIMER, and TISIDB. The molecular regulation of P4HA3 in COAD was further explored by gene set enrichment analysis (GSEA) using the R package clusterProfiler. The results indicated that expression levels of P4HA3 are significantly increased in COAD tissues compared with non-tumor tissues. Moreover, higher expression of P4HA3 predicted higher tumor stage and was associated with perineural invasion, lymphatic invasion, and poor prognosis. In addition, there was a significant relationship between the expression levels of P4HA3 and levels of immune cell infiltration, immunomodulators, chemokines, and microsatellite status in COAD. Finally, the results of this study suggest that high expression of P4HA3 is associated with lower response rates to traditional ICIs, such as anti-PD-L1, in patients of the IMvigor210 immunotherapy cohort. These observations emphasize the vital role of P4HA3 in COAD development and suggest that P4HA3 may play an important part in regulating the TIME and response to immunotherapy in COAD.
Materials and Methods
Patient Selection and Tissue Collection
Paired tumor tissues and cancer-adjacent tissues of 24 patients with COAD diagnosed and confirmed by histology were collected at the First Affiliated Hospital of Guangxi Medical University, Nanning, China, between April 2022 and August 2022. None of the participating patients had undergone chemotherapy or radiotherapy before surgery and patients who had other known tumors were excluded. The clinical characteristics of the 24 COAD patients are presented in . This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (NO.2022-KY-E-(240)), and all patients had provided written informed consent.
Table 1 Clinical Features of 24 Patients with Colon Adenocarcinoma for Quantitative Real-Time Polymerase Chain Reaction (PCR)
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (PCR)
RNAiso Plus solution (9109, Takara), isopropanol, and chloroform were used to extract and purify total RNA from tissues, and a PrimeScript RT Reagent Kit with gDNA Eraser (RR047A, Takara) was then used to reverse transcribe RNA. The expression of the P4HA3 gene was normalized to GAPDH expression. The mRNA expression levels of GAPDH and P4HA3 were determined by amplification with GoTaq qPCR Master Mix (A6002, Promega) and a 7500 instrument. Levels of mRNA were normalized relative to GAPDH. The relative mRNA expression levels of the target genes were calculated by the 2−ΔΔCT method.
The specific primer base sequences were as follows:
P4HA3-F: 5′-GTGGAGCAAGACCTTCCAGC-3′
P4HA3-R: 5′-GTGGAGCAAGACCTTCCAGC-3′
GAPDH-F: 5′-GCACCGTCAAGGCTGAGAAC-3′
GAPDH-R: 5′-TGGTGAAGACGCCAGTGGA-3′.
Acquisition and Bioinformatics Analysis of High-Throughput RNA Sequencing Data
High-throughput sequencing RNA data of 478 COAD patients and matching clinicopathological features were downloaded from TCGA (https://cancer-genome.nih.gov/) through Xiantao academic database (https://www.xiantaozi.com), including 480 COAD tissues and 41 normal tissues from healthy individuals. We obtained the RNA sequencing data in transcripts per million reads format from TCGA and further converted the data to log2 form for comparison of expression among normal, cancer-adjacent normal, and cancer samples. TCGA database is available to the public in accordance with specific guidelines, which ensure that all written informed consent is obtained prior to data collection.
Analysis of Correlation of P4HA3 Expression with Clinicopathological Features and Prognosis of COAD Patients
Clinicopathological characteristics obtained from TCGA included age, gender, body mass index (BMI), stage, T stage, N stage, M stage, perineural invasion and lymphatic invasion. The median expression level of P4HA3 was used as the threshold to stratify COAD patients into high and low P4HA3 expression groups, and the differences in clinicopathological features between the two groups were analyzed. Then, Kaplan–Meier (K–M) curves were constructed to analyze the differences in OS, disease-specific survival (DFS), and progression-free interval (PFI) between the groups, using R packages survival and survminer. The association between P4HA3 and survival outcomes in COAD patients was further validated in GEPIA (http://gepia.cancer-pku.cn/index.html) databases, and receiver operating characteristic (ROC) curves were used to analyze the diagnostic value of P4HA3 in COAD patients via R package pROC.
P4HA3 Correlated Genes Enrichment Analysis
Correlations between gene expression values of P4HA3 versus other genes in COAD were examined by Spearman correlation (two-sided) analysis using the R package stat. P <0.05 was considered to indicate statistical significance. Genes with a correlation coefficient of 0 were excluded, and all genes with significant correlation coefficients were included for further study. Based on the correlation coefficients, the genes were ranked and subjected to GSEA using the R package clusterProfiler. The significance threshold was set at adjusted P <0.05 and false discovery rate <0.25. Finally, the GSEA results were visualized using a bubble chart.
Immunological Role of P4HA3 in COAD
First, we evaluated the infiltration enrichment of 24 common immune cell types using the single-sample GSEA (ssGSEA) method in R package GSVA. The ESTIMATE method in R package ESTIMATE was used to evaluate the differences in stromal score, ESTIMATE score and immune score between the high and low P4HA3 expression groups. Next, we assessed the differences in immune checkpoint expression between the high and low P4HA3 expression groups. The relationships of P4HA3 expression levels with immune cell abundance and levels of corresponding specific markers were further confirmed in TIMER database (http://timer.cistrome.org/) and TISIDB database(http://cis.hku.hk/TISIDB/in-dex.php). Then, we used TISIDB to determine the correlations between P4HA3 expression levels and immune components including lymphocytes, immune modulators, and chemokines.
Analysis the Immunotherapy Efficacy of P4HA3
First, we explored the mutation status of the P4HA3 gene in COAD patients using the cBioPortal database (https://www.cbioportal.org/). Mutation annotation files for COAD were downloaded from TCGA and the tumor mutational burden (TMB) was calculated for each COAD tissue sample. Microsatellite state data of CRC patients, including microsatellite stable (MSS), MSI-L, and MSI-H status, were downloaded from The Cancer Immunome Atlas (TCIA) database (https://tcia.at/home). Subsequently, we analyzed the relationships between P4HA3 gene expression and TMB and microsatellite status. Tumor Immune Dysfunction and Exclusion (TIDE) is a computational framework that simulates the two main mechanisms of tumor immune evasion and predicts the outcome of ICI immunotherapy; this approach can better evaluate the efficacy of ICI therapy compared with TMB, PD-L1 level, and other recognized immunotherapy biomarkers. The TIDE score and T cell dysfunction score were obtained from the TIDE portal (http://tide.dfci.harvard.edu) based on the normalized transcriptome data of the TCGA-COAD data set. Concomitantly, another immunotherapeutic cohort was included to analyze the relationship between P4HA3 gene expression and immunotherapy outcome, consisting of advanced solid tumor patients treated with anti-PD-L1 antibody (IMvigor210, n = 298), whose clinical information was obtained using R package IMvigor210; patients with unclear immunotherapy efficacy were excluded.
Statistical Analysis
The statistical analysis was performed using R-3.6.3 and GraphPad Prism 8. Wilcoxon test was used if the normality test was not satisfied between the two groups; otherwise, t-test was used. K–M curves with log-rank difference test were used to analyze survival differences. The correlations of gene expression and immune cells were determined by Spearman correlation and statistical significance. The diagnostic significance of P4HA3 for COAD was evaluated using ROC curves. P <0.05 was considered to indicate statistical significance.
Results
Overexpression of P4HA3 in COAD
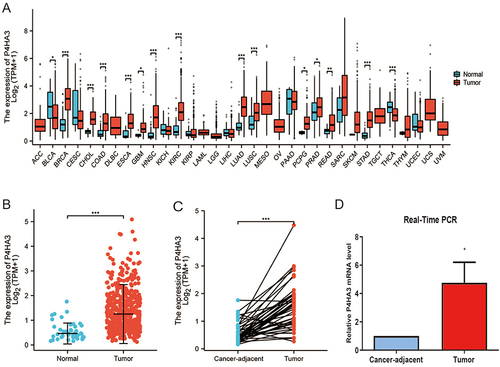
The results of the pan-cancer analysis suggested that P4HA3 was abnormally expressed in most tumors. We observed increased mRNA expression levels of P4HA3 in breast invasive carcinoma, cholangiocarcinoma, esophageal carcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, kidney renal clear cell carcinoma, lung squamous cell carcinoma, pheochromocytoma and paraganglioma, prostate adenocarcinoma, rectum adenocarcinoma, lung adenocarcinoma, stomach adeno-carcinoma, and COAD, whereas a reduced expression was found in a few tumor types including bladder urothelial carcinoma and thyroid carcinoma (P < 0.05) (). The expression levels of P4HA3 were higher in COAD tissues than in colon tissue from healthy individuals (P < 0.05) (). Paired t-test results showed increased expression of P4HA3 in COAD tissues compared with cancer-adjacent tissues (P < 0.05) (). The real-time PCR results further confirmed the increased expression of P4HA3 in COAD ().
Figure 1 Expression of P4HA3 in colon adenocarcinoma (COAD). (A) Expression status of P4HA3 in 33 tumor types versus normal colon tissues of healthy individuals obtained from TCGA pan-cancer data. (B) P4HA3 expression status in normal colon tissues of healthy individuals and COAD tumor tissues from TCGA. (C) Comparison of expression of P4HA3 between COAD tumor tissues and paired cancer-adjacent tissues. (D) Relative P4HA3 mRNA levels in COAD tumor tissues and paired cancer-adjacent tissues as determined by polymerase chain reaction (PCR). ***P<0.001; **P<0.01; *P<0.05.
Correlation Analysis of Clinicopathological Characteristics and Prognosis
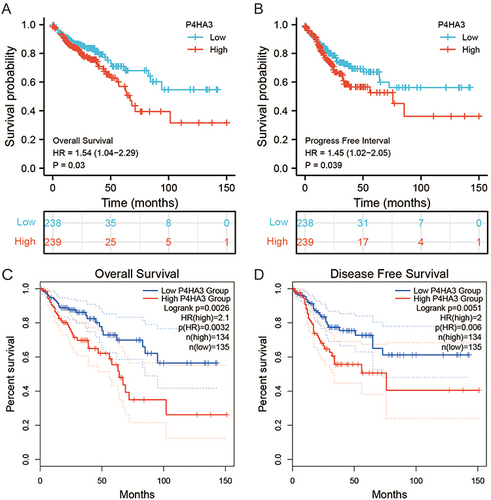
We obtained the clinicopathological characteristics of 478 COAD patients from TCGA, including age, gender, BMI, TNM stage, pathological stage, perineural infiltration, and lymphatic infiltration. The details are presented in . According to the results of the Wilcoxon test, there were statistically significant differences in P4HA3 expression by age, pathological stage, T stage, N stage, perineural infiltration, and lymphatic infiltration (P < 0.05). Expression of P4HA3 was significantly increased in patients aged below 65 years, and those with pathologic stages III and IV, T stages T3 and T4, N stages N1 and N2, perineural invasion, and lymphatic invasion (). There were no significant differences according to BMI, gender, and M stage (P > 0.05) (). The area under ROC curve was 0.837 (), indicating that the ability to discriminate between normal and malignant tissues was moderate. To further analyze the performance of P4HA3 in distinguishing normal individuals from patients with different stages or grades, we performed ROC analysis in patients with each stage or grade; the results all suggested that P4HA3 had moderate predictive performance (). Moreover, with increasing tumor grade, the accuracy of diagnosis showed a trend of improvement. The combined results indicate that P4HA3 could predict the progression of early or late COAD. According to the results of the K–M survival analysis, lower expression of P4HA3 was significantly associated with prolonged OS (HR = 1.54, P = 0.03) and PFI (HR = 1.45, P = 0.039) ( and ), consistent with online analysis using GEPIA ( and ).
Table 2 Correlation Analyzed Between P4HA3 Expression and Clinical Features in Colon Adenocarcinoma Based on TCGA
Figure 2 P4HA3 expression is associated with clinicopathological characteristics in colon adenocarcinoma (COAD) patients. Wilcoxon rank sum test was applied to analyze the association of P4HA3 expression with age (A), pathological stage (B), T stage (C), N stage (D), perineural infiltration (E), lymphatic infiltration (F), M stage (G), gender (H), and body mass index (BMI) (I). TPM, transcripts per million. ***P<0.001; **P<0.01; *P<0.05.
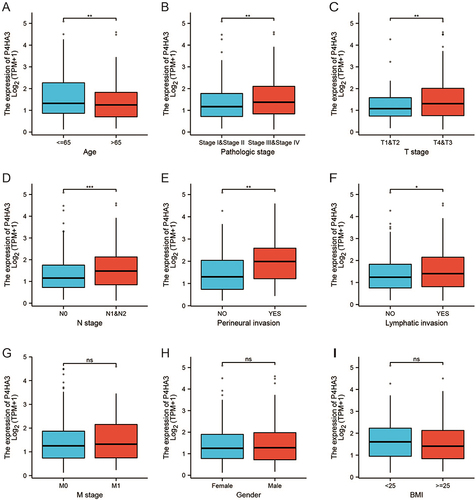
Figure 3 Assessment of the diagnostic performance of P4HA3 by receiver operating characteristic (ROC) curve analysis in colon adenocarcinoma (COAD) patients. ROC curve analysis was used to determine the ability of P4HA3 as a classifier to distinguish normal subjects from COAD patients (A) and COAD patients with different TNM stages (B–J) and pathological stages (K–N).
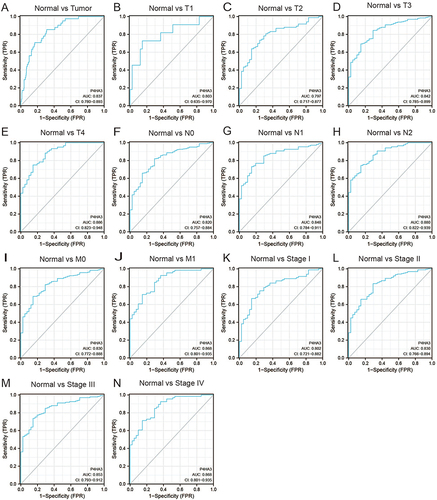
Figure 4 The prognostic value of P4HA3 in patients with colon adenocarcinoma (COAD). (A) Overall survival (OS) and (B) progression-free interval (PFI) of COAD patients based on P4HA3 expression data from TCGA. (C) OS and (D) disease-free survival (DFS) of COAD patients based on P4HA3 expression from GEPIA database.
P4HA3 Correlated Genes Enrichment Analysis
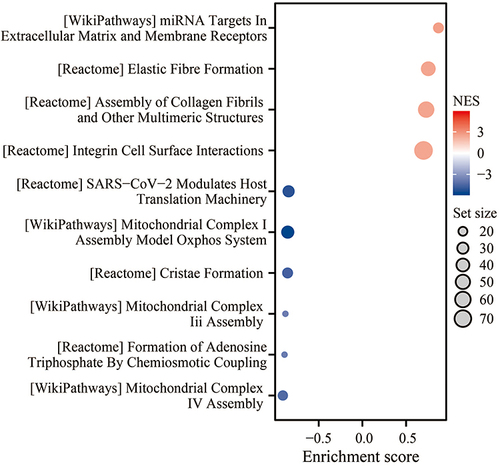
We obtained 12,352 genes related to P4HA3 expression through correlation analysis. The results of the GSEA of the P4HA3 correlated genes shown in and Table S1 indicated that these genes are primarily involved in miRNA targets in extracellular matrix and membrane receptors, elastic fibre formation, assembly of collagen fibrils and other multimeric structures, integrin cell surface interactions, SARS−CoV−2 modulates host translation machinery, mitochondrial complex I assembly model oxidative phosphorylation system, cristae formation, mitochondrial complex iii assembly, formation of adenosine triphosphate by chemiosmotic coupling, mitochondrial complex IV assembly, etc.
Relationship Between P4HA3 and Immune Cell Infiltration in COAD
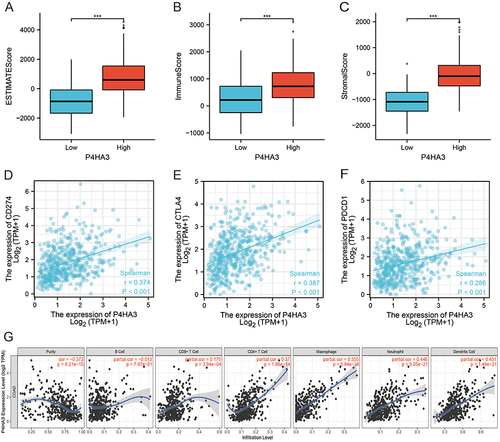
We investigated the abundances of 24 types of immune cells in colon tissues of COAD patients from the TCGA cohort by ssGSEA using the R package GSVA. The results showed that P4HA3 expression was positively correlated with abundances of macrophages, natural killer cells (NK cells), dendritic cells (DC), mast cells, immature DCs, neutrophils, Th1 cells, T effector memory cells, plasmacytoid DCs, regulatory T cells (Tregs), T follicular helper cells, T gamma delta cells, eosinophils, cytotoxic cells, activated DCs, NK CD56dim cells, CD8 T cells, T cells, B cells, T central memory (Tcm) cells, and T helper cells, but negatively correlated with abundance of Th17 cells (P < 0.05) (, Figure S1). R package ESTIMATE enables calculation of scores indicating the degree of immune cell and stromal cell infiltration within the TIME based on the expression levels of specific immune cell and stromal cell genes. The results suggested that COAD samples with high expression of P4HA3 had higher ESTIMATE scores, immune scores, and stromal scores (P < 0.05) (). Moreover, P4HA3 expression in COAD was positively correlated with common immune checkpoints including CD274 (PD-1), CTAL-4, and PDCD1 (P < 0.05) (). Analysis of the TIMER data further verified that P4HA3 was related to immune cell infiltration in COAD, with P4HA3 expression significantly negatively associated with tumor purity (Cor=−0.372, P = 8.21e−15) and significantly positively associated with infiltration of CD8+ T cells (partial cor = 0.175, P = 3.94e−04), CD4+ T cells (partial cor = 0.370, P = 1.86e−14), macrophages (partial cor = 0.555, P = 5.94e−34), neutrophils (partial cor = 0.446, P = 5.05e−21), and DC (partial cor = 0.451, P = 1.49e−21). The results are shown in . In addition, as shown in , correlation analysis of the relationship between expression of P4HA3 and immune cell-specific markers in the TIMER data confirmed our results. However, the association between P4HA3 expression and B cell infiltration was not significant (partial cor=−0.013, p = 7.97e−01).
Table 3 Relationships Among Infiltration Levels of 24 Immune Cell Types and P4HA3 Expression Levels by Spearman’s Analysis
Table 4 Correlation Between P4HA3 and Immune Cell-Specific Markers in Colon Adenocarcinoma
Figure 6 Correlations of tumor immune microenvironment component scores, immune checkpoints, and immune cell infiltration with P4HA3 expression in colon adenocarcinoma (COAD) patients. (A–C) Relationships of tumor immune microenvironment component scores, including ESTIMATE scores, immune scores, and stromal scores, with P4HA3 expression levels by Spearman’s analysis. (D–F) Relationships of immune checkpoints, including CD274, CTAL4, and PDCD1, with P4HA3 expression levels by Spearman’s analysis. (G) Relationships between P4HA3 expression levels and immune cell infiltration levels based on the TIMER database. ***P<0.001.
Relationship Between P4HA3 and Immune Molecules in COAD
To gain a deeper understanding of the role of P4HA3 in the TIME, the relationships between P4HA3 expression levels and immune components including lymphocytes, immune modulators, and chemokines in COAD patients were further explored using TISIDB.
First, to determine which types of tumor-infiltrating lymphocytes (TILs) might be regulated by P4HA3, we studied a study on the relationships between the abundances of various TILs and expression levels of P4HA3. The results showed that P4HA3 expression levels was positively correlated with abundances of macrophages (rho = 0.628), mast cells (rho = 0.580), NK cells (rho = 0.691), NKT cells (rho = 0.646), Tcm CD4 cells (rho = 0.544), and Tregs (rho = 0.624) (all P < 0.05; ).
Figure 7 Relationships of P4HA3 expression with immune components including lymphocytes, immunomodulators, and chemokines in patients with colon adenocarcinoma (COAD) base on the TISIDB database. (A) Relationship between P4HA3 expression levels and lymphocytes. (B–D) Relationships between P4HA3 expression levels and immune-modulators. (E and F) Correlations between P4HA3 expression levels and chemokines.
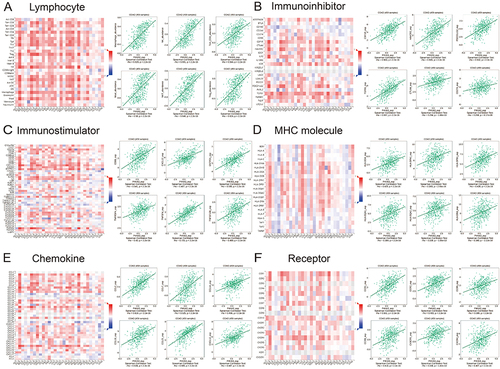
Next, the relationships between P4HA3 expression and levels of three different types of immunomodulators (immunoinhibitors, immunostimulators, and major histocompatibility complex (MHC) molecules) were investigated. P4HA3 expression was significantly positively correlated with levels of immunoinhibitors including CSF1R (rho = 0.563), HAVCR2 (rho = 0.565), PDCD1LG2 (rho = 0.532), TGFB1 (rho = 0.597), CTLA4 (rho = 0.294), and CD274 (rho = 0.268) (). The expression of P4HA3 was also closely associated with immunostimulators including CD86 (rho = 0.541), CXCL12 (rho = 0.487), ENTPD1 (rho = 0.595), TNFRSF4 (rho = 0.430), TNFSF4 (rho = 0.725), and TNFSF13B (rho = 0.489) (). There were also significant positive correlations between P4HA3 expression and levels of MHC including HLA-DOA (rho = 0.428), HLA-DPA1 (rho = 0.369), HLA-DPB1 (rho = 0.436), HLA-DQA1 (rho = 0.384), HLA-DQA2 (rho = 0.339), and HLA-DRA (rho = 0.348) (all P < 0.05; ).
Finally, we considered the relationships of chemokines and receptors with P4HA3 expression. According to the results, P4HA3 expression had significantly positive correlations with chemokines including CCL2 (rho = 0.619), CCL7 (rho = 0.525), CCL14 (rho = 0.466), CCL18 (rho = 0.539), CCL21 (rho = 0.695), and CXCL12 (rho = 0.487) (). P4HA3 expression was also positively correlated with most of the receptors, including CCR1 (rho = 0.551), CCR2 (rho = 0.403), CCR5 (rho = 0.389), CCR8 (rho = 0.418), CX3CR1 (rho = 0.338), and CXCR4 (rho = 0.407) (all P < 0.05; ).
The above results demonstrate that P4HA3 affects various immune molecules in the TIME and influences immune cell infiltration through various pathways.
Analysis of P4HA3 Gene Mutation and Its Relationship with Immunotherapy Response
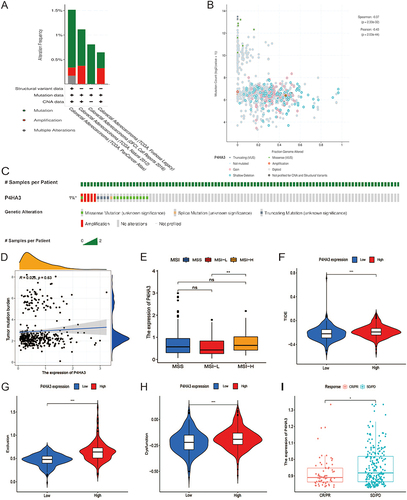
We explored the association between genetic mutation status and P4HA3 expression in patients with CRC using the cBioPortal database. Five in the cBioPortal datasets, including 2129 colorectal adenocarcinoma samples, showed rates of genetic alteration ranging from 0.64% to 1.52% (). The alterations included amplifications, missense mutations, splice mutations, and truncating mutations ( and ). However, further study indicated that P4HA3 expression was not correlated with TMB (P = 0.63, ). Compared with the group with low P4HA3 expression, the high P4HA3 expression group had a higher proportion of MSI-H COAD patients and higher TIDE score (); however, in the IMvigor210 immunotherapy cohort, there was a lower response rate to anti-PD-L1 treatment in the high P4HA3 expression group ().
Figure 8 Analysis of P4HA3 gene mutation and its relationship with immunotherapy. (A) Alteration frequency of P4HA3 in different colon adenocarcinoma (COAD) studies. (B) Methylation level of P4HA3 based on 2129 COAD samples. (C) Percentages of alterations of P4HA3 in COAD. (D) Relationship between P4HA3 and tumor mutational burden (TMB). (E–H) Microsatellite status, TIDE score, and T cell exclusion and dysfunction scores in COAD patients with low and high P4HA3 expression. (I) Distribution of P4HA3 expression in patients with distinct responses to anti-PD-L1 treatment in IMvigor210 immunotherapy cohort. ***P<0.001; **P<0.01; *P<0.05.
Discussion
COAD is the main histological subtype of CRC, accounting for more than 90% of colon cancers.Citation2 Radical resection is the preferred treatment for COAD. However, owing to tumor location and depth of invasion, it is difficult to completely remove cancer cells in this way; postoperative recurrence and metastasis often occur and are the main reasons for failure of surgical treatment.Citation24,Citation25 In recent years, as understanding of the mechanisms of molecular biology and tumor immunology has increased, comprehensive multi-disciplinary and neoadjuvant therapy strategies have improved greatly, effectively improving the survival of patients with COAD.
Immunotherapy effectively improves survival in patients with advanced tumors and has emerged as a promising new treatment option for COAD.Citation5–7,Citation26 The key challenge in immunotherapy is to identify cancer patients who may benefit from immunotherapy. The search for predictive biomarkers will help to maximize the clinical benefits of this treatment. In 2017, the US Food and Drug Administration (FDA) approved immunotherapy for dMMR-MSI-H CRC patients.Citation7 Unfortunately, fewer than a quarter of CRC patients presenting with MSI benefit from immunotherapy, and most of the remainder have MSS. Difference in the TIME have been confirmed to make a substantial contribution to the differences in responses to ICIs between MSI and MSS tumors. A TIME with MSI has higher levels of immune stimulators and chemokines, including IFNG, IL-15, CCL3, CXCL16 and GNLY, than one without.Citation27 Thus, how to induce activation of the TIME, screen populations that may benefit from immunotherapy, and select the best combination therapies are important directions in clinician research.
With the rapid development of transcriptome data analysis, there have been increasing numbers of studies aiming to mine immune genes related to tumor prognosis and those that can be used to predict potential beneficiaries of immunotherapy.Citation28,Citation29 In the present study, we found that overexpression of P4HA3 was associated with poor prognosis in COAD, as well as being involved in the regulation of the TIME.
First, we analyzed the expression of P4HA3 in various cancer tissues and normal tissues using public TCGA data. Compared with normal tissues, P4HA3 was amplified in most tumor tissues. Further analysis showed that overexpression of P4HA3 was also observed in COAD tissues, and that patients with high levels of P4HA3 had worse prognosis. These findings were consistent with previous reports suggesting that P4HA3 may function as an oncogene that promotes the development and progression of COAD.Citation23 Subsequently, we investigated the relationship between P4HA3 and clinical characteristics of COAD patients. High levels of P4HA3 were significantly associated with age, pathologic stage, perineural invasion and lymphatic invasion in COAD patients. There have been few reports on the functional role of P4HA3 in COAD. Here, we found indications that P4HA3 will be an intriguing issue for further examination, not only in terms of epigenetic changes but also with respect to pathogenesis and progression of cancer, as well as the interaction between P4HA3 and TIME.
The available evidence suggests that P4HAs regulate the occurrence and development of tumors in collagen-dependent or non-collagen-dependent ways.Citation13 Collagen can interact with specific downstream molecules to promote EMT and proliferation of cancer cells.Citation20,Citation30 Furthermore, collagen deposition creates an environment that is conducive to tumor growth and invasion. P4HAs can influence the behavior of cancer cells by regulating collagen production. In addition, P4HAs regulate the malignant behavior of tumor cells by altering the stability of hypoxia-inducible factor 1 stability and regulating tumor cell glycolysis.Citation16,Citation31 Nonetheless, very little is known about the non-transcriptional and transcriptional roles of P4HA3 in cancer. In the present study, we found strong correlations of P4HA3 expression levels with the clinical prognosis of COAD patients, as well as associations with the TIME and immune cell infiltration. As a result, we propose that P4HA3 may function as an oncogene in COAD.
Next, we carried out GSEA of the P4HA3 correlated genes indicated that these genes are primarily involved in miRNA targets in extracellular matrix and membrane receptors, elastic fibre formation, assembly of collagen fibrils and other multimeric structures, integrin cell surface interactions, SARS−CoV−2 modulates host translation machinery, mitochondrial complex I assembly model oxidative phosphorylation system, cristae formation, mitochondrial complex iii assembly, formation of adenosine triphosphate by chemiosmotic coupling, mitochondrial complex IV assembly, etc. Our GSEA analysis confirms the functional localization of the P4HA3 gene, suggesting a potential impact on the progression of COAD by influencing the composition of the TIME. Further investigation is necessary to elucidate the precise molecular regulatory mechanisms involved. Additionally, it is critical to explore the relationship between P4HA3 and COAD and investigate its contribution to the development and progression of COAD.
The TIME, which consists of extracellular matrix molecules, stromal cells, immune cells, and cytokines, has a profound impact on tumor growth and proliferation and is increasingly recognized to be involved in the regulation of immunotherapeutic response of tumors.Citation32,Citation33 Tumors that are rich in immune cell infiltration and express neoantigens that activate anti-tumor immune responses are termed hot tumors and tend to respond well to immunotherapy, whereas cold tumors respond poorly to immunotherapy. Evidence suggests that immune checkpoint receptors, such as PD-1 or CTLA-4, are highly expressed in activated CD4 and CD8 T cells and are often activated in the TIME. They are responsible for the suppression of T-cell-mediated immune responses, which have a critical role in tumor control; these findings support the use of checkpoint inhibitors as therapeutic agents.Citation34 In addition, tumor immunotherapy depends on the abundance of tumor blood vessels; normal blood vessels are conducive to the infiltration of immune cells and also enable antibody drugs to enter the tumor to activate immune cells.Citation35 Cytokine therapy in combination with other immunotherapies or chemotherapy has also achieved encouraging results. All current FDA-approved cytokine-targeting immunotherapy drugs for melanoma, renal cell carcinoma, and CRC target vascular endothelial growth factor receptors or epidermal growth factor receptors.Citation36 MSI COADs are hot tumors, and their TIMEs have high levels of immune stimulators and chemokines, including IFNG, IL-15, CCL3, CXCL16 and GNLY, as well as high lymphocyte infiltration. However, there are still many unknown aspects of the regulatory mechanism of the TIME and of immunotherapy in COAD, and further exploration is needed to find more appropriate immunotherapy markers.
In this study, we discovered a robust relationship between P4HA3 expression and the infiltration of diverse immune cell types in the TIME of COAD. Comprehensive analysis of the results of mining of multiple databases showed that P4HA3 expression was significantly positively correlated with infiltration of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DC but not with that of B cells, suggesting heterogeneity in the immune cells recruited in the TIME. High levels of T lymphocyte infiltration in tumor tissue have been reported to indicate better prognosis.Citation37–39 possibly owing to the important role of tumor-specific CD8+ and CD4+ T cells in antitumor activity. Tumor-associated macrophages (TAMs) are also important components of the TIME. They can coordinate with inflammatory mechanisms and play important parts in the promotion of tumor development, invasion, metastasis, immunosuppression, angiogenesis, and drug tolerance by secreting various cytokines and chemokines.Citation40–43 We also found that the P4HA3 expression levels were correlated with levels of lymphocytes, immune modulators, and chemokines, as well as being significantly correlated with expression of CD8A, CD68, CD163, NOS2, IL10, and other specific markers, which further confirmed the relationship between the expression of P4HA3 and immune cells. Increased P4HA3 expression was also positively correlated with ESTIMATE, immunological, and stromal scores. However, little is known about how P4HA3 promotes recruitment of immune cells in COAD, further study is required to elucidate this mechanism. Nonetheless, the results of the present study suggest an important role of P4HA3 the recruitment and regulation of COAD-infiltrating immune cells.
Another major result of our study was evidence of a clear correlation between abnormal P4HA3 expression and cytokines and MHC molecules in COAD patients, which was consistent with the characteristics of the microenvironment in MSI tumor patients. Furthermore, overexpression of P4HA3 was significantly associated with MSI-H COAD, providing further validation that this oncogene is likely to be more critical in the background of the MSI phenotype compared with other phenotypes. Yu et alCitation44 established a model of five genes including P4HA3 in a pan-cancer analysis and confirmed that these five genes were associated with the extracellular matrix and immune invasion score, especially in cold tumors. In addition, our correlation analysis results suggested that P4HA3 expression was positively correlated with levels of commonly used immune checkpoints and TIDE score, and patients with overexpression of P4HA3 in the IMvigor210 immunotherapy cohort had a low response rate to ICIs such as anti-PD-L1 treatments, suggesting that P4HA3 may synergistically regulate immune responses in the TIME with traditional immune checkpoints. These findings open up new possibilities for combination therapies for COAD. For example, combinations of P4HA3 inhibitors with conventional ICI therapy may help to overcome the problem of low response rates to conventional ICI therapy alone. Therefore, the available evidence suggests that P4HA3 in the TIME has vital roles in tumor progression and immunotherapy in COAD patients. However, further studies are needed to confirm these findings.
The above findings reveal the biological function and clinical significance of P4HA3, suggesting that P4HA3 could be used as a potential prognostic indicator and immunotherapy target, and that it may interact with TIME during the progression of COAD. However, the present study had some limitations. First, it was a retrospective study based on bioinformatics analysis, and the results need to be interpreted with caution. Second, analysis results from multiple online databases were integrated in this study, however, the available data were relatively limited, and there was heterogeneity in the results across different databases, which could have caused systematic bias. Third, although the association between P4HA3 and TIME in COAD patients was examined in our study, we lacked direct evidence on the influence of P4HA3 on prognosis and immunotherapy via effects on the TIME. Future research should be used in vivo and in vitro experiments to further explore the molecular mechanism and role of P4HA3 and tumor–immune interactions in COAD.
Conclusion
In conclusion, overexpression of P4HA3 in COAD is closely related to poor prognosis and enhancement of TIME components including stromal cells, immune cells, and immune molecules. P4HA3 may play an important part in regulation of the TIME and could represent a potential reliable biomarker for prognosis and immunotherapy response in COAD patients for use in future clinical studies.
Data Sharing Statement
The datasets used in our study can be found from the TCGA, GEPIA, TIMER, TISIDB, and TCIA database, which are publicly available. The names of the repository/repositories and accession number(s) can be found in the article.
Ethics Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest.
Acknowledgments
We are very grateful for TCGA and Xiantao database.
Additional information
Funding
References
- Tian Y, Kharazmi E, Brenner H, et al. Calculating the starting age for screening in relatives of patients with colorectal cancer based on data from large nationwide data sets. Gastroenterology. 2020;159(1):159–168.e3. doi:10.1053/j.gastro.2020.03.063
- Zhou XG, Huang XL, Liang SY, et al. Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis. Onco Targets Ther. 2018;11:2815–2830. doi:10.2147/OTT.S163891
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. doi:10.3322/caac.21565
- Liu N, Wu C, Jia R, et al. Traditional Chinese medicine combined with chemotherapy and cetuximab or bevacizumab for metastatic colorectal cancer: a randomized, double-blind, placebo-controlled clinical trial. Front Pharmacol. 2020;11:478. doi:10.3389/fphar.2020.00478
- Andre T, Shiu KK, Kim TW, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218. doi:10.1056/NEJMoa2017699
- Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, Phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi:10.1016/s1470-2045(17)30422-9
- Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi:10.1038/s41575-019-0126-x
- Harding JJ, Dika IE, Abou-Alfa GK. Immunotherapy in hepatocellular carcinoma: primed to make a difference? Cancer. 2016;122(3):367–377. doi:10.1002/cncr.29769
- Liu Y, Qiao L, Zhang S, et al. Dual pH-responsive multifunctional nanoparticles for targeted treatment of breast cancer by combining immunotherapy and chemotherapy. Acta Biomater. 2018;66:310–324. doi:10.1016/j.actbio.2017.11.010
- Tannir NM, Signoretti S, Choueiri TK, et al. Efficacy and safety of nivolumab plus ipilimumab versus sunitinib in first-line treatment of patients with advanced sarcomatoid renal cell carcinoma. Clin Cancer Res. 2021;27(1):78–86. doi:10.1158/1078-0432.CCR-20-2063
- Rangachari D, Costa DB. From hope to reality: durable overall survival with immune checkpoint inhibitors for advanced lung cancer. J Clin Oncol. 2019;37(28):2511–2513. doi:10.1200/JCO.19.01207
- Gorres KL, Raines RT. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol. 2010;45(2):106–124. doi:10.3109/10409231003627991
- Shi R, Gao S, Zhang J, et al. Collagen prolyl 4-hydroxylases modify tumor progression. Acta Biochim Biophys Sin. 2021;53(7):805–814. doi:10.1093/abbs/gmab065
- Kukkola L, Hieta R, Kivirikko KI, Myllyharju J. Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. J Biol Chem. 2003;278(48):47685–47693. doi:10.1074/jbc.M306806200
- Zhou T, Erber L, Liu B, Gao Y, Ruan HB, Chen Y. Proteomic analysis reveals diverse proline hydroxylation-mediated oxygen-sensing cellular pathways in cancer cells. Oncotarget. 2016;7(48):79154–79169. doi:10.18632/oncotarget.12632
- Murugesan M, Premkumar K. Systemic multi-omics analysis reveals amplified P4HA1 gene associated with prognostic and hypoxic regulation in breast cancer. Front Genet. 2021;12:632626. doi:10.3389/fgene.2021.632626
- Toss MS, Miligy IM, Gorringe KL, et al. Prolyl-4-hydroxylase alpha subunit 2 (P4HA2) expression is a predictor of poor outcome in breast ductal carcinoma in situ (DCIS). Br J Cancer. 2018;119(12):1518–1526. doi:10.1038/s41416-018-0337-x
- Ning Y, Zheng H, Zhan Y, et al. Overexpression of P4HA1 associates with poor prognosis and promotes cell proliferation and metastasis of lung adenocarcinoma. J Cancer. 2021;12(22):6685–6694. doi:10.7150/jca.63147
- Zhang Q, Yin Y, Zhao H, et al. P4HA1 regulates human colorectal cancer cells through HIF1alpha-mediated Wnt signaling. Oncol Lett. 2021;21(2):145. doi:10.3892/ol.2020.12406
- Cao Y, Han Q, Li J, Jia Y, Zhang R, Shi H. P4HA2 contributes to cervical cancer progression via inducing epithelial-mesenchymal transition. J Cancer. 2020;11(10):2788–2799. doi:10.7150/jca.38401
- Zhu M, Peng R, Liang X, et al. P4HA2-induced prolyl hydroxylation suppresses YAP1-mediated prostate cancer cell migration, invasion, and metastasis. Oncogene. 2021;40(41):6049–6056. doi:10.1038/s41388-021-02000-3
- Song H, Liu L, Song Z, Ren Y, Li C, Huo J. P4HA3 is epigenetically activated by slug in gastric cancer and its deregulation is associated with enhanced metastasis and poor survival. Technol Cancer Res Treat. 2018;17:1533033818796485. doi:10.1177/1533033818796485
- Zhou H, Zou J, Shao C, et al. Prolyl 4-hydroxylase subunit alpha 3 facilitates human colon cancer growth and metastasis through the TGF-beta/Smad signaling pathway. Pathol Res Pract. 2022;230:153749. doi:10.1016/j.prp.2021.153749
- Huang Z, Lai H, Liao J, et al. Upregulation of ADAM12 is associated with a poor survival and immune cell infiltration in colon adenocarcinoma. Front Oncol. 2021;11:729230. doi:10.3389/fonc.2021.729230
- Yang H, Lin HC, Liu H, et al. A 6 lncRNA-based risk score system for predicting the recurrence of colon adenocarcinoma patients. Front Oncol. 2020;10:81. doi:10.3389/fonc.2020.00081
- Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi:10.1200/JCO.2017.76.9901
- Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44(3):698–711. doi:10.1016/j.immuni.2016.02.025
- Dong X, Lv S, Zhang X, Hao R. Upregulation of LAGE3 correlates with prognosis and immune infiltrates in colorectal cancer: a bioinformatic analysis. Int Immunopharmacol. 2020;85:106599. doi:10.1016/j.intimp.2020.106599
- Pan JH, Zhou H, Cooper L, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6. doi:10.3389/fimmu.2019.00006
- Wang T, Wang YX, Dong YQ, Yu YL, Ma K. Prolyl 4-hydroxylase subunit alpha 3 presents a cancer promotive function in head and neck squamous cell carcinoma via regulating epithelial-mesenchymal transition. Arch Oral Biol. 2020;113. doi:10.1016/j.archoralbio.2020.104711
- Li Q, Wang Q, Zhang Q, Zhang J, Zhang J. Collagen prolyl 4-hydroxylase 2 predicts worse prognosis and promotes glycolysis in cervical cancer. Am J Transl Res. 2019;11(11):6938–6951.
- Wang M, Zhao J, Zhang L, et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8(5):761–773. doi:10.7150/jca.17648
- Mersakova S, Lasabova Z, Strnadel J, et al. Genomic profile and immune contexture in colorectal cancer-relevance for prognosis and immunotherapy. Clin Exp Med. 2021;21(2):195–204. doi:10.1007/s10238-020-00649-w
- Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front Immunol. 2020;11:369. doi:10.3389/fimmu.2020.00369
- Huang Y, Kim BYS, Chan CK, Hahn SM, Weissman IL, Jiang W. Improving immune-vascular crosstalk for cancer immunotherapy. Nat Rev Immunol. 2018;18(3):195–203. doi:10.1038/nri.2017.145
- Bess SN, Greening GJ, Muldoon TJ. Efficacy and clinical monitoring strategies for immune checkpoint inhibitors and targeted cytokine immunotherapy for locally advanced and metastatic colorectal cancer. Cytokine Growth Factor Rev. 2019;49:1–9. doi:10.1016/j.cytogfr.2019.10.002
- Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi:10.1038/nrclinonc.2017.101
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi:10.1038/nm.3909
- Wang H, Tian T, Zhang J. Tumor-associated macrophages (TAMs) in colorectal cancer (CRC): from mechanism to therapy and prognosis. Int J Mol Sci. 2021;22(16). doi:10.3390/ijms22168470
- Kim J, Bae JS. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm. 2016;2016:6058147. doi:10.1155/2016/6058147
- Yang Q, Guo N, Zhou Y, Chen J, Wei Q, Han M. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm Sin B. 2020;10(11):2156–2170. doi:10.1016/j.apsb.2020.04.004
- Ge Z, Ding S. The crosstalk between tumor-associated macrophages (TAMs) and tumor cells and the corresponding targeted therapy. Front Oncol. 2020;10:590941. doi:10.3389/fonc.2020.590941
- Yu C, You M, Zhang P, Zhang S, Yin Y, Zhang X. A five-gene signature is a prognostic biomarker in pan-cancer and related with immunologically associated extracellular matrix. Cancer Med. 2021;10(13):4629–4643. doi:10.1002/cam4.3986