Abstract
Introduction
Severe pneumonia progresses rapidly, so early assessment of the severity and prognosis is crucial for reducing mortality rates.
Objective
We explore the role of serum microRNA-24 (miR-24) and microRNA-223 (miR-223) in the prognosis of severe pneumonia.
Methods
There were a total of 96 patients with general pneumonia, 94 patients with severe pneumonia, and 93 healthy people, who were enrolled in this study. The levels of serum miR-24 and miR-223 were detected by real-time fluorescent quantitative PCR in all groups.
Results
The serum miR-223 level in the severe group was higher than that in the common group and the control group, and the miR-24 level was lower than that in the common group and the control group (P<0.05). The serum miR-223 levels and APACHEII scores in the death group were higher than those in the survival group on the first, third, and seventh day after admission, while the miR-24 levels were lower than those in the survival group (P<0.05). The proportion of patients with mechanical ventilation in the death group was higher than that in the survival group (P<0.05). The level of serum miR-24 was negatively correlated with APACHEII score and mechanical ventilation in patients who died of severe pneumonia (P<0.05), and miR-223 was positively correlated with APACHEII score and mechanical ventilation (P<0.05). The AUC predicted by serum miR-24, miR-223, and APACHEII scores alone and jointly were 0.867, 0.839, 0.791, and 0.952, respectively. MiR-24 and miR-223 are protective and independent risk factors for mortality in severe pneumonia patients, respectively (P<0.05). MiR-24 was a protective factor affecting the death of patients with severe pneumonia, and miR-223 was an independent risk factor affecting the death of patients with severe pneumonia (P<0.05).
Conclusion
The combination of serum miR-24 and miR-223 levels on the first day after admission and APACHEII score can effectively predict prognosis.
Introduction
Severe pneumonia is a common critical disease of respiratory system that can involve the functions of multiple organs other than the lung. With a high fatality rate, it is a major disease among patients in intensive care unit.Citation1,Citation2 It is important to implement disease monitoring and assess prognosis early, which helps determine the therapy and plays a key role in saving patients’ lives and improving prognosis. In previous studies, acute physiology and chronic health evaluation scoring system II (APACHE II) was used to evaluate the disease severity of pneumonia patients. Nevertheless, with more involved contents, strong subjectivity, and low evaluation efficiency, it cannot provide effective reference for doctors to grasp the optimal treatment opportunity.Citation3,Citation4 In recent years, studies have shown that microRNA (miRNA) and other biological indexes are closely related to the occurrence and development of pneumonia, which can offer reliable information for pneumonia diagnosis and treatment.Citation5–7 MiR-24 is a miRNA associated with inflammation, lung injury, and pneumonia progression.Citation8 Research by Houshmandfar et al shows that miR-223, as an inflammatory regulator and NLRP3 inflammatory body, participates in the immune inflammatory mechanism of COVID-19.Citation9 Accordingly, this study aims to investigate the expression and clinical significance of miR-24 and miR-223 in severe pneumonia patients.
Data and Methods
General Data
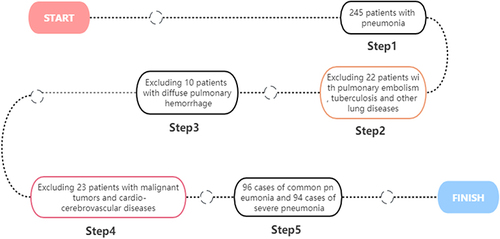
The clinical data of 190 pneumonia patients admitted to our hospital from December 2020 to November 2022 were retrospectively analyzed. According to whether the criterion for severe pneumonia was met, the patients were divided into 96 common pneumonia cases (common group) and 94 severe pneumonia cases (severe group). The common group and the severe group had an age of 18–76 years and 18–78 years, respectively. According to whether patients died 28 days after admission, severe pneumonia patients were divided into survival group (62 cases) and death group (32 cases). In the same period, 93 healthy subjects aged from 18 to 75 years old were selected as the control group. The study was approved by the ethics committee of our hospital (GZSRMYY2019070309) and gained informed consent from the patients and their families. The case collection flow chart is shown in .
Inclusion criteria: ① Pneumonia patients met the diagnostic criteriaCitation10; ② Hospital stay lasted more than 28 d; (3) Patients had complete clinical data, and informed consent was obtained from family members or the patient. Exclusion criteria: ① Patients with severe cardiovascular and cerebrovascular diseases; ② Patients with malignant tumors, abnormal coagulation, or blood system diseases; ③ Patients with diffuse pulmonary hemorrhage; ④ Patients with pulmonary embolism, pulmonary tuberculosis, and other lung diseases.
Research Methods
General Data Collection
Data were collected from severe pneumonia patients, including gender, age, body mass index, smoking history, hypertension history, diabetes history, primary disease, mechanical ventilation, partial pressure of carbon dioxide, APACHE II score.
Determination of Serum miR-24 and miR-223 Levels
A total of 3 mL peripheral venous blood was collected from each group at the time of inclusion (the first, third, and seventh days. after admission) and centrifuged at 3000 r/min at 4°C for 15 min. The upper serum was collected and stored at −80°C in ultra-low temperature refrigerator. Total serum RNA was extracted using miRNA extraction kit (Shanghai Yaji Biotechnology Co., LTD., 51076) and reverse transcription kit (Jiangxi IBIO Biotechnology Co., LTD., IBO1164G). Template cDNA was obtained by reverse transcription. Specific procedures followed the instructions. Quantitative real-time PCR (qRT-PCR) was performed using cDNA as a template. The reaction system was as follows: qPCR SYBR® Green Master Mix (Beijing Bovols Biotechnology Co., LTD., G3325-01) 10 µL, template cDNA (50 ng/µL) 1 µL, upstream and downstream primers (10 µmol/L) 0.8 µL each, ddH2O supplemented to 20 µL. Reaction conditions were as follows: predenaturation at 95°C for 5 min; denaturation at 95°C for 30s, annealing at 58°C for 30s, extension at 72°C for 30s, for a total of 40 cycles. The relative expression levels of serum miR-24 and miR-223 were calculated by 2−∆∆CT method, with primer sequences shown in .
Table 1 Primer Sequence
Statistical Analysis
SPSS 25.0 statistical software was used to analyze and process the data. The measurement data conformed to normal distribution and were expressed as mean ± standard deviation (). Independent sample t-test was used for comparison between the two groups, univariate analysis of variance was used for comparison of multiple groups at the same time point, while repeated measurement analysis of variance was used for comparison of multiple time points. The count data was represented by n (%), and χ2 test was used for comparison between groups. Pearson or Spearman correlation analysis was performed to examine the correlation between serum miR-24 and miR-223 levels and APACHEII scores, and mechanical ventilation in severe pneumonia death cases. Receiver operating curve (ROC) was plotted to test the predictive value of serum miR-24 and miR-223 levels and APACHEII score for prognosis of severe pneumonia patients. Multivariate logistic regression analysis was performed to examine the factors influencing the prognosis of severe pneumonia patients. P < 0.05 indicated statistically significant difference.
Results
Comparison of Serum miR-24 and miR-223 Levels in the Three Groups
The severe group had higher miR-223 level and lower miR-24 level than the normal group and control group (P < 0.05), as shown in .
Table 2 Comparison of Serum miR-24 and miR-223 Levels in the Three Groups ()
Comparison of Serum miR-24 and miR-223 Levels and APACHEII Scores in Severe Pneumonia Patients with Different Prognosis at Different Time Points
The survival group had lower serum miR-223 level and APACHEII score and higher miR-24 level on day 3 and 7 after admission than on day 1 after admission (P < 0.05). The death group had higher serum miR-223 level and APACHEII score and lower miR-24 level on day 3 and 7 after admission than on day 1 after admission (P < 0.05). In addition, the death group had higher serum miR-223 level and APACHEII score and lower miR-24 level than the survival group at all time points (P < 0.05), as shown in .
Table 3 Comparison of Serum miR-24 and miR-223 Levels and APACHEII Scores Between Survival Group and Death Group at Different Time Points ()
Univariate Analysis of Prognosis in Severe Pneumonia Patients
There were no statistically significant differences between the survival group and the death group in terms of gender, age, body mass index, smoking history, hypertension history, diabetes history, primary diseases, and partial pressure of carbon dioxide (P > 0.05). The death group had a higher proportion of mechanical ventilation patients than the survival group (P < 0.05), as shown in .
Table 4 Univariate Analysis of Prognosis in Severe Pneumonia Patients [() /n (%)]
Correlation Between Serum miR-24 and miR-223 Levels and APACHEII Scores, Mechanical Ventilation in Severe Pneumonia Death Cases
Pearson and Spearman correlation analyses revealed that serum miR-24 level was negatively correlated with APACHEII score and mechanical ventilation in severe pneumonia death cases (P < 0.05), while miR-223 was positively correlated with APACHEII score and mechanical ventilation (P < 0.05), as shown in .
Table 5 Correlation Between Serum miR-24 and miR-223 Levels and APACHEII Score, Mechanical Ventilation in Severe Pneumonia Death Cases
Predictive Value of Serum miR-24 and miR-223 Levels and APACHEII Scores for the Prognosis of Severe Pneumonia Patients
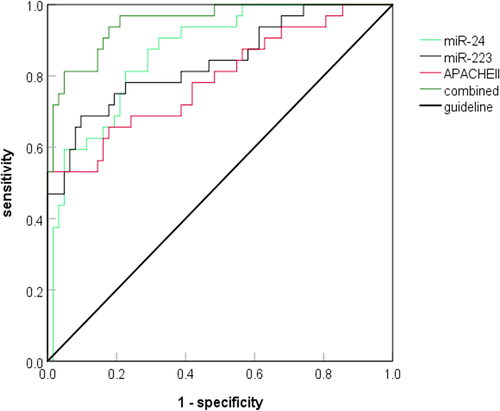
Serum miR-24 and miR-223 levels and APACHEII score on the first day after admission were used as test variables, and whether severe pneumonia patients died was used as state variable to draw the ROC curve. The results showed that, separately, combined predicted AUC of serum miR-24, miR-223, and APACHEII scores were 0.867, 0.839, 0.791, and 0.952, respectively, with combined predicted AUC significantly higher than separately predicted AUC (Z=1.998, 2.235, 2.871, P < 0.05), as shown in and .
Table 6 Predictive Value of Serum miR-24 and miR-223 Levels and APACHEII Scores for the Prognosis of Severe Pneumonia Patients
Multivariate Logistic Regression Analysis on Factors Affecting the Prognosis of Severe Pneumonia Patients
Whether severe pneumonia patients died was used as the dependent variable, statistically significant miR-24, miR-223, APACHEII score, and mechanical ventilation in the above univariate analysis were used as independent variables to perform multivariate logistic regression analysis. The results showed that miR-24 was a protective factor affecting death of severe pneumonia patients, while miR-223 was an independent risk factor in death of severe pneumonia patients (P < 0.05), as shown in .
Table 7 Multivariate Logistic Regression Analysis on Factors Affecting Prognosis of Severe Pneumonia Patients
Discussion
Pneumonia is a pulmonary inflammatory respiratory disease mainly due to pathogenic microbial infection, immune system damage, drugs, allergies, and so on.Citation11 For common pneumonia mainly involving the pulmonary respiratory system, good therapeutic effects are possible through clinical anti-inflammatory programs. Nonetheless, the prevalence rate of severe pneumonia tends to grow due to aggravating aging population, antibacterial drug abuse, and so on.Citation12 Inflammatory cytokines infiltrate the lungs in severe pneumonia patients, thereby causing endothelial cell dysfunction and greater pulmonary capillary permeability, which in turn causes pulmonary vascular microthrombus and acts as the main factor in respiratory failure and acute respiratory distress syndrome.Citation13 Hence, it is particularly important to choose positive and correct diagnosis and treatment methods in clinical practice.
miRNA, a class of highly conserved endogenous non-coding single-stranded RNA with a length of 21–25 nt, can mediate gene silencing after transcription.Citation14 miRNA takes part in many biological processes of metabolism, cell differentiation, apoptosis, and disease control, involving multiple systems, including respiratory system, cardiovascular system, nervous system, and hematopoietic system.Citation15 Excessive studies have confirmed the correlation of miRNA with the growth and development of lung, as well as onset, development, and outcome of pulmonary fibrosis, pneumonia, lung cancer, chronic obstructive pulmonary disease, and so on.Citation16,Citation17 Lin et alCitation8 demonstrated that miR-24 inhibited inflammatory response in neonatal rats with lipopolysaccharide-induced acute lung injury by targeting NLRP3. Ren et alCitation18 found that miR-24-3p exhibited down-regulated expression in patients with acute lung injury complications caused by severe COVID-19. This study found that severe pneumonia patients had lower serum miR-24 level than common pneumonia patients and healthy people. Studies have shown that severe pneumonia patients are in a state of systemic inflammatory stress with unbalanced anti-inflammatory and pro-inflammatory effects, and inflammatory response mediated by inflammatory mediators is involved in disease progression.Citation19 From previous studies, it is speculated that pneumonia patients infected with pathogenic microorganisms have unbalanced anti-inflammatory and pro-inflammatory effects, and a variety of pathological factors lead to lower serum miR-24 level, so it is unable to effectively inhibit the inflammatory response in acute lung injury, which in turn aggravates patients’ disease.
miR-223 is a hematopoietic cell-derived miRNA involved in the regulation of monocyte-macrophage differentiation, neutrophil recruitment, and pro-inflammatory response.Citation20 Zhang et alCitation21 found that miR-223 secretion was significantly enhanced in the pathogenesis of pulmonary macrophage-mediated inflammatory response, which could be detected in bronchoalveolar fluid and serum. In this study, serum miR-223 was highly expressed in severe pneumonia patients, suggesting that miR-223 may also play a pro-inflammatory role in the progression of severe pneumonia. miR-223 acts as a pro-inflammatory factor in lung infiltration, and inflammatory cascade response mediates endothelial cell dysfunction, as well as disease progression in patients. In addition, this study showed that both miR-24 and miR-223 were factors influencing death in severe pneumonia patients, whose detection levels could be used to predict death. Where, miR-223 has a specificity of up to 90.3%. In clinical practice, miR-24 and miR-223 can be used as auxiliary biological indexes for the evaluation of severe pneumonia to increase accuracy in prognosis assessment.
Conclusions
To sum up, the low expression of serum miR-24 and high expression of miR-223 in severe pneumonia patients are correlated factors affecting the prognosis of severe pneumonia patients, which can effectively predict the prognosis and should be clinically popularized. The changes of miR-24 and miR-223 levels can reflect the progression of pneumonia and predict the prognosis. In clinical practice, by closely monitoring the two levels and taking timely targeted measures, it is possible to improve the physical function of patients and therefore reduce mortality. However, the sample size of this study is small, whether miR-24 and miR-223 can serve as prognostic biological indexes of severe pneumonia waits to be confirmed in future large-sample clinical studies.
Research Involving Human Participants
This retrospective study involving human participants was in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by Ganzhou People’s Hospital’s ethics review board.
Consent for Publication
All authors give consent for publication.
Disclosure
The authors declare no conflicts of interest in this work.
Additional information
Funding
References
- Guo K, Cai W, Chen Y, Shi Y, Xu Z, Chen C. Skeletal muscle depletion predicts death in severe community-acquired pneumonia patients entering ICU. Heart Lung. 2022;52:71–75. doi:10.1016/j.hrtlng.2021.11.013
- Stafylaki D, Maraki S, Vaporidi K, et al. Impact of molecular syndromic diagnosis of severe pneumonia in the management of critically ill patients. Microbiol Spectr. 2022;10(5):e0161622. doi:10.1128/spectrum.01616-22
- Eldaboosy S, Almoosa Z, Saad M, et al. Comparison between physiological scores SIPF, CURB-65, and APACHE II as predictors of prognosis and mortality in hospitalized patients with COVID-19 pneumonia: a multicenter study, Saudi Arabia. Infect Drug Resist. 2022;15:7619–7630. doi:10.2147/IDR.S395095
- Wei Q, Chen X, Chen X, Yuan Z, Wang C. Contribution of IL-38 in lung immunity during pseudomonas aeruginosa-induced pneumonia. Shock. 2022;57(5):703–713. doi:10.1097/SHK.0000000000001919
- Zhang F, Zhou Y, Ding J. The current landscape of microRNAs (miRNAs) in bacterial pneumonia: opportunities and challenges. Cell Mol Biol Lett. 2022;27(1):70. doi:10.1186/s11658-022-00368-y
- Fernández-Pato A, Virseda-Berdices A, Resino S, et al. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerg Microbes Infect. 2022;11(1):676–688. doi:10.1080/22221751.2022.2038021
- Li J, Luu LDW, Wang X, et al. Metabolomic analysis reveals potential biomarkers and the underlying pathogenesis involved in Mycoplasma pneumoniae pneumonia. Emerg Microbes Infect. 2022;11(1):593–605. doi:10.1080/22221751.2022.2036582
- Lin Y, Yang Y. MiR-24 inhibits inflammatory responses in LPS-induced acute lung injury of neonatal rats through targeting NLRP3. Pathol Res Pract. 2019;215(4):683–688. doi:10.1016/j.prp.2018.12.028
- Houshmandfar S, Saeedi-Boroujeni A, Rashno M, Khodadadi A, Mahmoudian-Sani M-R. miRNA-223 as a regulator of inflammation and NLRP3 inflammasome, the main fragments in the puzzle of immunopathogenesis of different inflammatory diseases and COVID-19. Naunyn Schmiedebergs Arch Pharmacol. 2021;394(11):2187–2195. doi:10.1007/s00210-021-02163-6
- Losier A, Dela Cruz CS. New testing guidelines for community-acquired pneumonia. Curr Opin Infect Dis. 2022;35(2):128–132. doi:10.1097/QCO.0000000000000824
- Muhammad W, Zhai Z, Wang S, Gao C. Inflammation-modulating nanoparticles for pneumonia therapy. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022;14(2):e1763. doi:10.1002/wnan.1763
- Rathbun KP, Bourgault AM, Sole ML. Oral microbes in hospital-acquired pneumonia: practice and research implications. Crit Care Nurse. 2022;42(3):47–54. doi:10.4037/ccn2022672
- Dadhwal K, Stonham R, Breen H, Poole S, Saeed K, Dushianthan A. Severe COVID-19 pneumonia in an intensive care setting and comparisons with historic severe viral pneumonia due to other viruses. Clin Respir J. 2022;16(4):301–308. doi:10.1111/crj.13482
- Li S, Zhang J, Feng G, et al. The emerging role of extracellular vesicles from mesenchymal stem cells and macrophages in pulmonary fibrosis: insights into miRNA delivery. Pharmaceuticals. 2022;15(10):1276. doi:10.3390/ph15101276
- Fan X, Zou X, Liu C, et al. Identify miRNA-mRNA regulation pairs to explore potential pathogenesis of lung adenocarcinoma. Aging. 2022;14(20):8357–8373. doi:10.18632/aging.204341
- Qin L, Zhong M, Adah D, et al. A novel tumour suppressor lncRNA F630028O10Rik inhibits lung cancer angiogenesis by regulating miR-223-3p. J Cell Mol Med. 2020;24(6):3549–3559. doi:10.1111/jcmm.15044
- Nouws J, Wan F, Finnemore E, et al. MicroRNA miR-24-3p reduces DNA damage responses, apoptosis, and susceptibility to chronic obstructive pulmonary disease. JCI Insight. 2021;6(2). doi:10.1172/jci.insight.134218
- Ren J, Guo W, Feng K, Huang T, Cai Y. Identifying MicroRNA markers that predict COVID-19 severity using machine learning methods. Life. 2022;12(12). doi:10.3390/life12121964
- Li K, Wu J, Wu F, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;55(6):327–331. doi:10.1097/RLI.0000000000000672
- Roffel MP, Maes T, Brandsma C-A, et al. MiR-223 is increased in lungs of patients with COPD and modulates cigarette smoke-induced pulmonary inflammation. Am J Physiol Lung Cell Mol Physiol. 2021;321(6):L1091–L1094. doi:10.1152/ajplung.00252.2021
- Zhang D, Lee H, Wang X, et al. A potential role of microvesicle-containing miR-223/142 in lung inflammation. Thorax. 2019;74(9):865–874. doi:10.1136/thoraxjnl-2018-212994