Abstract
Background
Human serum has the potential to become the most informative source of novel biomarkers, but its study is very difficult due to the incredible complexity of its molecular composition. We describe a novel tool based on biodegradable nanoporous nanoparticles (NPNPs) that allows the harvesting of low-molecular-weight fractions of crude human serum or other biofluids. NPNPs with a diameter of 200 nm and pore size of a few nm were obtained by ultrasonication of nanoporous silicon. When incubated with a solution, the NPNPs harvest only the molecules small enough to be absorbed into the nanopores. Then they can be recovered by centrifugation and dissolved in water, making the harvested molecules available for further analyses.
Results
Fluorescence microscopy, gel electrophoresis, and mass spectrometry were used to show the enrichment of low-molecular-weight fraction of serum under physiological conditions, with a cut-off of 13 kDa and an enrichment factor >50.
Conclusion
From these findings, we conclude that ability to tune pore size, combined with the availability of hundreds of biomolecule cross-linkers, opens up new perspectives on complex biofluid analysis, discovery of biomarkers, and in situ drug delivery.
Background
The search for novel tools for early diagnosis is one of the major issues in medical research. The discovery of biomarkers in biological fluids and blood is especially challenging due to the tremendous number of biomolecular species, which differ by many orders of magnitude in their relative abundance.Citation1,Citation2 Of the several approaches proposed in the last few years, the study of the proteome seems to hold the greatest potential.Citation2,Citation3 Proteomics is a quickly developing area of biochemical investigation. The basic aim of proteomic analysis is the identification of specific protein patterns from cells, tissues, and biological fluids related to physiological or pathological condition.Citation2,Citation4 It provides a different view from that of gene expression profiling, which does not evaluate post-transcriptional and post-translational modifications, or protein compartmentalization and half-life changes (eg, ubiquitination and proteosome-driven degradation). All these characteristics make the protein profile much more complex but more informative than gene expression profiling. Several approaches can be used to perform proteomic analysis; among these, the most common are methods based on 2D-polyacrylamide gel electrophoresis (2D-PAGE) and mass spectrometry (MS).Citation5–Citation9
Table 1 Proteins identified by in-gel digestion and nanoLC-MS/MS of SDS-PAGE-isolated bands
It is now well accepted that the low-molecular-weight (LMW), low-abundance fraction of biological fluids might contain the most informative source of novel biomarkers. The conventional analytical methods mentioned above do not seem to reach a sufficient degree of resolution and sensitivity to reliably detect and identify LMW and low-abundance peptides. Therefore, research has recently focused on the development of innovative devices that are able to enrich this fraction of body fluid proteome, making it available for use in common analytical tools.
Harvesting and enrichment of candidate biomarkers from complex protein mixtures can be pursued in different ways. Usually, in the plasma or serum, in which albumin and immunoglobulin alone account for >90% of total protein content, these species are conventionally removed prior to 2D-PAGE and MS by immunoaffinity depletion columns. However, it must be emphasized that many potentially informative biomarkers, represented by very small peptides noncovalently associated with the carrier protein albumin, are lost following this procedure. Many commercial albumin removal kits based on different methods are available, but it has been demonstrated that they can cause loss of several low-abundance proteins, including albuminome.Citation10–Citation12 Moreover, most of these sieving/filtering systems do not allow sufficient flexibility of the whole process. Therefore, alternative approaches are urgently needed. Nanotechnology methods appear to offer a promising and powerful strategy for overcoming these limitations.Citation13–Citation16
Discovered over 40 years ago, porous silicon (PSi) has attracted increasing attention in many fields of research for its interesting features. In particular, the demonstration of its biodegradability in physiological environments has opened up new perspectives for biomedical application.Citation17 The desired dissolution rate can be obtained through the accurate control of the morphology, pore size, and pH. The typical dissolution rate in alkaline conditions ranges from a few minutes to up to a few days. Moreover, the well-known functionalization processes of the porous surface provide further control of bioreactivity and hydrophobicity.Citation18–Citation21 Various surface derivatization has been reported in the literature and hundreds of different cross-linking agents are now available to selectively bind the target molecules.Citation22
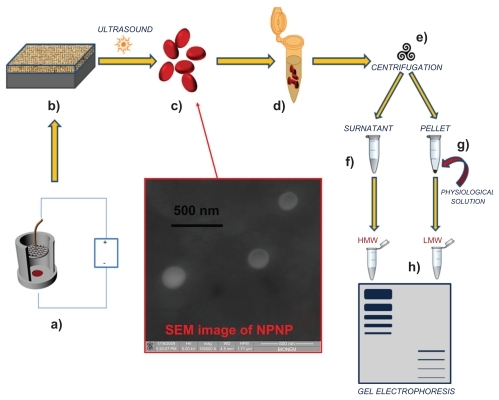
In this paper, we present a direct approach to harvesting the LMW fraction of a complex solution that relies on 3 important properties of silicon nanoporous nanoparticles (NPNPs): (a) they can act as nanosponges and absorb small molecules depending on nanopore size; (b) they can be separated from solution through efficient centrifugation (the nanoparticle density is higher than that of the solvent); (c) they can be dissolved in water. This last property is relevant because the filtration process can be carried out in physiological solution, without the need for introducing particular solvents, which can contaminate, denaturate, or degrade the potential biomarkers. The whole process is depicted in ( fabrication process and harvesting process). The starting solution is incubated with NPNPs which, bcause of size-exclusion, can absorb only the LMW fraction into the nanopore. Afterwards, NPNPs can easily be recovered from solution by means of centrifugation, and resuspended in water, or other solvents in which they can be dissolved. The harvested molecules are then available, in their native state, for further analyses. This easy, cheap, and fast process enables the harvesting of peptides <13 kDa from raw serum.
Figure 1 Pictorial description of the whole process from nanoparticles fabrication to splitting: (a) anodization of silicon wafer to produce porous silicon film; (b) porous silicon film on silicon substrate; (c) nanoporous nanoparticles (NPNPs) fabricated by ultrasonication; (d) incubation of the nanoparticles with biological fluid; (e) centrifugation and wash; (f) supernatant; (g) pellet (physiological solution is added to dissolve it); (h) low-molecular-weight (LMW) harvesting and enrichment is demonstrated on a gel electrophoresis. The middle panel shows a scanning electron microscope image of nanoparticles.
Abbreviation: HMW, high molecular weight.
Modern biology demands not only fast and easy techniques but also full compatibility with existing protocols. The present approach can be mixed and matched with the majority of current investigation protocols.
Material and methods
NPNPs were fabricated by ultrasonication of a thin film of nanoporous silicon.Citation13–Citation24 PSi was obtained by anodization of a boron-doped silicon wafer (resistivity 5–10 Ωcm) of [100] crystal orientation, using an electrolyte binary mixture of hydrofluoric acid (25%), water (25%), and ethanol (50%). Applied constant current density was 10 mA/cm2 for 5 minutes at 25°C. Samples were rinsed in deionized water, then in ethanol and pentane. The PSi film was oxidized in an oven at 200°C for 2 hours. In order to obtain NPNPs, the PSi film was sonicated in dimethylformamide for about 60 minutes and then, after washes in ethanol, ultrasonicated (5 W) in water for 10 minutes at a constant temperature of 4°C, and finally filtered to eliminate impurities >500 nm.
An ad hoc protein mixture was prepared by mixing 50% (v/v) human serum albumin (MW 66,000 Da; Sigma-Aldrich, St. Louis, Missouri, USA); 30% (v/v) bovine plasma gamma globulin (heavy chain MW 45,000 Da; light chain MW 30,000 Da; Biorad, Berkeley, California, USA); and 20% (v/v) aprotinin (6,500 Da; Sigma-Aldrich). All proteins were dissolved at a concentration of 1 mg/ mL in 100 mM sodium phosphate buffer and 9% (w/v) sodium chloride pH 7.4 (PBC) to reproduce physiological conditions.
Human serum was obtained from a healthy anonymous male donor and collected in accordance with Human Proteome Organization (HUPO) plasma proteome project guidelines.Citation25 Approximately 8 mL of blood were drawn by venipuncture and collected in tubes without additive and allowed to clot at room temperature for 40 minutes. The sample was centrifuged within 2 hours of collection at 1300 × g for 10 minutes, aliquoted into silicon tubes, and stored at −80°C.
Results and discussion
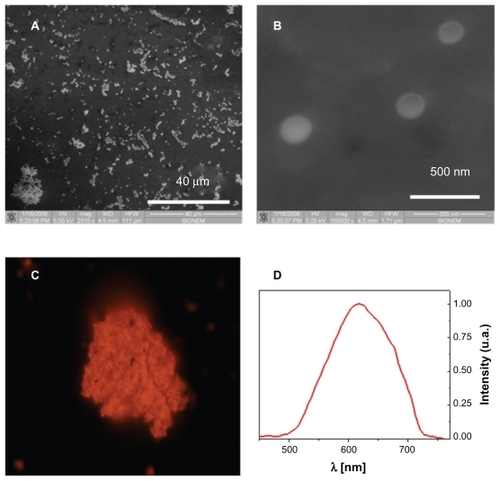
The fabricated NPNPs were deposited onto a glass substrate and characterized using scanning electron microscopy (SEM) and fluorescence microscopy. The results are reported in . The pores of the particles are too small to be shown by SEM (), but the typical emission spectrum peak at around 620 nm () indicates a pore size of about 2–3 nm (excitation wavelength 408 nm). The nanoparticle diameter is about 200 nm, and it can be adjusted by changing the power and duration of the sonication process.
Figure 2 Nanoporous nanoparticles characterization. Scanning electron microscope images of nanoporous nanoparticles dried on a slide (panels A and B). The pores of the particles are too small to be shown, but the typical emission spectrum in red-orange band (panels C and D) indicates a pore size of about 2–3 nm (excitation wavelength 408 nm).
We studied the interactions of the nanoparticles with complex biological fluids in different environmental conditions. Here we report 3 experiments with fluids of increasing complexity in order to show the splitting capability of the NPNPs:
Experiment 1. Interaction with 1 component solution: small dyes of different MWs.
Experiment 2. Interaction of a complex mixture of proteins with a wide range of MWs simulating a biological fluid.
Experiment 3. Interaction with crude human serum.
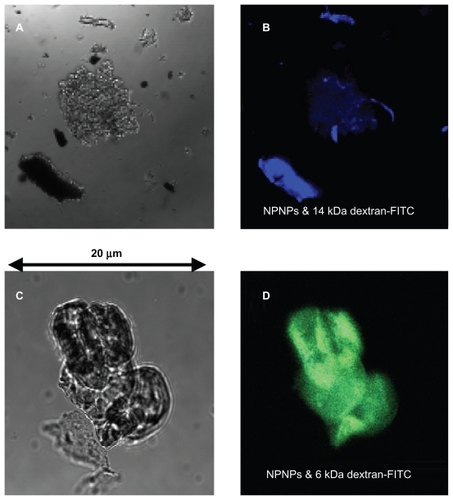
In the first experiment, NPNPs were incubated with 2 solutions of fluorescent polymer of different MWs (6 kDa and 14 kDa dextran-fluorescein isothiocyanate [FITC] 10 mg, NPNP 5 mg, water 10 mL, 1 hour). After incubation, the NPNPs were separated from supernatant (centrifugation), dropped on a slide, dried, and analyzed with fluorescence microscopy. The results are summarized in . Optical and fluorescence images collected on the NPNPs incubated with polymers of 14 and 6 kDa are reported in upper and lower panels, respectively; for higher MW polymer there is no trace of absorption, and only a weak blue fluorescence coming from salt residue is visible. In contrast, green fluorescence emitted by NPNPs incubated with the lower MW polymer indicates good absorption.
Figure 3 Optical and fluorescence images of the nanoporous nanoparticles (NPNPs) after incubation with 2 fluorescent polymers of different molecular weights (MWs): dextran-FITC 14 kDa (panels A and B), and dextran-FITC 6 kDa (panels C and D). The adsorption of the lighter polymer is clearly indicated by the green fluorescence emitted from the nanoparticles (panel D). In contrast, no green fluorescence can be observed being emitted from the heavier polymer (the blue fluorescence comes from salt residues), confirming the MW cut-off.
After incubation and centrifugation, the recovered NPNPs can be dissolved in water at a rate depending on the temperature and acidity of the medium. At pH 8 and 90°C, the dissolution takes a few minutes, but it can be also carried out at room temperature in a few hours, if nondenaturing conditions are needed. At pH <5, the NPNPs do not dissolve, allowing their long-term storage in water at or above room temperature.
In experiment 1, the amount of harvested fraction of dextran-FITC (MW 6 kDa) can be evaluated by comparing the total fluorescence intensity of 2 solutions (harvested vs supernatant). The 2 solutions show very similar fluorescence intensity values, indicating that, under the experimental conditions described above, 50% of the molecules were harvested, whereas the remaining 50% were left in solution. This result shows that the loading capacity is very high, about 1 mg of harvested molecules for each mg of NPNPs.
In the second experiment, the ability of NPNPs to enrich the LMW fraction of a complex mixture was tested with an ad hoc protein mixture (see Materials and methods). For this purpose, the effects of several parameters such as pH, osmolarity, temperature, and incubation time were studied, and conditions were optimized, and, finally, a volume of 200 mL of protein mixture was incubated with 5 mg of NPNPs (about 1015 particles) at room temperature for 1 hour. NPNPs were subsequently separated from supernatant by centrifugation, and washed once with PBC buffer. The sodium dodecyl sulfate-PAGE (SDS-PAGE) in (panel A) clearly shows that nanoparticles selectively retain small molecules as aprotinin (MW 6,500 Da), whereas proteins with higher MW are completely excluded from the nanopores. We noted that during the incubation a small fraction of the nanoparticles dissolve releasing silicic acid into the solution (about 8% of NPNP volume under our conditions).Citation26 This amount can be decreased by lowering the incubation temperature, pH, or incubation time when possible.
Figure 4 (A) The sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of protein mixture before and after incubation with nanoporous silicon particles. 100 mL of protein mixture (see text for details) was subjected to incubation with nanoparticles. Aliquots of the mixture before and after incubation were subjected to tristricine SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1 A: molecular weight markers; lane 2 A: protein mixture before incubation; lane 3 A: protein mixture following nanoparticle incubation (supernatant); lane 4 A, low-molecular-weight (LMW) protein fraction enriched (pellet). (B) SDS-PAGE analysis of human serum before and after incubation with nanoporous silicon particles. Serum was diluted to 1:2 with 100 mM sodium phosphate buffer, 9% (p/v) sodium chloride, pH 7.4, and incubated with nanoparticles. Aliquots of serum before and after incubation were subjected to tristricine SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1B, molecular weight markers; lane 2B, crude human serum; lane 3B, human serum following nanoparticle incubation (supernatant); lane 4B, LMW serum fraction enriched using (nondenaturing conditions) nanoporous silicon particles (pellet).
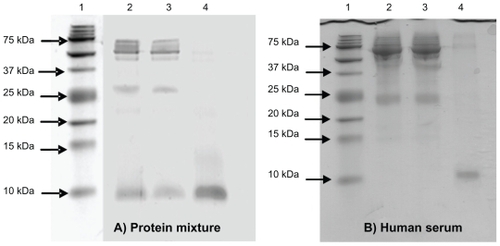
In the third experiment, the same protocol for experiment 2 was applied to raw human serum to demonstrate that NPNPs are able to selectively enrich LMW serum proteome (LMWP). After 1 hour of incubation (100 mL of serum sample diluted 1:2 with PBC buffer, 5 mg of NPNPs), the LMW fraction was recovered by dissolving the nanoparticles in PBC buffer for 12 hours at 37°C. We noted that the dissolution process can be hastened by heating the solution and adding a base (to increase pH) or other solvent. An aliquot of serum, before and after incubation, was analyzed by SDS-PAGE using a 16.5% ready prepared tris-tricine/peptide gel. The efficiency of NPNPs to selectively enrich the LMWP is clearly demonstrated in the SDS-PAGE analysis shown in (panel B): in lines 2 and 3, crude human serum and supernatant are represented. In line 4, the LMW fraction of human serum extracted from NPNPs is visible: no molecules >12 kDa are still present, except for a very small trace of albumin. From densitometric analysis of lane 4, it was estimated that >50% of total pixel volume was from LMW species, whereas in lane 2 (unprocessed serum) the same area represented <1% of the total pixel volume. The enrichment factor was thus estimated to be >50.
In order to characterize the LMW protein species extracted by incubation with NPNPs, gel bands from SDS-PAGE-separated extracts (, lane 4) were cut and processed for tryptic digestion according to the protocol shown in the Supplementary Information, section 1. Based on MW marker information, the 3 processed gel bands corresponded to MW intervals of 15–20 kDa, 10–15 kDa and <10 kDa. Tryptic peptides were analyzed by nanoscale liquid chromatography interfaced with tandem MS (nanoLC-MS/MS, Supplementary Information, section 2).Citation27,Citation28 In an alternative approach, proteins extracted by incubation with NPNPs were directly digested in solution by trypsin and analyzed by nanoLC-MS/MS. Validated protein identifications are reported in 2 tables (Supplementary Information, section 3). Besides abundant LMW serum proteins, such as apolipoprotein A-II and transthyretin, a database search identified a number of LMW serum proteins that are not considered abundant, such as tetranectin, platelet basic protein, and dermcidin. Furthermore, LC-MS/MS analysis identified several high-MW proteins in the in-gel-digested SDS-PAGE protein bands. Such identifications confirm that the LMW proteome is also populated with fragments of abundant high-MW serum proteins.Citation29
A comparison of the results of these experiments with those reported in the literature shows many remarkable characteristics of the NPNPs:
easy and cheap production.
controllable pore size and well known surface chemistry.
small molecules can be absorbed with a tuneable MW cut-off.
biocompatibility and biodegradability in physiological solution.
easy recovery by centrifugation,
long-term storage wet (water solution of pH <5) or dry.
We note that such properties are very attractive also for drug delivery applications, for which the use of nanocarriers is attracting a lot of interest.
Conclusion
We report a straightforward tool relying on water-soluble silicon NPNPs used to harvest the LMW molecules in their native state from a complex fluid. The method is based on the porosity of the nanoparticles, which act as a molecular sieve, and their solubility in a physiological environment. The proposed approach can be mixed and matched with currently available techniques and protocols, and does not require high temperature, denaturing solvents, or other contaminants. A cut-off of about 13 kDa was demonstrated for crude human serum. The ability to tune pore size, combined with the availability of hundreds of biomolecule cross-linkers, opens up new perspectives on complex biofluid analysis, discovery of biomarkers, and in situ drug delivery.
Supplementary information
In-gel digestion
Gel bands below 25 kDa were excised and digested by trypsin incubation; selected bands were punched out manually and placed in a silicon Eppendorf tube. Gel pieces were washed once with 150 mL of deionized water and then destained by 3 washes of 150 mL 35% acetonitrile in 25 mM NH4HCO3 buffer. After destaining, trypsin digestion was performed overnight at 37°C with modified trypsin (Sigma-Aldrich, St Louis, MO, USA) 0.2 mg/mL. The resulting tryptic peptides were acidified, purified by Ziptips C18 (Millipore, Billerica, MA) according to the manufacturer’s procedure and eluted with 2 μL of a 1:1 mixture of acetonitrile and 0.1% trifluoro acetic acid (v/v). 28 μL of loading pump solvent (see below) were added, and 10 μL were injected for nanoLC-MS/MS analysis.
In-solution digestion
Protein extracts were reduced by 2 mM DTT (1 hour at 37°C). 500 ng of sequencing grade modified trypsin (Sigma-Aldrich) were added, and digestion was allowed to proceed for 16 h. The resulting tryptic peptides were fractionated by Off-Gel eletrophoresis before nanoscale LC-MS/MS analysis, in order to achieve a 2-dimensional fractionation of the peptide mixture. An Agilent 3100 Off-Gel fractionator (Agilent Technologies, Santa Clara, CA) was used. Off-Gel isoelectric focusing was essentially peformed according to the manufacturer’s instructions. The peptide mixture was diluted with carrier ampholyte mixture (3.6 mL final volume, 10% carrier ampholyte concentration). The sample was then loaded on separate ImmobilineTM DryStrip, linear pH range 3.0–10, 18 cm long, purchased from GE Healthcare (Chalfont St. Giles, UK). Peptides were focused at a constant temperature of 20°C, and at constant current intensity of 50 μA. After focusing was complete, fractions were collected. In order to improve peptide recovery, sample wells were washed with 100 μL of a water/methanol/formic acid mixture, 49:50:1 (v/v/v). The wash solution was added to each well and allowed to incubate for 90 minutes before being collected and pooled with the corresponding fraction supernatant. Pooled supernatants were reduced to a volume of approximately 10 μL in a vacuum centrifuge.
A 1:4 mixture of concentrated eluates and loading pump solvent of the nanoLC-MS/MS system (see below) was injected for nanoLC-MS/MS analysis. Considering the injection volume of 10 μL, approximately 1/5 of each OGE fractions was injected for nanoLC-MS/MS.
Nanoscale LC-MS/MS analysis
Chromatography was performed on an Ultimate nanoLC system from Dionex (Sunnyvale, CA, USA), using a valveless setup.Citation1,Citation2 The peptide extracts were redissolved in 30 μL of loading pump solvent (see below) and 10 μL were loaded onto an in-house packed 100 μm i.d., Integra FritTM (New Objective, Cambridge, MA) trapping column (packing bed length 1.5 cm) at 10 μL/min of loading pump solvent, consisting of H2O/acetonitrile/trifluoroacetic acid (TFA) 97.95:2:0.05 (v/v/v). After 4 minutes of column washing, the trapping column was switched on-line to the analytical column: an in-house packed 50 μm i.d., Pico FritTM column (New Objective), filled with the same stationary phase used for the trapping column packing: 3 μm C18 silica particles (Dr Maisch, Entringen, Germany). Peptide separation started at 100 nL/min using a binary gradient. Mobile phase A was H2O/acetonitrile/formic acid/TFA 97.9:2:0.09:0.01 (v/v/v/v); mobile phase B was H2O/acetonitrile/formic acid/TFA 29.9:70:0.09:0.01 (v/v/v/v). Gradient was from 5 to 45% B in 40 minutes. After 10 minutes at 95% B, the column was re-equilibrated at 5% B for 20 minutes before the following injection.
MS detection was performed on a QSTAR XL hybrid LC-MS/MS from Applied Biosystems (Foster City, CA, USA) operating in positive ion mode, with nESI potential at 1300 V, curtain gas at 15 units, CAD gas at 3 units. Information-dependent acquisition (IDA) was performed by selecting the 2 most abundant peaks for MS/MS analysis after a full TOF-MS scan from 400 to 1600 m/z lasting 4 seconds. Both MS/MS analyses were performed in enhanced mode (3 seconds/scan). Threshold value for peak selection for MS/MS was 30 counts.
Data analysis
MS/MS data were converted to Mascot Generic Format (mgf) by the Analyst software 1.1 (Applied Biosystems). Data were searched on the Mascot search engine (www.matrixscience.com), version 1.9, against the International Protein Index database (IPI version 3_38) using the following parameters: MS tolerance 30 ppm; MS/ MS tolerance 0.2 Da; variable modifications methionine oxidized; enzyme trypsin; max. missed cleavages 1. MS/ MS identifications were validated by using the transproteomics pipeline.Citation3 Peptide identifications with a minimum probability score of 0.7 were retained (4% false discovery rate). Proteins identified with a minimum of 2 peptides were retained (protein probability score >0.9). Protein identifications based on a single peptide were manually validated.
References
- MeiringHDvan der HeeftEten HoveGJde JongAPJMSepJScience200225557568
- GaspariMAbbonanteVCudaGSepJScience20073022102216
- KellerAEngJZhangNLiXJAebersoldRMol Syst Biol20051 msb4100024-E4100021-msb4100024E4100028 (2005)
Disclosure
The authors declare that they have no competing interests. Giovanni Cuda and Enzo Di Fabrizio equally share the senior authorship. They conceived the study, and participated in its design and coordination and helped to draft the manuscript. Antonella Pujia was involved in the fabrication of NPNPs and carried out the harvesting experiments; Francesco De Angelis contributed to the design of NPNPs and to the drafting of the manuscript; Domenica Scumaci carried out gel electrophosesis experiments; Marco Gaspari was involved in the mass spectrometry analysis; Carlo Liberale contributed to the design and fabrication of NPNPs; and Patrizio Candeloro contributed to the design of NPNPs and to the analysis of experimental data.
References
- AndersonNLAndersonNGThe human plasma proteome: history, character, and diagnostic prospectsMol Cell Proteomics200211184586712488461
- HanashSMPitteriSJFacaVMMining the plasma proteome for cancer biomarkersNature200845257157918385731
- IssaqHJXiaoZVeenstraTDSerum and plasma proteomicsChem Rev2007107360162017636887
- KulasingamVDiamandisEPStrategies for discovering novel cancer biomarkers through utilization of emerging technologiesNature Clin Pract Oncol2008558859918695711
- MindenJSDowdSRMeyerHEDifference gel electrophoresisElectrophoresis200930Suppl 1S15616119517495
- GörgADrewsOLückC2-DE with IPGsElectrophoresis200930Suppl 1S12213219441019
- DiamandisEPHanashSMLopezMProtein quantification by mass spectrometry: is it ready for prime timeClin Chem2009551427143019443560
- MotoyamaAYatesJRMultidimensional LC separations in shotgun proteomicsAnal Chem200880187193
- LiaoLMcClatchyDBYatesJRShotgun proteomics in neuroscienceNeuron200963122619607789
- SahabZJIczkowskiKASangQXAnion exchange fractionation of serum proteins vs albumin eliminationAnal Biochem20073681243217618595
- RothemundDLLockeVLLiewADepletion of the highly abundant protein albumin from human plasma using the GradiflowProteomics2003327928712627381
- YocumAKYuKOeTEffect of immunoaffinity depletion of human serum during proteomic investigationsJ Proteome Res2005451722173116212426
- LuchiniLGehoDHBishopBSmart hydrogel particles: biomarker harvesting: one-step affinity purification, size exclusion, and protection against degradationNano Lett20088135036118076201
- FuJMaoPHanJArtificial molecular sieves and filters: a new paradigm for biomolecule separationTrends Biotechnol20082631132018430480
- ChengMMCudaGBunimovichYLNanotechnologies for biomolecular detection and medical diagnosticsCurr Opin Chem Biol200610111916418011
- ChunxiongLQiangFHaoLPDMS microfluidic device for optical detection of protein immunoassay using gold nanoparticlesLab Chip2005572672915970965
- CanhamLTBioactive silicon structure fabrication through nanoetching techniquesAdv Mater1995710331037
- AndersonSHCElliottHWallisDJDissolution of different forms of partially porous silicon wafers under simulated physiological conditionsPhys Stat Sol (a)2003197331335
- VaccariLCantonDZaffaroniNPorous silicon as drug carrier for controlled delivery of doxorubicin anticancer agentMicroelectronic Eng20068315981601
- CanhamLTReevesCLNeweyJPDerivatized mesoporous silicon with dramatically improved stability in simulated human blood plasmaAdv Mater19991115051507
- SongJHSailorMJChemical modification of crystalline porous silicon surfacesComm Inorg Chem1999216984
- Tinsley-BownAMCanhamLTHollingsMTuning the pore size and surface chemistry of porous silicon for immunoassaysPhysica Status Solidi A Appl Res2000182547553
- AkcakirOTherrienJBelomoinGDetection of luminescent single ultrasmall silicon nanoparticles using fluctuation correlation spectroscopyAppl Phys Lett20007618571859
- FronerEAdamoRGaburrZoLuminescence of porous silicon derived nanocrystals dispersed in water: dependence on initial porous silicon oxidationJ Nanopart Res2006810711074
- TirumalaiRSChanKCPrietoDACharacterization of the low molecular weight human serum proteomeMol Cell Proteomics200321096110312917320
- AnglinEJChengLFreemanWRPorous silicon in drug delivery devices and materialsAdv Drug Deliv Rev2008601266127718508154
- MeiringHDvan der HeeftEten HoveGJNanoscale LC-MS(n): technical design and applications to peptide and protein analysisJ Sep Sci200225557568
- GaspariMAbbonanteVCudaGGel-free sample preparation for the nanoscale LC-MS/MS analysis and identification of low-nanogram protein samplesJ Sep Sci2007302210221617763504
- RaiAJGelfandCAHaywoodBCHUPO plasma proteome project specimen collection and handling: towards the standardization of parameters for plasma proteome samplesProteomics20055133262327716052621