Abstract
Phage display technology has been demonstrated to be a powerful tool for screening useful ligands that are capable of specifically binding to biomarkers on the surface of tumor cells. The ligands found by this technique, such as peptides, have been successfully applied in the fields of early cancer diagnostics and chemotherapy. In this study, a novel nonsmall cell lung cancer-targeting peptide (LCTP, sequence RCPLSHSLICY) was screened in vivo using a Ph.D.-C7C™ phage display library. In order to develop a universal tumor-targeting drug carrier, the LCTP and fluorescence-labeled molecule (FITC) were conjugated to an acetylated polyamidoamine (PAMAM) dendrimer of generation 4 (G4) to form a PAMAM–Ac–FITC–LCTP conjugate. The performance of the conjugate was first tested in vitro. In vitro results of cell experiments analyzed by flow cytometry and inverted fluorescence microscopy indicated that PAMAM–Ac–FITC–LCTP was enriched more in NCI-H460 cells than in 293T cells, and cellular uptake was both time- and dose-dependent. The tissue distribution of the conjugate in athymic mice with lung cancer xenografts was also investigated to test the targeting efficiency of PAMAM–Ac–FITC–LCTP in vivo. The results showed that LCTP can effectively facilitate the targeting of PAMAM–Ac–FITC–LCTP to nonsmall cell lung cancer cells and tumors. These results suggest that the LCTP-conjugated PAMAM dendrimer might be a promising drug carrier for targeted cancer diagnosis and treatment.
Introduction
Nonspecific anticancer chemotherapy has no tumor targeting effect and will kill normal and cancer cells, causing severe side effects in many patients. To solve this problem, targeted drug delivery systems have received great attention.Citation1 In this system, drugs can be specifically targeted to the tumors, increase drug solubility,Citation2,Citation3 prolong time of drugs in the circulation, protect drugs from degradation, and improve their metabolic kinetics.Citation4,Citation5 To date, liposomes,Citation6 micelles,Citation7,Citation8 and dendrimersCitation9 conjugated with specific ligands have been identified as being able to target drugs to cancer cells.
Polyamidoamine (PAMAM) dendrimers have attracted considerable research interest as drug carriers due to their unique properties, ie, hyperbranched and monodispersed tree-like structures with multifunctional surfaces, enabling the dendrimers to encapsulate or conjugate drug molecules. Moreover, the terminal groups of PAMAM can be easily modified (by, eg, acetylation or poly(ethyleneglycol) modification [PEGylation]) to improve their solubility and biocompatibility. In order to improve the targeting capability of PAMAM, many targeting molecules, such as antibodies,Citation10 folic acid,Citation11,Citation12 biotin,Citation13 peptides,Citation14–Citation17 and carbohydratesCitation18,Citation19 have been connected to PAMAM dendrimers. Finally, chemotherapy drugs have been covalently bound to these dendrimersCitation20,Citation21 or encapsulated in the internal cavities of dendritic moleculesCitation11,Citation22 to generate targeted drug delivery systems.
A key factor of a targeted drug delivery system is the finding of targeting molecules that can specifically recognize tumor and cancer cells. Phage display technology is a powerful approach to screening targeting molecules, such as peptides, for cancer cells or tumor blood vessel endothelial cells.Citation23,Citation24 Many novel angiogenic vessels and homing peptides have been isolated recently using this method.Citation17,Citation25,Citation26 Furthermore, in vivo phage display can screen cancer-binding peptides regardless of whether the receptor is known or not. Therefore, this technique can quickly screen cancer-specific peptides.Citation27,Citation28 In addition to drug delivery systems, tumor-targeting peptides are also employed in diagnosis or radiotherapy by delivering radionuclide.Citation29–Citation31 Some peptides discovered by phage display are even able to inhibit cancer growth and induce cancer cell apoptosis.Citation32,Citation33
In order to develop a universal drug carrier for nonsmall cell lung cancer chemotherapy, a Ph.D.-C7C™ (New England Biolabs, Beverly, MA) phage display library was utilized to screen peptides specific for nonsmall cell lung cancer. It has become generally accepted that organs have their own special markers. Such heterogeneity was reported after it became apparent that endothelial cells in different organs display organ-specific markers. The phage display library includes all possible peptide sequences, and some random peptides displayed by phage can bind specifically with tumor-specific markers displayed by nonsmall cell lung cancer, based on protein–protein interactions. The phage displaying specific peptides would home to the tumor site, after intravenous injection. The tumor is then harvested and the phage therein collected and amplified. After four to five rounds, the phage including the tumor-specific targeting peptide can be isolated and the inserted peptide sequence translated by the phage DNA sequence. In our study, a novel peptide, lung cancer-targeting peptide (LCTP; RCPLSHSLICY), was discovered which can specifically target nonsmall cell lung cancer. The LCTP was then conjugated with fluoresceinisothiocyanate (FITC, a fluorescence labeling agent used as a tracer) and acetylated PAMAM to generate a targeted drug delivery carrier (PAMAM–Ac–FITC–LCTP). The performance of this drug carrier was evaluated by in vitro culturing NCIH460 and 293T cells and in vivo using athymic mice with lung cancer xenografts. Our results showed that the modified PAMAM peptide dendrimer could be easily taken up by NCI-H460 cells in vitro and by tumors in vivo.
Materials and methods
Materials
Phenyl methyl sulphonyl fiuoride (PMSF), isopropyl β-D-1-thiogalactopyranoside (IPTG), aprotinin, leupeptin, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal), FITC, 5,5-dithiobis-2-nitrobenzoic acid (DTNB), and polyamidoamine (G 4) were purchased from Sigma-Aldrich (St Louis, MO). Dialysis membrane (MWCO, 3500) was purchased from BBI Inc (Shanghai, China). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), all cell culture media, and supplies were purchased from Gibco Corporation (Grand Island, NE).
The NCI-H460 cell line was kindly provided by Dr Yong Wang (Institute of Radiation Medicine, Chinese Academy of Medical Sciences, Tianjin, China). The 293T cell line was purchased from Keygen Co (Nanjing, China). Phage DNA sequencing was performed by Shanghai Sangon Corp (Shanghai, China). Peptides were synthesized by GL Biochem Ltd (Shanghai, China). BALB/c-nu/nu athymic mice (female, 4–6 weeks old) were purchased from the Laboratory Animal Center of The Academy of Military Medical Sciences (Beijing, China). The animal studies were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (Tianjin, revised in June 2004) and adhered to the Guiding Principles in the Care and Use of Animals of the American Physiological Society. The Ph.D.-C7C™ phage display peptide library kit was used to screen specific peptides binding to human lung cancer xenografts on BALB/c-nu/nu athymic mice.
The phage display library kit is based on a combinatorial library of random peptide 7-mers fused to the N terminus of the minor coat protein (cpIII) of M13 phage. The random sequence is flanked by a pair of cysteine residues. Under nonreducing conditions, the cysteines spontaneously form a disulfide crosslink, resulting in phage display of cyclized peptides. The titer of the library is 2 × 1013 plaque-forming units (pfu). The library consists of 1.2 × 109 electroporated sequences (compared with 20Citation7 = 1.28 × 109 possible seven-residue sequences). Extensively sequencing the naive library has revealed a wide diversity of sequences with no obvious positional biases. The Escherichia coli host strain ER2738 (Arobust F+ strain with a rapid growth rate; New England Biolabs) was used for M13 phage propagation.
In vivo phage display screening and peptide synthesis
BALB/c-nu/nu athymic mice (female, 4–6 weeks) were used, and 2 × 107 NCI-H460 cells in 0.2 mL of Roswell Park Memorial Institute 1640 (RPMI-1640) medium were subcutaneously injected in the right flank to produce lung cancer xenografts. A Ph.D.-C7C phage display peptide library, which contained 2 × 1011 pfu was mixed with 200 μL of Dulbecco’s modified Eagle’s medium (DMEM), and injected into the mice through the tail vein when the tumor grew up to about 0.5 cm in diameter. After circulation for 5 minutes, the mice were sacrificed, and perfused by injection of 50 mL phosphate-buffered saline through the heart to wash the unbounded phage. Then the tumors were harvested, weighed, and ground in 1 mL of DMEM-PI (DMEM containing the protease inhibitors PMSF [1 mM], aprotinin [20 μg/mL], and leupeptin [1 μg/mL]). The tissue was washed five times with ice-cold DMEM-PI containing 1% bovine serum albumin. After centrifugation, the phage particles were amplified by ER2738 bacteria overnight at 37°C. The phages were titered on agar plates in the presence of IPTG/X-gel (1 mg/L) and tetracycline (40 μg/mL). The amplified phage was injected into the mice to repeat the above procedures. This screening was repeated for five rounds.Citation17,Citation27,Citation34 At the end of the fifth round, the phage was eluted and titered on LB/IPTG/X-gel plates. The phage clones were randomly selected, and the inserted DNA sequence was determined using primer 5′-CCCTCATAGTTAGCGTAACG-3′ (New England Biolabs). The phage-displayed peptides were translated and synthesized according the DNA sequences.
Synthesis of acetylated PAMAM dendrimer
The acetylated PAMAM dendrimer (PAMAM–Ac) was synthesized according to the method described in the literature.Citation13,Citation16,Citation35 Briefly, 1.012 g (MW 14215, 71.2 μmol) of PAMAM dendrimer in 120 mL anhydrous methanol (MeOH) was reacted with 0.214 g (2.110 mmol) of acetic anhydride overnight at room temperature with stirring in the presence of 0.352 g (3.454 mmol) of triethylamine. After evaporation, the product was dialyzed for 3 days against phosphate-buffered saline and double-distilled water using the dialysis membrane (MWCO = 3500 Da), then lyophilized, and 1.165 g of PAMAM–Ac was obtained. The average number of acetyl groups grafted to each PAMAM molecule was determined by 1H nuclear magnetic resonance calibration.
Conjugation of FITC to acetylated PAMAM dendrimer
FITC (38.2 mg, 98.1 μmol) was added dropwise into 40 mL of dimethyl sulfoxide (DMSO) solution containing 0.401 g (MW 15433, 25.9 μmol) of PAMAM–Ac. The reaction mixture was allowed to stir overnight. The product was dialyzed against phosphate-buffered saline and double-distilled water using dialysis membrane (MWCO = 3500 Da) for 3 days in a dark room, and the adsorbance at 500 nm of the eluent was detected to ensure that all free FITC was removed. The product was then lyophilized in a dark room, and 0.387 g of PAMAM–Ac–FITC was obtained. The average number of FITC molecules conjugated to each PAMAM molecule was determined by 1H nuclear magnetic resonance calibration and ultraviolet-visible adsorbance (Varian Cary 100 bio ultraviolet-visible spectrophotometer).
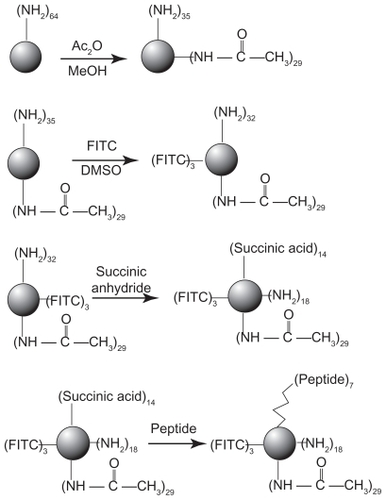
Conjugation of peptides to FITC-labeled dendrimer
The solution containing 9.1 mg (90.9 μmol) of succinic anhydride in 6 mL of anhydrous MeOH was added dropwise into a solution of 0.103 g (MW 16601, 6.2 μmol) of PAMAM–Ac–FITC and 24.5 mg of (0.242 mmol) triethanolamine (TEA) in 90 mL of anhydrous MeOH with stirring. After overnight reaction at room temperature, the solvent was evaporated, and then the crude product was dialyzed (MWCO = 3500 Da) and lyophilized using the aforementioned method; 12.8 mg (MW 18001, 0.71 μmol) of the above product was then reacted with 2.0 mg (10.4 μmol) of 1-[3-(dimethylamino)-propyl]-3-ethyl-carbodimide HCl (EDC) in 4 mL of water for 3 hours. Then 9.2 mg (7.1 μmol) of the LCTP in 1 mL of DMSO was added dropwise to the above solution and stirred overnight. The crude product was dialyzed (MWCO = 3500 Da) using the aforementioned method, and the adsorbance at 275 nm of the elute was detected to ensure that all free peptide was removed. Then the product was lyophilized in a dark room. The average number of peptides conjugated to the dendrimer was determined using the Ellman assay.Citation36 A standard curve was plotted with free LCTP. During the process, an excess of sodium sulfite was added to the peptide solution at pH 9.0 to cleave the disulfide bond, and the reaction was carried out in a dark room. The adsorbance at 412 nm was used to calculate the number of peptides conjugated to the dendrimer according to the standard curve.
Laser light-scattering assay
Samples of PAMAM–Ac, PAMAM–Ac–FITC, and PAMAM–Ac–FITC–LCTP were filtered (pore size, 0.45 mm) and analyzed with a light-scattering spectrometer (BI-200SM; Brookhaven Instruments, Holtsville, NY), equipped with a digital correlator (BI-9000AT; Brookhaven Instruments) to determine the granulometric distribution of the conjugates at 532 nm.
Cell culture
NCI-H460 cells were cultured in RPMI-1640 medium and 293T cells in DMEM medium. The medium was supplemented with 10% fetal bovine serum, 1% penicillin, and 100 μg/mL streptomycin. Cells were grown at 37°C in 5% CO2.
Cytotoxicity assay
The cytotoxicity of the PAMAM, PAMAM–Ac, PAMAM–Ac–FITC, and PAMAM–Ac–FITC–LCTP were evaluated by the MTT assay. The 293T cells were seeded into 96-well plates at a density of 104 cells/well and incubated for 24 hours. The filtrate of the samples (0.22 μm) was added to the cells at different concentrations (0, 0.125, 0.25, 0.5, 1.0, 2.0, and 4.0 μM). After incubation for another 24 hours, 25 μL of MTT solution (5 mg/mL in phosphate-buffered saline) was added to each well. Four hours later, the medium containing MTT was removed, and the samples in the wells were air dried. Acidic isopropanol (100 μL, 0.04 M HCl in absolute isopropanol) was added to dissolve the formazen crystals. The optical density of the solution was measured at 570 nm using a microplate reader (Labsystem; Multiskan, Ascent, Finland).
In vitro cellular uptake assay
293T and NCI-H460 cells were seeded in 24-well plates at a density of 104 cells/well. After 24 hours, the cells were incubated with PAMAM–Ac–FITC or PAMAM–Ac–FITC–LCTP at different concentrations ranging from 0.05 to 1.0 μM for 4 hours, or incubated with 0.25 μM of PAMAM–Ac–FITC–LCTP at different intervals ranging from 0.5 to 8.0 hours. To compare the uptake efficiency between 293T cells and NCI-H460 cells, the two cell lines were incubated with 0.5 μM of the conjugates for 4 hours. To examine the competitive uptake, 293T cells and NCI-H460 cells were preincubated with 20 μM free LCTP for 1 hour before the conjugates were added. At the end of incubation, the cells were trypsinized, washed with physiologic saline three times, and we then proceeded to fluorescence-activated cell sorting (FACS) analysis (Altra FCM; Beckman Coulter, Miami, FL). The uptake efficiency was expressed as the percentage of FITC-positive cells. The cells were also viewed under a confocal microscope (DMI6000B; Leica, Wetzlar, Germany) and an inverted fluorescence microscope. Images were recorded using CCD (ECLIPSE TE2000-U; Nikon, Tokyo, Japan).
In vivo targeting assay
BALB/c-nu/nu athymic mice with lung cancer xenografts were prepared as described earlier. Mice were used for experiment when the diameter of tumor xenografts reached 0.4–0.6 cm. The animals were randomly divided into two groups. A 0.3 mL solution of PAMAM–Ac–FITC or PAMAM–Ac–FITC–LCTP (10 μM in phosphate-buffered saline) was injected through the tail vein of the animal. Four hours later, the tumors and main organs (heart, liver, lung, spleen, kidney, thyroid, brain, and ovary) were harvested and proceeded to imaging (KODAK IS in vivo FX; Kodak, New Haven, CT). The relative fluorescence intensity per unit area was calculated using KODAK 5.1 software (KODAK IS in vivo FX).
Statistical analysis
The unpaired Student’s t-test was used to evaluate the significance between experimental groups. A value of P < 0.05 was considered to be statistically significant.
Results
Screening and peptide synthesis
After five rounds of screening, 40 random phage clones were amplified and the DNA sequences were determined. The phage displayed peptide sequences were translated (see ).
Table 1 The displayed peptide sequences of screening phage from in vivo lung cancer xenografts
Preparation of PAMAM–Ac–FITC–LCTP conjugate
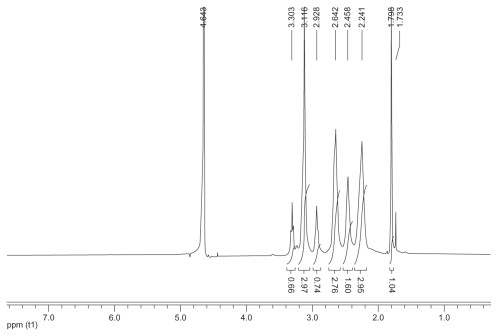
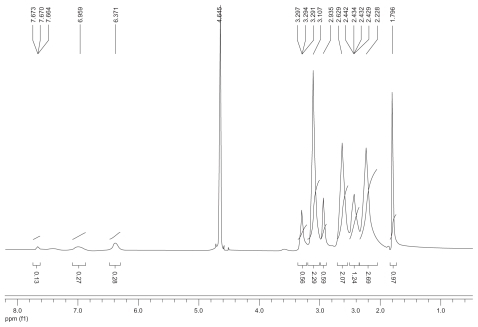
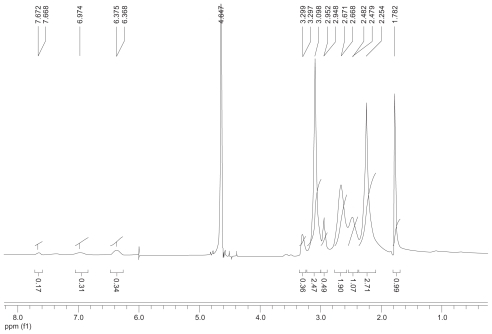
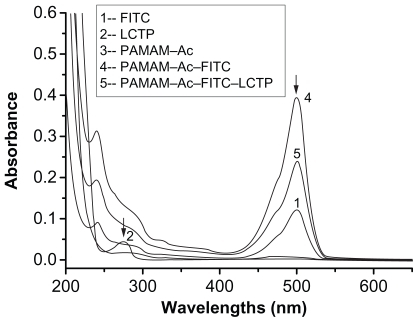
The synthetic route of PAMAM–Ac–FITC–LCTP conjugate is shown in . The degree of PAMAM–Ac acetylation was measured by 1H nuclear magnetic resonance (300 mHz, D2O); the aliphatic peaks appear at 1.80 ppm, 2.24 ppm, 2.46 ppm, 2.64 ppm, 2.93 ppm, 3.12 ppm, 3.23 ppm, and 3.30 ppm. The integration ratio of methylene protons signal of –CH2C(O)– at δ2.24 ppm (248 H) and methyl protons signal of –C(O)CH3 at δ1.80 ppm was 2.84, indicating that about 29 acetyl groups were grafted to each PAMAM molecule (PAMAM–Ac29). Conjugation of FITC to PAMAM–Ac was characterized by 1H nuclear magnetic resonance (300 mHz, D2O) and ultraviolet–visible spectrum. The aliphatic peaks appear at 1.80 ppm, 2.23 ppm, 2.44 ppm, 2.63 ppm, 2.94 ppm, 3.11 ppm, 3.29 ppm, and 3.30 ppm. The aromatic peaks at 6.37 ppm, 6.96 ppm, 7.42 ppm, and 7.67 ppm corresponded to the protons in FITC. FITC has a characteristic absorbance peak at around 500 nm and not for PAMAM–Ac and the peptide (). The number of FITC conjugated to PAMAM–Ac was about 3–4, which was determined based on the standard calibration curve of free FITC. The number of LCTP grafted to PAMAM–Ac–FITC was 7–8 by Ellman assay (see supporting information for the 1H nuclear magnetic resonance spectra of PAMAM–Ac, PAMAM–Ac–FITC, and PAMAM–Ac–FITC–LCTP).
Cytotoxicity assay
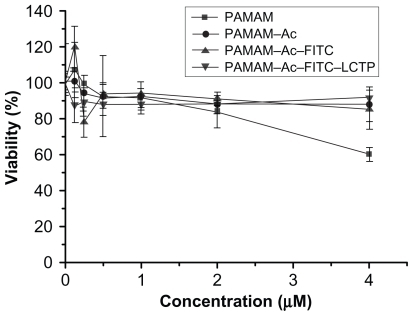
As shown in , when the concentration of the polymers was higher than 2 μM, the cytotoxicity of PAMAM–Ac, PAMAM–Ac–FITC, and PAMAM–Ac–FITC–LCTP was lower than that of unmodified PAMAM. The cells viability was over 88%, which suggests that the PAMAM–Ac–FITC–LCTP conjugate has good biocompatibility.
Figure 3 MTT assay for cellular toxicity of 293T cells. Cells were incubated with polyamidoamine, fluorescence-labeled acetylated polyamidoamine, acetylated polyamidoamine and fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide, at varied concentrations. The viability of the nontreated cells was arbitrarily defined as 100%. Data are expressed as mean ± standard deviation (n = 6).
In vitro cellular uptake assay
shows the uptake efficiencies of the two conjugates for 293T cells over 4 hours. The uptake efficiencies of both conjugates were nearly of equal value and were concentration-dependent. However, although the uptake efficiencies of the two conjugates by NCI-H460 cells were also concentration-dependent, but the uptake efficiency of the PAMAM–Ac–FITC–LCTP was much higher than that of PAMAM–Ac–FITC (). The cellular uptake of the conjugates by both cell lines was time-dependent, but reached saturation after incubation for 4 hours (). Under the same conditions, the uptake of PAMAM–Ac–FITC–LCTP by NCI-H460 cells was threefold higher than that of PAMAM–Ac–FITC, and more than threefold higher than the uptake of the two conjugates by 293T cells. When preincubated with free LCTP, uptake of PAMAM–Ac–FITC–LCTP by NCI-H460 cells was effectively decreased, but for 293T cells and PAMAM–Ac–FITC there was almost no change ().
Figure 4 In vitro evaluation of dose-dependent uptake of the conjugates (fluorescence-labeled acetylated polyamidoamine and fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide) by 293T cells (A) and NCI-H460 cells (B) determined by fluorescence-activated cell sorting. The cells were incubated with various concentrations of the conjugates for 4 hours at 37°C. C) Time-dependent uptake of the conjugates by NCI-H460 cells. The NCI-H460 cells were incubated with the conjugates at a concentration of 0.25 μM for different time. D) Uptake of the conjugates by 293T and NCI-H460 cells. The incubation conditions were 0.5 μM and four hours.
The uptake of PAMAM–Ac–FITC and PAMAM–Ac–FITC–LCTP by the cells was directly visualized under the inverted fluorescence microscope. As shown in , the fluorescence signal of the PAMAM–Ac–FITC–LCTP-treated NCI-H460 cells () was much stronger than those from the two conjugates-treated 293T cells and the PAMAM–Ac–FITC-treated NCI-H460 cells. A clear image was observed under confocal microscopy. shows the image of PAMAM–Ac–FITC–LCTP-treated NCI-H460 cells.
Figure 5 In vitro evaluation of the conjugates (fluorescence-labeled acetylated polyamidoamine and fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide) targeting to 293T and NCI-H460 cells determined by inverted fluorescence microscopy. The cells were incubated with the conjugates (0.5 μM) for 4 hours. 100 × (A) 293T cells incubated with fluorescence-labeled acetylated polyamidoamine. B) 293T cells incubated with fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide. C) NCI-H460 cells incubated with fluorescence-labeled acetylated polyamidoamine. D) NCI-H460 cells incubated with fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide. E) local amplification of D.
In vivo targeting assay
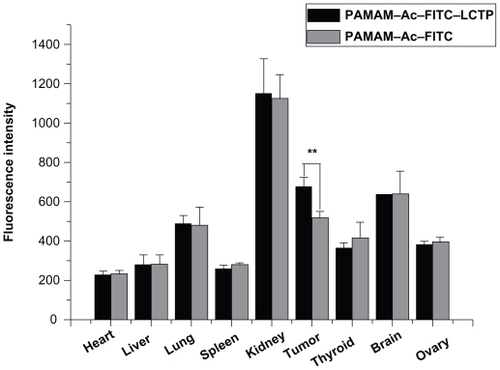
The distribution of PAMAM–Ac–FITC and PAMAM–Ac–FITC–LCTP in different organs was evaluated by measuring the fluorescence intensity of each organ at 4 hours after intravenous injection. showed the fluorescence density of the main organs. The highest density of the two conjugates was found in the kidney next in the tumors, followed by the brain and the lung. There was no difference in the distribution of the two conjugates among all the other organs, except in the tumors where the distribution of PAMAM–Ac–FITC–LCTP was significantly higher than that of PAMAM–Ac–FITC (P < 0.01).
Figure 6 Distribution of fluorescence-labeled acetylated polyamidoamine and fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide 4 hours postinjection in athymic mice bearing nonsmall cell lung cancer tumor xenografts, the relative fluorescence intensity per unit area data are presented as mean ± standard deviation (n = 6, **P < 0.01 fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide versus fluorescence-labeled acetylated polyamidoamine in tumor).
Discussion
In vivo phage display has been proven to be a useful method in isolating organ- or tumor-specific binding probes. The random sequence oligonucleotides were inserted into a specific site of phage DNA, then random sequence peptides or proteins were expressed and exposed on the phage surface. Because organs and tumors carry unique marker molecules,Citation27,Citation37 phage display can be used to screen organ- or tumor-specific peptide sequences for targeted drug delivery. The present study used an in vivo phage display method to screen peptides. Cyclic peptides have higher stability and binding capability,Citation38 so the Ph.D.-C7C phage display peptide library kit was used, in which the inserted peptide were cyclized by two cysteine residues. As shown in the Table, the phage inserted ACPLSHSLIC sequence was amplified and enriched. Among the total of 37 phage displayed peptide sequences, 28 were the same, which was the ACPLSHSLIC peptide.
The cationic PAMAM has hemolytic toxicity.Citation39 Several methods to decrease the hemolytic toxicity of PAMAM have been reported, one of which is the PEG modification.Citation40–Citation42 PEGylation can inhibit the nonspecific interactions between PAMAM and biomolecules and improve the biocompatibility of PAMAM, and can also decrease the immunogenicity and antigenicity of PAMAM. Another method is acetylation, which can decrease the positive charge density of PAMAM. In our studies, partially acetylated PAMAM displayed higher cell viability than PAMAM by the MTT assay. Further conjugation of FITC and the LCTP to the PAMAM–Ac had no influence on the biocompatibility of the dendrimer ().
The uptake of PAMAM–Ac–FITC–LCTP by both 293T and NCI-H460 cells was found to be time- and dose-dependent. However, the uptake efficiency of PAMAM–Ac–FITC–LCTP by NCI-H460 cells was much higher than that of PAMAM–Ac–FITC. In contrast, the uptake of both conjugates by 293T cells was very low. Similar results were previously reported by Hill et alCitation16 and Shukla et al.Citation43 In their studies, RGD peptide drove PAMAM conjugates specifically to MDPC-23 and HUVEC cancer cells, and enhanced cellular uptake. The free LCTP could greatly decrease NCI-H460 cell uptake of PAMAM–Ac–FITC–LCTP, but had no effect on NCI-H460 cell uptake of PAMAM–Ac–FITC and 293T cell uptake of PAMAM–Ac–FITC–LCTP and PAMAM–Ac–FITC. These observations suggest that LCTP played an important role in driving the conjugates highly and selectively to bind the membrane of NCI-H460 cells, thus facilitating the endocytosis process.
Because the penetration of FITC fluorescence through the body of mice is weak, in vivo imaging of live athymic mice bearing nonsmall cell lung cancer tumors cannot provide an exact fluorescence intensity. In this study, the tumors and organs were harvested and imaged ex vivo. The distribution of PAMAM–Ac–FITC and PAMAM–Ac–FITC–LCTP conjugates in each organ was almost the same except that the tumors had significantly higher uptake of PAMAM–Ac–FITC–LCTP than PAMAM–Ac–FITC (P < 0.01). This result suggests that the targeting effect of LCTP might enhance the permeability of PAMAM–Ac–FITC–LCTP in nonsmall cell lung cancer tumor. Indeed, the difference of tumor uptake for the two conjugates was not remarkable, and more experiments need to be done to improve the targeting effect of the conjugate. The distribution of the conjugates among different organs was found in the following order: kidney > tumor ≥ brain > lung > thyroid ≥ ovary> liver ≥ spleen ≥ heart (). The highest distribution of the conjugates was found in the kidney 4 hours after injection which is probably due to the fact that kidney is the main organ that excretes waste out of the body. And four hours after the injection, part of the conjugates had moved to the metabolic system, which is a phenomenon often observed in drug pharmacokinetic studies.Citation15,Citation25 It was interesting that the distribution of the conjugate in the brain was similar to that in the tumors, indicating that this PAMAM conjugate can penetrate the blood–brain barrier and enter the brain.Citation44,Citation45
Although antibody, folic acid, and biotin are often used as targeting agents, the peptides screened here by an in vivo phage display method could target the unique receptors expressed by cancer cells, thus remarkably improved the targeting capability for chemotherapy.
Conclusion
In summary, NCI-H460 cell-specific LCTP was screened by the phage display method and conjugated to the acetylated PAMAM dendrimer. The specific LCTP-modified PAMAM dendrimer could be easily taken up by NCI-H460 cells in vitro and by tumors in vivo. These results indicate that the LCTP–dendrimer conjugate might be a useful drug carrier for cancer clinical diagnosis and treatment.
Acknowledgments
This study was financially support by the National Natural Science Foundation of China (30700178 and 20774050), the National Outstanding Youth Fund (30725030), Tianjin Natural Science Foundation (09JCYBJC13400), and the Development Foundation of the Institute of Radiation Medicine, Chinese Academy of Medical Sciences and Peking Union Medical College (SF0627, SF0823).
Supplementary Material
Figure S2 1H nuclear magnetic resonance spectrum (300 mHz,D2O) of the fluorescence-labeled acetylated polyamidoamine.
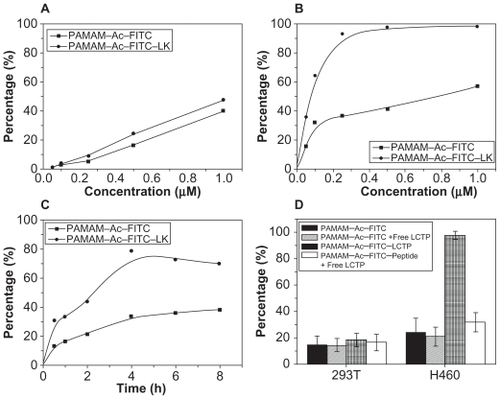
Figure S3 1H nuclear magnetic resonance spectrum (300 mHz, D2O) of the fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide.
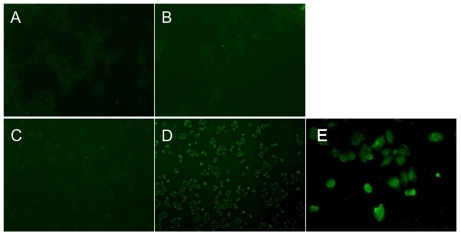
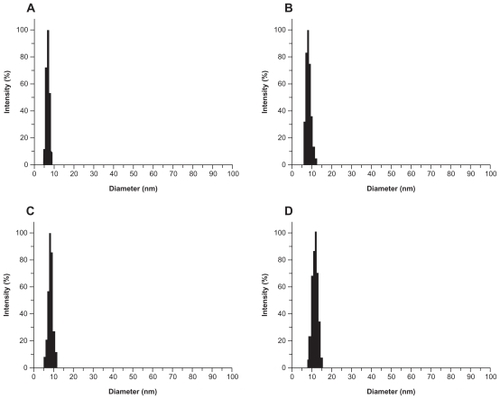
Figure S4 Laser light-scattering assay results. The polyamidoamine particle size was about 7 nm (A) and the particle size of acetylated polyamidoamine (B) fluorescence-labeled acetylated polyamidoamine (C), and fluorescence-labeled acetylated polyamidoamine lung cancer targeting peptide (D) becomes bigger and bigger.
Disclosure
The authors report no conflicts of interest in this work.
References
- LammersTHenninkWEStormG Tumour-targeted nanomedicines: Principles and practice Br J Cancer 2008 3 99 392 397 18648371
- PrajapatiRNTekadeRKGuptaUGajbhiyeVJainNK Dendimer-mediated solubilization: Formulation development and in vitro-in vivo assessment of piroxicam Mol Pharm 2009 6 3 940 950 19231841
- SvensonSChauhanAS Dendrimers for enhanced drug solubilization Nanomed 2008 3 5 679 702
- PatilMLZhangMBetigeriSTaratulaOHeHMinkoT Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery Bioconjug Chem 2008 19 7 1396 1403 18576676
- KaminskasLMKellyBDMcLeodVM Pharmacokinetics and tumor disposition of PEGylated, methotrexate conjugated poly-l-lysine dendrimers Mol Pharm 2009 6 4 1190 1204 19453158
- PanwarPPandeyBLakheraPCSinghKP Preparation, characterization, and in vitro release study of albendazole-encapsulated nanosize liposomes Int J Nanomedicine 2010 5 101 108 20309396
- GaucherGMarchessaultRHLerouxJC Polyester-based micelles and nanoparticles for the parenteral delivery of taxanes J Control Release 2010 143 1 2 12 19925835
- MatsumotoSChristieRJNishiyamaN Environment-responsive block copolymer micelles with a disulfide cross-linked core for enhanced siRNA delivery Biomacromolecules 2009 10 1 119 127 19061333
- SamadAAlamMISaxenaK Dendrimers: A class of polymers in the nanotechnology for the delivery of active pharmaceuticals Curr Pharm Des 2009 15 25 2958 2969 19754372
- ShuklaRThomasTPPetersJL HER2 specific tumor targeting with dendrimer conjugated anti-HER2 mAb Bioconjug Chem 2006 17 5 1109 1115 16984117
- SinghPGuptaUAsthanaAJainNK Folate and folate-PEG-PAMAM dendrimers: Synthesis, characterization, and targeted anticancer drug delivery potential in tumor bearing mice Bioconjug Chem 2008 19 11 2239 2252 18950215
- SwansonSDKukowska-LatalloJFPatriAK Targeted gadolinium-loaded dendrimer nanoparticles for tumor-specific magnetic resonance contrast enhancement Int J Nanomedicine 2008 3 2 201 210 18686779
- YangWChengYXuTWangXWenLP Targeting cancer cells with biotin-dendrimer conjugates Eur J Med Chem 2009 44 2 862 868 18550227
- YangHKaoWJ Synthesis and characterization of nanoscale dendritic RGD clusters for potential applications in tissue engineering and drug delivery Int J Nanomedicine 2007 2 1 89 99 17722516
- BoswellCAEckPKReginoCA Synthesis, characterization, and biological evaluation of integrin alphavbeta3-targeted PAMAM dendrimers Mol Pharm 2008 5 4 527 539 18537262
- HillEShuklaRParkSSBakerJRJr Synthetic PAMAM-RGD conjugates target and bind to odontoblast-like MDPC 23 cells and the predentin in tooth organ cultures Bioconjug Chem 2007 18 6 1756 1762 17970585
- LeeTYLinCTKuoSYChangDKWuHC Peptide-mediated targeting to tumor blood vessels of lung cancer for drug delivery Cancer Res 2007 67 22 10958 10965 18006841
- ChabreYMRoyR Recent trends in glycodendrimer syntheses and applications Curr Top Med Chem 2008 8 14 1237 1285 18855708
- BhadraDYadavAKBhadraSJainNK Glycodendrimeric nanoparticulate carriers of primaquine phosphate for liver targeting Int J Pharm 2005 295 1 2 221 233 15847986
- WuGBarthRFYangWKawabataSZhangLGreen-ChurchK Targeted delivery of methotrexate to epidermal growth factor receptor-positive brain tumors by means of cetuximab (IMC-C225) dendrimer bioconjugates Mol Cancer Ther 2006 5 1 52 59 16432162
- KurtogluYEMishraMKKannanSKannanRM Drug release characteristics of PAMAM dendrimer-drug conjugates with different linkers Int J Pharm 2010 384 1 2 189 194 19819318
- BharaliDJKhalilMGurbuzMSimoneTMMousaSA Nanoparticles and cancer therapy: A concise review with emphasis on dendrimers Int J Nanomedicine 2009 4 1 7 19421366
- SmithGP Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface Science 1985 228 4705 1315 1317 4001944
- ScottJKSmithGP Searching for peptide ligands with an epitope library Science 1990 249 4967 386 390 1696028
- HuiXHanYLiangS Specific targeting of the vasculature of gastric cancer by a new tumor-homing peptide CGNSNPKSC J Control Release 2008 131 2 86 93 18700158
- YangWLuoDWangS TMTP1, a novel tumor-homing peptide specifically targeting metastasis Clin Cancer Res 2008 14 17 5494 5502 18765541
- PasqualiniRRuoslahtiE Organ targeting in vivo using phage display peptide libraries Nature 1996 380 6572 364 366 8598934
- PasqualiniRKoivunenERuoslahtiE Alpha v integrins as receptors for tumor targeting by circulating ligands Nat Biotechnol 1997 15 6 542 546 9181576
- HowellRCRevskayaEPazoVNosanchukJDCasadevallADadachovaE Phage display library derived peptides that bind to human tumor melanin as potential vehicles for targeted radionuclide therapy of metastatic melanoma Bioconjug Chem 2007 18 6 1739 1748 17907769
- ZitzmannSMierWSchadA A new prostate carcinoma binding peptide (DUP-1) for tumor imaging and therapy Clin Cancer Res 2005 11 1 139 146 15671538
- ZitzmannSKrämerSMierW Identification and evaluation of a new tumor cell-binding peptide, FROP-1 J Nucl Med 2007 48 6 965 972 17504878
- ChenBCaoSZhangY A novel peptide (GX1) homing to gastric cancer vasculature inhibits angiogenesis and cooperates with TNF alpha in anti-tumor therapy BMC Cell Biol 2009 10 63 19740430
- WuXYanQHuangY Isolation of a novel basic FGF-binding peptide with potent antiangiogenetic activity J Cell Mol Med 2010 14 1 2 351 356
- LaakkonenPPorkkaKHoffmanJARuoslahtiE A tumor-homing peptide with a targeting specificity related to lymphatic vessels Nat Med 2002 8 7 751 755 12053175
- LesniakWGKariapperMSNairBM Synthesis and characterization of PAMAM dendrimer-based multifunctional nanodevices for targeting alphavbeta3 integrins Bioconjug Chem 2007 18 4 1148 1154 17566976
- ThannhauserTWKonishiYScheragaHA Analysis for disulfide bonds in peptides and proteins Methods Enzymol 1987 143 115 119 3657523
- LeeGGeB Cancer cell expressions of immunoglobulin heavy chains with unique carbohydrate-associated biomarker Cancer Biomark 2009 5 4 177 188 19729827
- PakkalaMHekimCSoininenP Activity and stability of human kallikrein-2-specific linear and cyclic peptide inhibitors J Pept Sci 2007 13 5 348 353 17436344
- MalikNWiwattanapatapeeRKlopschR Dendrimers: Relationship between structure and biocompatibility in vitro, and preliminary studies on the biodistribution of 125I-labelled polyamidoamine dendrimers in vivo J Control Release 2000 65 1 2 133 148 10699264
- SinghPGuptaUAsthanaAJainNK Folate and folate-PEG-PAMAM dendrimers: Synthesis, characterization, and targeted anticancer drug delivery potential in tumor bearing mice Bioconjug Chem 2008 19 11 2239 2252 18950215
- ToshinariTEijiYChieKAtsushiHKenjiK Synthesis of a polyamidoamine dendron-bearing lipid having sugar moieties and its use for preparation of nonviral gene vectors Res Chem Intermed 2009 35 8 9 1005 1014
- DuanYYangCZhangZLiuJZhengJKongD Poly(ethylene glycol)-grafted polyethylenamine modified with G250 monoclonal antibody for tumor gene therapy Hum Gene Ther 2010 21 2 191 208 19788387
- ShuklaRThomasTPPetersJKotlyarAMycABakerJRJr Tumor angiogenic vasculature targeting with PAMAM dendrimer-RGD conjugates Chem Commun (Camb) 2005 14 46 5739 5741 16307130
- SarinHKanevskyASWuH Effective transvascular delivery of nanoparticles across the blood-brain tumor barrier into malignant glioma cells J Transl Med 2008 6 80 19094226
- PrietoMJSchilrreffPTesorieroMVMorillaMJRomeroEL Brain and muscle of Wistar rats are the main targets of intravenous dendrimeric sulfadiazine Int J Pharm 2008 360 1 2 204 212 18583072