Abstract
Background
Circulating tumor cells (CTCs) detection, an approach considered to be “liquid biopsy”, is a potential alternative method in clinical use for early diagnosis of solid tumor progression.
Methods
In this study, we developed a poly (lactic-co-glycolic acid) (PLGA) – nanofiber (PN)-NanoVelcro chip as an efficient device for simple and rapid capture of CTCs from peripheral blood. We evaluated the device performance by assessing the capture efficiency and purity. Single CTC was isolated via laser microdissection system for subsequent genetic analysis, with an aim to find the concordance of epidermal growth factor receptor (EGFR) mutations between tumor tissue and CTCs.
Results
PN-NanoVelcro chip exhibits great performance in capture efficiency and high purity. The genetic analysis results showed that most EGFR mutation in tumor tissue could also be detected in CTCs.
Conclusion
Compared to computed tomography image results, CTC detection can be implemented throughout the course of diseases and provides an accurate and earlier diagnosis of tumor progression, which make it possible for patients to acquire suitable and timely treatment.
Introduction
Lung cancer is the main cause of cancer-related death due to its late prognosis and poor therapy, leading to 1.6 million deaths annually.Citation1 Non-small cell lung cancer (NSCLC), accounting for ~80% of lung cancers, can also be divided into two subtypes: adenocarcinoma and squamous cell carcinoma. Lung adenocarcinoma (LADC), representing about one-half of all lung cancers, is the most frequent histologic subtype of NSCLC.Citation2 Although significant advancement has been achieved in chemotherapy, targeted therapy and surgical technique, the average 5-year survival rate of NSCLC was just 18% due to its late diagnosis and lack of timely and efficient treatment.Citation3 As known to all, metastasis mostly contributes to the progression of cancer, so early detection of metastasis is of great significance for early intervention and individual treatment.
During the progression of cancer metastasis, circulating tumor cells (CTCs) are shed from the solid primary tumor, enter into the blood stream and travel to remote tissues. In recent years, numerous researches have demonstrated that CTC analysis could be used in detecting early tumor progression of various kinds of cancer, including breast, colorectal, prostate cancers and so on.Citation4–Citation7 Thus, CTC analysis is now regarded as “liquid biopsy” and has been used in clinical application to predict the clinical outcomes and monitor treatment responses throughout the course of diseases. Although Cell Search™ had been allowed by the US Food and Drug Administration to detect CTCs in the clinical application in 2004, it was still constrained by its low sensitivity in epithelial cell adhesion molecule (EpCAM)-negative cases.Citation8 With the development of technology, various approaches have been found for capturing rare CTCs in peripheral blood more conveniently and efficiently, including capture agent-labeled magnetic beads,Citation9,Citation10 microfluidic deviceCitation11 and microfilter device.Citation12,Citation13 However, due to the extremely low number of CTCs in peripheral blood and limitation of technology, enrichment and detection approaches still need to be further improved for the sake of identifying and characterizing CTCs.
Herein, we developed polymer nanofiber-based microchips to capture CTCs and implemented Laser Microdissection System to isolate single CTC, followed by subsequent genetic analysis. Recent advances in amplification refractory mutation system polymerase chain reaction (ARMS-PCR) have made it possible to investigate the genomic alterations in CTCs.Citation14 With the assistance of polymer nanofiber-based microchips and genetic analysis technologies, CTC detection could be used in timely detection of progression of cancer.
Epidermal growth factor receptor (EGFR), a member of the ErbB family, is a transmembrane tyrosine kinase receptor.Citation15 Accumulating researches had demonstrated that EGFR mutation occurred frequently in non-smoking female Asian patients with LADC.Citation16–Citation18 Gefitinib, an EGFR tyrosine kinase inhibitor (TKI), is widely used in clinical application as a molecular-targeting drug in the treatment of NSCLC. Unlike those patients with EGFR wild-type tumors, patients with EGFR mutations (exon 21 L858R mutation or exon 19 deletions) benefit much from EGFR-TKI treatment.Citation19 However, although patients responded well to EGFR-TKI treatment initially, they would finally develop resistance and about 50% of this resistance could be attributed to T790M mutation, the emergence of EGFR-resistant mutation.Citation20 Currently, tumor tissues obtained from surgery are used as a standard sample for EGFR mutation detection in the clinical practice, but it still has several limitations.Citation21 Herein, our present study aims to develop CTC analysis as a promising, noninvasive alternative for EGFR mutation detection in clinical application.
Materials and methods
Polymer nanofiber-based microchips preparation and surface coating
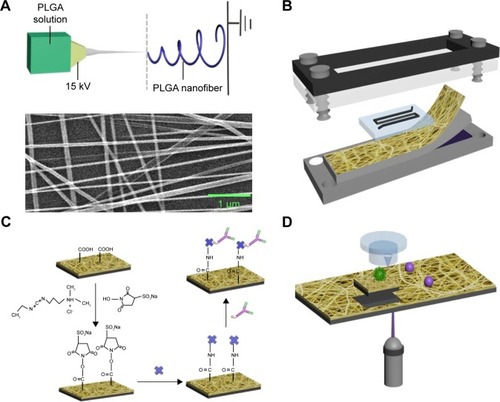
PLGA was fabricated onto the substrates by using electrospinning technology (). The polymer nanofiber-based microchip comprises two functional components, including an overlaid polydimethylsiloxane (PDMS) chaotic mixer and a transparent poly(lactic-co-glycolic acid) (PLGA) – nanofiber (PN)-NanoVelcro substrate (). PLGA, a polymeric material with extremely good compatibility, was electrospun onto laser capture microdissection (LCM) glass slide. PLGA nanofibers were covalently conjugated with streptavidin through 1-ethyl-3-(3-dimethylamino-propyl)carbodiimide/N-hydroxy-succinimide chemistry, which was employed to activate the carboxylic acid groups of PLGA. Then, streptavidin was conjugated with anti-EpCAM, which especially binds to EpCAM on the CTCs ().
Figure 1 The configuration of PLGA nanofiber microchip for the high-purity isolation of CTCs.
Notes: (A) A transparent PN-NanoVelcro substrate was prepared by depositing electrospun PLGA nanofibers onto a commercial LMD glass slide (with a pre-deposited 1.2 µm thick PPS membrane). The SEM image of the electrospun PLGA nanofibers is shown. (B) A custom-designed chip holder is used to sandwich a microchip that is composed of an overlaid PDMS chaotic mixer chip. (C) Schematic representation of the chemistry of EpCAM-modified PLGA for cancer cell capture applications. (D) Single CTC isolation by laser microdissection.
Abbreviations: CTCs, circulating tumor cells; EpCAM, epithelial cell adhesion molecule; LMD, laser microdissection; PLGA, poly(lactic-co-glycolic acid); SEM, scanning electron micrograph.
Patient characteristics and clinical samples
A total of 72 NSCLC patients were enrolled in the study, consisting of 27 males (37.50%) and 45 females (62.50%), including 18 smokers and 54 non-smokers. The patients could be divided into four groups according to the clinical stages. Other characteristics are clearly listed in . Written informed consent was obtained from every patient.
Table 1 Clinical characteristics of 72 NSCLC patients
Blood samples and tissue samples from both patients with lung cancer and healthy donors were recruited from the First Affiliated Hospital of Sun Yat-sen University and were used to perform subsequent studies. The blood samples were drawn into vaccutainer collection tubes containing EDTA and processed within 12 h, while the tissue samples were stored in liquid nitrogen. This study was approved by the institutional ethics committee of the First Affiliated Hospital of Sun Yat-sen University.
CTCs enrichment procedure
One milliliter of whole blood sample was initially subjected to red blood cell lysis buffer followed by 20 min of incubation at room temperature until the red blood cells were completely lysed. Then the supernatant was removed after centrifugation at 200× g at 4°C for 5 min. After centrifugation, the cell pellets were resuspended by the same volume of 1× PBS. SK-BR-3 cells (American Type Culture Collection, Manassas, VA, USA), stained with fluorescent markers from Circulating Tumor Cell Control Kit (Cell Search, San Diego, CA, USA), were used to spike into the suspension solution as artificial CTC blood sample. The same number of SK-BR-3 cells was spiked into another 1 mL of whole blood to mimic the CTCs in peripheral blood. CTC enrichment was implemented through polymer nanofiber-based microchips at a flow rate of 1 mL/h at room temperature for 1 h.
Immunostaining and imaging
NCl-H1975 cells, a cell line of LADC which was purchased from Shanghai Institute of Cell Biology, were spiked in whole blood for the examination of performance of polymer nanofiber-based microchips. After washing with 1× PBS for three times, the captured cells were fixed with 4% paraformaldehyde for 20 min, permeated with 0.1% triton X-100 in 1× PBS for 5 min and then blocked with 5% bovine serum albumin for half an hour. Subsequently, the cells were stained with mouse anti-human pan cytokeratin (Abcam, Cambridge, UK) and rabbit anti-human CD45 (CST, MA, USA) overnight at 4°C, followed by washing with 1× PBS for three times. After washing with 1× PBS, the cells were stained with fluorescein isothiocyanate-labeled goat-anti-mouse secondary antibody and tetramethylrhodamine-labeled goat-anti-rabbit secondary antibody for 1 h at room temperature. After thorough washing, 4,6-diamino-l2-phenyl indole (DAPI) was used for nuclear staining. CTCs were identified under a fluorescence microscope.
Laser microdissection and CTCmutation analysis
After confirming the locations of the CTCs by immunostaining, the microchip was placed on the LCM microscope (ArcturusXT™), which is used for isolating a single cell.Citation12 Then, single CTC was dissected by laser () and collected in 200 µL tube caps, with 10 µL lysis solution (40 µg/mL proteinase K in PBS, 0.5% Triton X-100) on the inside surface of it.Citation14,Citation22 The purified CTCs were analyzed through subsequent targeting sequencing and whole genome amplification (WGA).
Concordance of EGFR mutations in primary tissues and CTCs
We recovered single CTC through white blood cell (WBC) depletion on the LCM device and analyzed EGFR mutations in the isolated CTCs in comparison with its primary tumor. Targeted sequencing technology was used to find out the similarity of EGFR mutations in CTCs and primary tissues. KAPA Hyper Prep Kit (Kapa Biosystems, Wilmington, MA, USA) and the SureSelect Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA) were used following the instructions to prepare sequencing libraries for targeted sequencing analysis. Then, targeted sequencing was performed on an Illumina HiSeq 2000 (Illumina, San Diego, CA, USA), and the results were analyzed by the Genomics Workbench software (Agilent Technologies).
Genomic DNA extraction and genetic analysis
Genomic DNA of primary tissue and CTCs were extracted by QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Genomic DNA was eluted with 10 µL nuclease-free water for subsequent analysis. 3 μL of the extracted DNA was used for WGA using the Ampli1™ WGA Kit (Silicon Biosystems, Zona Artigianale BO, Italy) according to manufacturer’s protocol, and 2 µL products were analyzed for quality control by utilizing the Ampli1 QC Kit (Silicon Biosystems).Citation23
Polymerase chain reaction (PCR) was used to separately amplify EGFR exons 20 and 21 fragments containing the mutation sites. The PCR steps were performed in a 25 µL reaction volume, containing 2 µL WGA products, 2 µL sense primer (10 µmol/L), 2 µL antisense primer (10 µmol/L), 2.5 µL 10× PCR buffer, 2.5 µL MgCl2 (25 mmol/L), 1 µL deoxyribonucleotide triphosphate (10 nmol/L), 0.5 µL Taq-polymerase and 12.5 µL ddH2O. The thermocycling conditions for exon 20 and exon 21 were as follows: 1 cycle of 95°C for 10 min, 40 cycles of 94°C for 30 s and 58°C for 1 min, followed by 4°C hold. PCR products were detected by gel electrophoresis on a 1% agarose gel.
L858R mutation in exon 20 and T790M mutation in exon 21 were detected using ARMS-PCR. AMPS-PCR was performed according to the protocol of the Human EGFR Gene Mutation Quantitative Detection Kit (fluorescent PCR; Beijing ACCB Biotech Ltd, Beijing, China). Also, the thermal cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s, and fluorescent signals were collected at the end of each cycle of 60°C for 60 s.Citation24
Relationship between T790M and gefitinib-induced acquired resistance
Computed tomography (CT) was performed to observe tumor progression, while ARMS-PCR was performed to detect the mutations of L858R and T790M during continuous gefitinib treatment, aiming to find out the relationship between specific EGFR mutation and gefitinib-induced acquired resistance.
Results
Fabrication of PLGA nanofibers
PLGA was fabricated onto the substrates by using electro-spinning technology. As shown in the scanning electron micrograph, the nanofibers were smooth, randomly orientated and uniform, without any defects of curved and cross-linked nanofibers by fabricating at 15 cm receiving distance, using 0.5 mL/h as the feeding rate ().
Cell capture in the nanofiber-embedded microchip
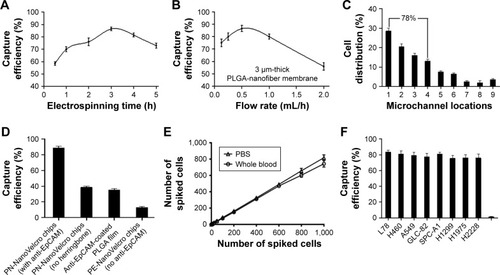
During the optimization test of electrospinning time, we discovered that 3 h of deposition led to better CTC capture efficiency (). The 3 µm thick PLGA nanofibers were fabricated through electrospining for 3 h for the following experiment. Apart from the electrospinning time, flow rate was another significant condition that could not be ignored. Flow rates ranging from 0.1 to 2 mL/h were used to optimize the flow rate for subsequent studies. As shown, 0.5 mL/h flow rate could achieve the best capture performance (). Also, the distribution of cells was mainly focused on the first four microchannels, accounting for about 78% (). In addition, PN-NanoVelcro chip design had an effect on the capture efficiency. As observed, the capture efficiency of PN-NanoVelcro chips with herringbone and modified with EpCAM could reach ~90%, much higher than PN-NanoVelcro chips with no herringbone, PLGA film coated with anti-EpCAM and PN-NanoVelcro chips with no anti-EpCAM (). In order to evaluate the capture efficiency for different CTC numbers, different numbers of SK-BR-3 cells prestained with DIO, ranging from 10 to 1,000, were spiked into both PBS and whole blood. Consistent recovery rates were observed at various numbers of spiked cells as low as 10 cells/mL (). Various kinds of cell lines of NSCLC were used to prove that PN-NanoVelcro chips could specifically capture cancer cells instead of WBCs ().
Figure 2 Optimization and validation of the PN-NanoVelcro chip using circulating tumor cell control kit and various cell lines.
Notes: (A) The cell capture efficiency of PN-NanoVelcro chip at different electrospinning time points. (B) The cell capture efficiency of PN-NanoVelcro chip in different electrospinning flow rates. (C) The distribution of cell in different microchannels at flow rate of 1.0 mL/h. (D) Comparison of the capture performance between PN-NanoVelcro chip and four different controls: 1) PN-NanoVelcro chip with herringbone and anti-EpCAM; 2) PN-NanoVelcro chip without herringbone structure; 3) anti-EpCAM-coated PLGA thin film with electrospinning; 4) PE-NanoVelcro chip without anti-EpCAM. (E) Capture efficiencies at different spiking cell numbers ranging from 10 to 1,000/mL. (F) Capture efficiencies of different cell lines of NSCLC and WBCs.
Abbreviations: EpCAM, epithelial cell adhesion molecule; NSCLC, non-small cell lung cancer; PLGA, poly(lactic-co-glycolic acid); WBC, white blood cell.
Immunostaining for CTC identification
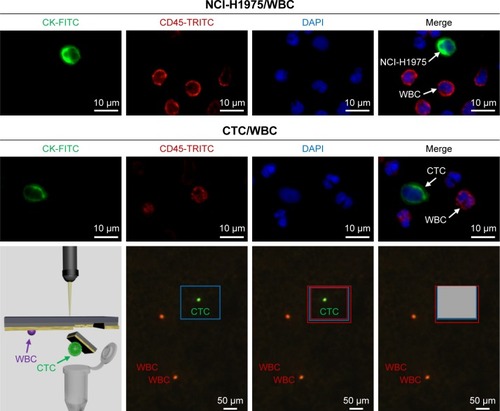
In order to confirm the feasibility of three-color immunostaining method for differentiating CTCs from WBCs, NCI-H1975 was chosen as a positive control. The CK+/CD45−/DAPI+ cells were defined as CTC candidates, while the CK−/CD45+/DAPI+ cells were considered as WBCs, and their locations were recorded and double-checked under 40× objectives for cellular morphology ().
Figure 3 Identification of CTC, based on a three-color immunostaining using FITC-conjugated anti-CK, TRICT-conjugated anti-CD45 (a marker of WBC) and DAPI (nuclear specific).
Notes: NCl-H1975 cells spiked in normal blood were used as a positive control. Isolating a single CTC was performed through laser microdissection after confirming the location by immunostaining.
Abbreviations: CTC, circulating tumor cell; DAPI, 4,6-diamino-l2-phenyl indole; FITC, fluorescein isothiocyanate; TRICT, tetramethylrhodamine; WBC, white blood cell.
EGFR mutations presented in both primary tissues and CTCs
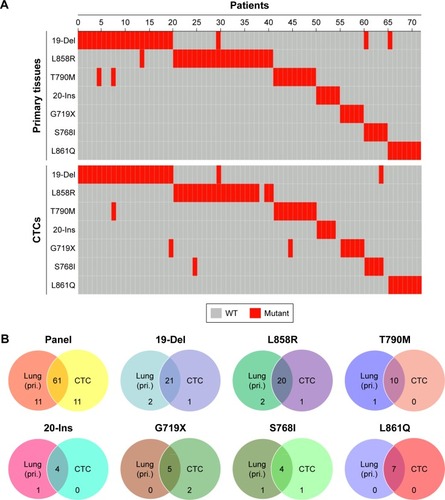
The mutational landscapes showed the specific mutation sites in primary tissues and CTCs of the 72 NSCLC patients, including 19-Del, L858R, T790M, 20-Ins, G719X, S768I and L861Q (). On comparing the mutation sites between primary tissues and CTCs, we found that the mutation sites were almost consistent to some extent. Also, the Venn diagrams demonstrated that EGFR mutations in CTCs were almost in concordance with those in primary tissues ().
Figure 4 Concordance of EGFR mutation sites detected in primary tissues and CTCs from NSCLC patients.
Notes: (A) Landscape of somatic mutations detected in primary tissues and CTCs from 72 patients with NSCLC. Seven EGFR alterations were detected by targeted sequencing and the results were computed in the mutational landscape. (B) The Venn diagrams were used to summarize the similarity of EGFR mutations in primary tissues and CTCs from 72 NSCLC patients.
Abbreviations: CTC, circulating tumor cell; EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer; pri., primary tissue; WT, wild type.
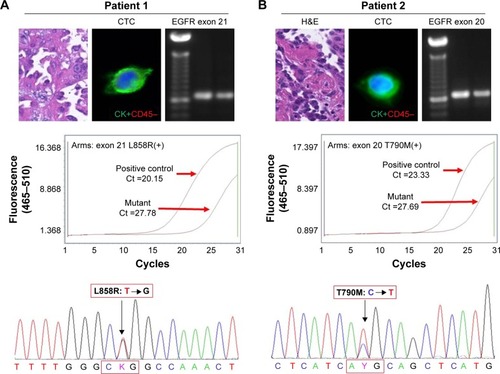
EGFR L858R (exon 21) and T790M (exon 20) sequencing of tissues and CTCs
PCR results also showed that mutations in both exon 20 and exon 21 could be detected in the primary tissue and CTCs, following which the ARMS-PCR results further confirmed L858R mutation in exon 21 and T790M mutation in exon 20 in the CTCs. Finally, Sanger sequencing technology was implemented to confirm the specific mutation sites of L858R and T790M. As shown in the results, we can primarily come to a conclusion that EGFR mutations found in primary tissues could also be found in CTCs, which may provide us with a new insight that CTC detection could be a substitute for primary tissue biopsy ().
Figure 5 EGFR L858R (exon 21) and T790M (exon 20) sequencing of primary tissues and CTCs.
Notes: (A) PCR of exon 21 was successfully performed and a band of proper size was found with 2% gel electrophoresis. ARM-PCR and Sanger sequencing were used to detect the mutation sites in L858R. (B) PCR of exon 20 was successfully performed and a band of proper size was found with 2% gel electrophoresis. ARM-PCR and Sanger sequencing were used to detect the mutation sites in T790M.
Abbreviations: ARM-PCR, amplification refractory mutation system PCR; CTCs, circulating tumor cells; EGFR, epidermal growth factor receptor; PCR, polymerase chain reaction.
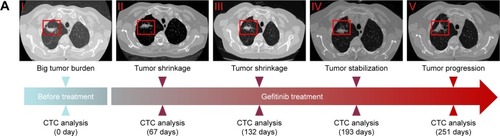
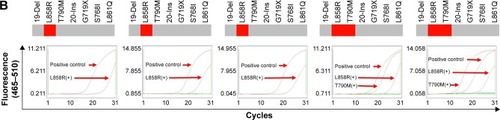
The correlation of T790M mutation and gefitinib-induced acquired resistance
As shown in the CT results () of primary tumor, the primary tumor appeared to shrink gradually after gefitinib treatment for 132 days. However, gefitinib seemed to be less effective after 193 days, and the tumor progressed after 251 days. CTCs were analyzed by ARMS-PCR at the same time during the time course. According to the AMRS-PCR results (), we discovered that only L858R mutation could be found in previous successful gefitinib treatment, while both L858R and T790M appeared in CTCs of NSCLC patients with acquired resistance. Comparing the results of ARMS-PCR to those of CT images, T790M mutation could be detected long before tumor progression, which means that CTC analysis can provide possibility for early prognosis.
Figure 6 The relationship between acquired resistance induced by continuous gefitinib treatment and EGFR mutation.
Notes: (A) Course of the disease with CT images and treatment history and driver mutation. (B) L858R and T790M mutation detection via ARMS-PCR.
Abbreviations: ARMS-PCR, amplification refractory mutation system polymerase chain reaction; CT, computed tomography; CTC, circulating tumor cell; EGFR, epidermal growth factor receptor.
Discussion
Lung cancer, the main cause of cancer-related death worldwide, has been a threat to health that could not be ignored.Citation25 Currently, tumor tissues used for EGFR mutations analysis are acquired through surgery. However, tumor tissues are unavailable in some cases, so a substitute method of biopsy is in great need. Taking the advantages into consideration, CTC detection is recognized to be a better choice not only for its acquisition without invasiveness, but also for its possibility to be conducted throughout the course of disease.
Our study aimed to develop a new method to capture CTCs with high efficiency and purity and implement genetic analysis of the isolated single CTC throughout the time course of disease. The evaluation results of optimization and validation of modified PN-NanoVelcro chip showed that the capture efficiencies of different cell lines of NSCLC could reach about 80%, which means that PN-NanoVelcro chip can be an effective method for capturing CTCs. Apart from developing a new approach to capture CTCs, we also found out the concordance of the EGFR mutation sites between tumor tissues and CTCs acquired from 72 NSCLC patients. In addition, the correlation between specific EGFR mutation sites and gefitinib-induced acquired assistance has also been preliminarily clarified, which may disclose the reasons for discouraging gefitinib treatment to some extent.
As known to all, EGFR mutation in the tumor tissues has a close relationship with NSCLC.Citation26 With a further study of the EGFR mutations in NSCLC, specific mutation sites with different functions have been discovered. Numerous researches have demonstrated that NSCLC patients with mutation sites such as 19-Del, L858R, 20-Ins, G719X, S768I and L861Q seemed to be sensitive to gefitinib and could undergo successful treatment.Citation27 According to the mutational spectrum in our study, we discovered similar mutation sites in primary tissues and CTCs, which makes observation of timely tumor progression possible.
As reported in clinical researches, NSCLC patients cannot benefit from EGFR-TKIs all along, as the majority of patients with EGFR mutation would eventually develop acquired resistance after 12–24 months of treatment.Citation28 T790M, a mutation of exon 20 in EGFR, has been reported to be closely related to acquired resistance induced by gefitinib.Citation29 Some researchers have found that T790M mutation reduced the affinity to EGFR-TKIs, which may be the cause of gefitinib-induced acquired resistance.Citation30 As shown in the CT results and ARMS-PCR results of L858R and T790M mutations during the time course of the disease, it is clear that NSCLC patients with L858R mutation can acquire obvious efficacy after initial treatment with gefitinib. However, gefitinib became less effective gradually or even lost its effect at last. Mutation analysis of tumor has shown that not only LR858 mutation but also T790M mutation appears in patients with acquired resistance. In general clinical application, L858R and T790M mutations are implemented through tumor tissues obtained from surgery. Compared with traditional tissue biopsy, CTC detection can be a better alternative to monitor patients’ state of illness for its noninvasive acquisition and simple procedure throughout the course of diseases. With our further research of CTCs in the following study, the mechanisms of NSCLC with and without EGFR mutations could be further clarified. Also, we would develop a technique to culture the isolated CTCs in vitro in the future, with an aim to find a suitable drug therapy for NSCLC patients and lay solid foundation for the realization of individual treatments.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30900650, 81372501, 81572260 and 81773299), the Guangdong Natural Science Foundation (2011B031800025, S2012010008378, 2015A030313036 and 2015A030313036) and the Guangzhou and Guangdong/Guangzhou Science and Technology Planning Program (2014J4100132, 2012B031800115, 2013B02180021, 2015A020214010, 2016A020215055, 201704020094, 16ykjc08 and 2015ykzd07).
Disclosure
The authors report no conflicts of interest in this work.
References
- ChengTYCrambSMBaadePDYouldenDRNwoguCReidMEThe International epidemiology of lung cancer: latest trends, disparities, and tumor characteristicsJ Thorac Oncol201611101653167127364315
- RocaEGurizzanCAmorosoVVermiWFerrariVBerrutiAOutcome of patients with lung adenocarcinoma with transformation to small-cell lung cancer following tyrosine kinase inhibitors treatment: a systematic review and pooled analysisCancer Treat Rev20175911712228806542
- TorreLASiegelRLJemalALung cancer statisticsAdv Exp Med Biol201689311926667336
- ZhangYLvYNiuYSuHFengARole of circulating tumor cell (CTC) monitoring in evaluating prognosis of triple-negative breast cancer patients in ChinaMed Sci Monit2017233071307928643770
- XuTShenGChengMXuWShenGHuSClinicopathological and prognostic significance of circulating tumor cells in patients with lung cancer: a meta-analysisOncotarget2017837625246253628977966
- WuWZhangZGaoXHClinical significance of detecting circulating tumor cells in colorectal cancer using subtraction enrichment and immunostaining-fluorescence in situ hybridization (SE-iFISH)Oncotarget2017813216392164928423493
- LiuWYinBWangXCirculating tumor cells in prostate cancer: precision diagnosis and therapyOncol Lett20171421223123228789337
- YangDWangLTianXApplication of circulating tumor cells scope technique on circulating tumor cell researchMol Cell Ther20142826056577
- KallergiGPolitakiEAlkahtaniSStournarasCGeorgouliasVEvaluation of isolation methods for circulating tumor cells (CTCs)Cell Physiol Biochem2016403–441141927889762
- VilaAAbalMMuinelo-RomayLEGFR-based immunoisolation as a recovery target for low-EpCAM CTC subpopulationPLoS One20161110e016370527711186
- LeeMKimEJChoYPredictive value of circulating tumor cells (CTCs) captured by microfluidic device in patients with epithelial ovarian cancerGynecol Oncol2017145236136528274569
- KangYTDohIByunJChangHJChoYHLabel-free rapid viable enrichment of circulating tumor cell by photosensitive polymer-based microfilter deviceTheranostics20177133179319128900503
- ZhouMDHaoSWilliamsAJSeparable bilayer microfiltration device for viable label-free enrichment of circulating tumour cellsSci Rep20144739225487434
- RanRLiLWangMWangSZhengZLinPPDetermination of EGFR mutations in single cells microdissected from enriched lung tumor cells in peripheral bloodAnal Bioanal Chem2013405237377738223828210
- LiYQLiuYSYingXWLentivirus-mediated disintegrin and metalloproteinase 17 RNA interference reversed the acquired resistance to gefitinib in lung adenocarcinoma cells in vitroBiotechnol Prog Epub2017927
- MitsudomiTKosakaTYatabeYBiological and clinical implications of EGFR mutations in lung cancerInt J Clin Oncol200611319019816850125
- LeeDHSrimuninnimitVChengRWangXOrlandoMEpidermal growth factor receptor mutation status in the treatment of non-small cell lung cancer: lessons learnedCancer Res Treat201547454955425943319
- LynchTJBellDWSordellaRActivating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinibN Engl J Med2004350212129213915118073
- XuMXieYNiSLiuHThe latest therapeutic strategies after resistance to first generation epidermal growth factor receptor tyrosine kinase inhibitors (EGFR TKIs) in patients with non-small cell lung cancer (NSCLC)Ann Transl Med2015379626015938
- LiangZChengYChenYEGFR T790M ctDNA testing platforms and their role as companion diagnostics: correlation with clinical outcomes to EGFR-TKIsCancer Lett201740318619428642172
- TakamochiKOhSMatsunagaTSuzukiKPrognostic impacts of EGFR mutation status and subtype in patients with surgically resected lung adenocarcinomaJ Thorac Cardiovasc Surg201715451768.e11774.e128826599
- CourtCMAnkenyJSShoSReality of single circulating tumor cell sequencing for molecular diagnostics in pancreatic cancerJ Mol Diagn201618568869627375074
- SchneckHBlasslCMeier-StiegenFAnalysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patientsMol Oncol20137597698623895914
- ZhangCWeiBLiPPrognostic value of plasma EGFR ctDNA in NSCLC patients treated with EGFR-TKIsPLoS One2017123e017352428333951
- SiegelRMaJZouZJemalACancer statistics, 2014CA Cancer J Clin201464192924399786
- LinCCHuangWLWeiFSuWCWongDTEmerging platforms using liquid biopsy to detect EGFR mutations in lung cancerExpert Rev Mol Diagn201515111427144026420338
- FuruyamaKHaradaTIwamaESensitivity and kinase activity of epidermal growth factor receptor (EGFR) exon 19 and others to EGFR-tyrosine kinase inhibitorsCancer Sci2013104558458923387505
- CappuzzoFMorabitoANormannoNEfficacy and safety of rechallenge treatment with gefitinib in patients with advanced non-small cell lung cancerLung Cancer201699313727565910
- LiWRenSLiJT790M mutation is associated with better efficacy of treatment beyond progression with EGFR-TKI in advanced NSCLC patientsLung Cancer201484329530024685306
- GiacconeGEGFR point mutation confers resistance to gefitinib in a patient with non-small-cell lung cancerNat Clin Pract Oncol20052629629716264986
