 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
To address the issue of delivery of proteins, a six-arm copolymer, six-arm poly (ε-caprolactone)–poly(ethylene glycol) (6S-PCL-PEG), was synthesized by a simple two-step reaction. Thereafter, the application of 6S-PCL-PEG as a protein carrier was evaluated.
Materials and methods
A six-arm copolymer, six-arm poly(ε-caprolactone) (6S-PCL), was synthesized by ring-opening polymerization, with stannous octoate as a catalyst and inositol as an initiator. Then, poly(ethylene glycol) (PEG) was linked with 6S-PCL by oxalyl chloride to obtain 6S-PCL-PEG. Hydrogen-1 nuclear magnetic resonance spectrum, Fourier-transform infrared spectroscopy, and gel-permeation chromatography were conducted to identify the structure of 6S-PCL-PEG. The biocompatibility of the 6S-PCL-PEG was evaluated by a cell counting kit-8 assay. Polymeric nanoparticles (NPs) were prepared by a water-in-oil-in-water double emulsion (W1/O/W2) solvent evaporation method. The size distribution and zeta potential of NPs were determined by dynamic light scattering. Transmission electron microscopy was used to observe the morphology of NPs. Drug-loading capacity, encapsulation efficiency, and the release behavior of ovalbumin (OVA)-loading NPs were tested by the bicinchoninic acid assay kit. The stability and activity of OVA released from NPs were detected and the uptake of NPs was evaluated by NIH-3T3 cells.
Results
All results indicated the successful synthesis of amphiphilic copolymer 6S-PCL-PEG, which possessed excellent biocompatibility and could formulate NPs easily. High drug-loading capacity and encapsulation efficiency of protein NPs were observed. In vitro, OVA was released slowly and the bioactivity of OVA was maintained for over 28 days.
Conclusion
6S-PCL-PEG NPs prepared in this study show promising potential for use as a protein carrier.
Introduction
Compared with many conventional treatments, therapeutics of peptides and proteins possess noteworthy advantages such as high effectiveness, excellent specificity, and good activity.Citation1 However, their clinical application has been limited by the protein-based drugs encountering several challenges, such as degradation by enzymes in the systemic circulation, poor bioavailability, and low cell permeability.Citation2 Therefore, to address these issues, extensive research efforts have gone into finding and characterizing suitable protein delivery carriers.Citation3–Citation5 Among these, many excellent carrier biomaterials have been discovered, such as poly(lactic-co-glycolic acid) (PLGA), poly(caprolactone) (PCL), mesoporous silica, chitosan, cyclodextrin, and carbon nanotubes.Citation6–Citation11 However, the challenges of low cell permeability and short biological half-life remain. The short biological half-life of protein-based drugs causes a short action time of drugs and affect the therapeutic effects.Citation12 Therefore, long-term sustained-release systems have emerged. Many researchers have put their efforts into this and various biodegradable protein delivery systems have been obtained. For example, Peng et al explored insulin–phospholipid complex-loaded poly(hydroxybutyrate-co-hydroxyhexanoate) nanoparticles (NPs) as a sustained drug delivery system to increase the biological half-life of insulin.Citation13 Xie et al used PLGA as a novel emulsifier to prepare hydrophilic protein-loaded solid lipid NPs to improve therapeutic efficiency.Citation14 Besides, amphiphilic polymers have many advantages and have been studied widely as well.Citation15–Citation18
Amphiphilic polymers have many advantages, such as a long circulation duration, high biocompatibility, and augmented loading capacity.Citation19–Citation21 Poly(ethylene glycol) (PEG) has been widely used in this regard. Conjugation of PEG has exhibited significantly improved hydrophilicity as well as prolonged drug circulation period in vivo, features which are considered beneficial for NP-based delivery systems.Citation22–Citation24 Therefore, various biodegradable amphiphilic block copolymers, such as poly(caprolactone)–poly(ethylene glycol) (PCL-PEG), poly(lactide)–poly(ethylene glycol) (PLA-PEG), and poly(lactide-glycolide)–poly(ethylene glycol) (PLGA-PEG) copolymers, have been synthesized and applied in drug delivery systems. Particularly noteworthy are star-shaped amphiphilic copolymers, which have attracted increasing attention because they exhibit many advantages over linear amphiphilic copolymers of the same molar mass, such as a smaller hydrodynamic diameter, lower solution viscosity, and higher stability.Citation25–Citation27 Moreover, several reports have demonstrated that star-shaped copolymers have other advantages in drug delivery compared with linear copolymers, such as prolonging sustained release and improving the drug-loading capacity.Citation28–Citation30
The overall aim of the present work was to develop a sustained-release star-shaped amphiphilic copolymer carrier [six-arm poly(ε-caprolactone)–poly(ethylene glycol) (6S-PCL-PEG)], which augments the encapsulation efficiency and the protein-loading capacity. Initial work in this study focused on the preparation of 6S-PCL-PEG by a two-step method which is simple and efficient.Citation31 Thereafter, ovalbumin (OVA) was used as a model protein and a double-emulsion solvent evaporation method was employed to prepare the NPs, followed by property analysis, in vitro stability, and release behavior studies of NPs. Further, the in vitro uptake of NPs by NIH-3T3 cells was performed to examine the potential of this formulation in delivering protein into living cells.
Materials and methods
Materials
ε-Caprolactone, stannous octoate, OVA, and polyvinyl alcohol (PVA) (Mn=30,000–70,000) were purchased from Sigma-Aldrich (St Louis, MO, USA). PEG (Mn=4,000 g/mol), inosi-tol, and oxalyl chloride (chemically pure) were bought from GuangFu Fine Chemical Research Institute (Tianjin, China). Calcium hydride, dichloromethane (CH2Cl2), methanol, and diethyl ether were procured from Tianjin Jiang Tian Chemical Technology Co. (Tianjin, China). Lipopolysaccharide (LPS), PBS, Tween 20, Roswell Park Memorial Institute (RPMI) 1640 (Hyclone), and fetal bovine serum (FBS) were supplied by Solarbio (Beijing, China). The MicroBCA™ protein assay kit was obtained from Thermo Fisher Scientific (Rockford, IL, USA). Anti-mouse enzyme-linked immunosorbent assay (ELISA) kits, tumor necrosis factor-α (TNF-α), and IL-6 were purchased from eBioscience (San Diego, CA, USA). ε-Caprolactone and dichloromethane were distilled with calcium hydride, while other reagents were used as received.
NIH-3T3 and RAW-264.7 cell lines were purchased from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China).
Six-week-old female C57BL/6 mice were bought from the Academy of Military Medical Sciences (Beijing, China). The Administrative Committee on Animal Research in the Institute of Biomedical Engineering, Chinese Academy of Medical Sciences, approved all the protocols for animal experiments. All the animal experiments were performed in compliance with the Guiding Principles for the Care and Use of Laboratory Animals, Peking Union Medical College, China.
Synthesis of 6S-PCL
Classical ring-opening polymerization was employed to produce 6S-PCL. In brief, 195.7 mg inositol (1.095 mmol; Aladdin, Shanghai, China) was charged into a two-neck round-bottom polymeric flask. Then, 23.38 mL ε-caprolactone (219 mmol) and 1.21 mL stannous octoate (0.0365 mmol) were added into the flask. This was followed by degassing for 10 min and argon-filling for 1 min three times to remove the oxygen in the flask. Thereafter, a vacuum was required and the reaction was maintained at 180°C for 48 h. The resulting product was cooled to ambient temperature and dissolved in CH2Cl2, then purified by precipitation with ice methanol three times. Finally, 6S-PCL polymer was obtained after filtering and drying in a vacuum for 24 h.
Synthesis of 6S-PCL-PEG
First, oxalyl chloride (1.50 g, 11.84 mmol) was dissolved in 10 mL anhydrous CH2Cl2 in a flask. Then, 15 mL anhydrous CH2Cl2 solution containing PEG (3.16 g, 0.789 mmol) and 0.240 g triethylamine (2.367 mmol) were added dropwise into the flask under stirring for 3 h at 4°C. Thereafter, the reaction solution was reevaporated and redissolved in CH2Cl2 three times. The final residue and triethylamine (0.053 g, 0.526 mmol) were co-dissolved in 20 mL dried CH2CI2. The anhydrous CH2Cl2 solution containing 6S-PCL (1 g, 0.044 mmol) was added into the above solution dropwise under stirring at ambient temperature in a nitrogen atmosphere for 48 h. After washing three times in 1 M HCl and saturated NaCl solution, the solution was dried with anhydrous MgSO4. After filtration, the filtered solution was precipitated in cold diethyl ether three times and washed in cold methanol to remove unreacted PEG. The resulting product was obtained after drying at 40°C in a vacuum for 24 h until achieving a constant weight.
Hydrogen-1 nuclear magnetic resonance (1H-NMR) spectroscopy measurements were performed using a 400 MHz spectrometer (Varian, Palo Alto, CA, USA) with CDCl3 as the solvent and tetramethylsilane as the internal reference, with a chemical shift of 0 ppm.Citation31 A Nicolet Nexus 470-ESP Fourier-transform infrared (FTIR) spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) was employed to analyze the main functional groups. The molecular weight of the copolymer was investigated by gel-permeation chromatography (GPC) with polymethyl methacrylate (PMMA) as the standard using a Hitachi L-2490 differential refraction detector and two Agilent PL gel particle columns (7.5×300 mm, 10 mm).
Preparation of NPs
NPs were prepared by a water-in-oil-in-water double emulsion (W1/O/W2) solvent evaporation method.Citation32 OVA, as a model protein, was encapsulated into the NPs. In brief, the aqueous solution containing 15 mg OVA was added to 25 mg/mL copolymer CH2CI2 solution to form a water-in-oil emulsion at 35% amplitude with a microtip probe sonicator (VCX-130-PB; Sonics & Materials, Newtown, CT, USA). Then, the primary emulsion was added into another aqueous solution containing 0.5% (w/v) PVA at 30% amplitude. Thereafter, the multiple emulsion was stirred in a fume hood to volatilize the organic solvent. Finally, the NPs were washed three times by centrifugation at 23,000 rpm for 30 min, then freeze-dried and stored at 4°C. Blank 6S-PCL-PEG NPs were produced in a similar manner by adding nothing to the first aqueous solution.
In vitro biocompatibility assay of NPs
A cell counting kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan) was used to detect the biocompatibility of blank NPs with NIH-3T3 cells and RAW-264.7 cells. In brief, cells were seeded in 96-well culture plate (5×103 cells/well) using RPMI 1640 medium for 24 h. Then, cells were treated with blank NPs at different concentrations (12.5, 25, 50, and 100 µg/mL) for another 24 h. Thereafter, the medium was removed and 200 µL mixture medium (RPMI 1640 with 10% FBS/CCK-8, 9/1, v/v) was added to each well and incubated for 2 h. The absorbance (AB) at 450 nm was detected with a Varioskan Flash Microplate Reader (Thermo Fisher Scientific, Waltham, MA, USA). The relative cell viability was calculated as follows and the results are represented as mean±SD (n=3).
To evaluate the biocompatibility of the blank NPs further, RAW-264.7 cells were co-cultured with blank NPs and the cytokine secretion of RAW-264.7 cells was measured by ELISA. In brief, RAW-264.7 cells were cultivated with RPMI 1640 containing 10% FBS and 1% penicillin streptomycin at 37°C in a 5% (v/v) carbon dioxide atmosphere. Then, RAW-264.7 cells were seeded into 24-well plates with a density of 5×105 cells/well and incubated for 24 h. Thereafter, cells were treated with different concentrations of blank NPs (12.5, 25, and 50 µg/mL) and incubated for another 24 h. The cytokines IL-6 and TNF-α in supernatants were analyzed. LPS was used as a positive control.Citation33
Characterization of NPs
Size, polydispersity, zeta potential, morphology, and stability of NPs
The size and zeta potential of the OVA-loaded NPs in aqueous solution were measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). Each sample was analyzed in triplicate. The morphology of the NPs was observed by transmission electron microscopy (TEM) (Tianjin Research Center of Basic Medical Science, Tianjin, China). A drop of freshly prepared NP suspension was deposited on to a copper grid. Then, the grid was dried at room temperature overnight before being examined by TEM. To evaluate the stability of NPs, changes in particle size and size distribution were closely monitored every 15 days for 1 month.
Encapsulation efficiency and loading capacity
The encapsulation efficiency (EE) and protein-loading capacity (LC) of NPs were detected by a bicinchoninic acid (BCA) assay kit (Beyotime, Shanghai, China). In brief, OVA-loaded NPs were dissolved in 1 M NaOH and incubated in a shaker at 37°C for 3 h. Then, an equal volume of 1 M HCl was added to neutralize the NaOH. OVA in the supernatant was determined using the BCA assay kit. The EE and LC were calculated by the following equations. The determinations were carried out in triplicate and the results are expressed as mean±SD (n=3).
In vitro OVA release
OVA was used as a model protein for determining protein release behavior from 6S-PCL-PEG NPs in vitro. The detection of OVA released from 6S-PCL-PEG NPs was carried out in PBS (pH 7.4) at 37°C. First, 20 mg of OVA-loaded NPs was suspended in 4 mL PBS (pH 7.4) and shaken at 150 rpm at 37°C for 28 days. The amount of released OVA was measured at set time points. After centrifugation, 2 mL of supernatant was collected and an equal volume of PBS was added. Total protein in the supernatant samples was measured with the BCA assay kit according to the manufacturer’s instructions. Absorbance at 562 nm was measured using a microplate reader (Varioskan LUX multimode reader; Thermo Fisher Scientific). Protein concentrations were calculated according to standard curves.
Stability and activity of OVA released from NPs
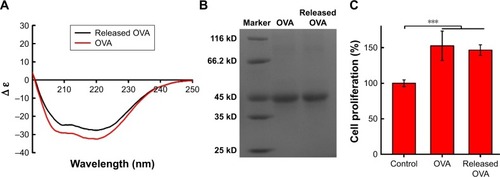
Secondary structures and molecular weight were used to evaluate the stability of protein before and after the preparation of NPs.Citation34 The changes in secondary structures of OVA encapsulated in NPs were monitored by far-ultraviolet circular dichroism (CD). All results of CD spectra were collected on a J-815 spectrometer (JASCO, Tokyo, Japan) with a 1 mm quartz cell. The background PBS solution was subtracted from each protein spectrum and native OVA as the control. A minimum of three protein spectra were collected per sample and averaged. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was employed to qualitatively identify the integrity of OVA which was encapsulated into 6S-PCL-PEG NPs.
Bioactivity is of importance for proteins. In this study, therefore, splenocyte proliferation was used to evaluate the OVA bioactivity.Citation35 In brief, 6-week-old female C57BL/6 mice (five mice/group) were immunized with naked OVA or OVA released from NPs (20 µg OVA/mouse) by subcutaneous injection three times at 2 week intervals. Splenocytes were isolated 7 days after the last immunization and splenocytes (1×106 cells/mL) were restimulated with soluble OVA (20 µg/mL) in a 96-well plate at 37°C for 72 h. Following this, splenocyte proliferation was detected by MTT at 570 nm using a Varioskan Flash Microplate Reader (Thermo Fisher Scientific).
In vitro cellular uptake studies
NIH-3T3 cells (5×104 cells/mL) were seeded in a confocal dish for 24 h then cultured with fluorescein isothiocyanate (FITC)-conjugated OVA (OVA-FITC) or OVA-FITC encapsulated NPs (OVA-FITC NPs) at 37°C for 4 h. Thereafter, the cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, St Louis, MO, USA) for 5 min. Florescent images were recorded by confocal laser scanning microscopy (CLSM 410; Zeiss, Jena, Germany).
For quantitative cellular uptake analysis, NIH-3T3 cells were cultured with OVA-FITC and OVA-FITC NPs at 37°C for 4 h and then washed. The uptake efficiency of NPs by NIH-3T3 cells was expressed as OVA-FITC-positive cells and mean fluorescence intensity, which were determined by a BD Accuri™ C6 flow cytometer (BD Biosciences, San Jose, CA, USA).
Results and discussion
Characterization of copolymer
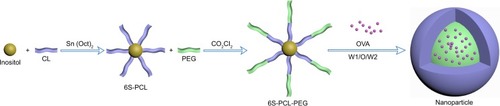
A schematic illustration of the copolymer is depicted in . First, inositol as the initiator and stannous octoate as the catalyst were used to synthesize 6S-PCL by ring-opening polymerization. Then, 6S-PCL was linked to PEG by an acylation reaction using oxalyl chloride to obtain 6S-PCL-PEG. Compared with the traditional synthesis method using dicyclohexylcarbodiimide (DCC) and N-hydroxysuccinimide (NHS), this synthetic method is simple and convenient.Citation30,Citation31,Citation36
Figure 1 Schematic illustration of the synthesis of 6S-PCL-PEG. The hydrophilic model drug OVA was incorporated into the hydrophilic core with a W1/O/W2 solvent evaporation method.
Abbreviations: 6S-PCL, six-arm poly(ε-caprolactone); CL, ε-caprolactone; PEG, poly(ethylene glycol); 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); OVA, ovalbumin; W1/O/W2, water-in-oil-in-water double emulsion.
1H-NMR was used to confirm the chemical structure of 6S-PCL and 6S-PCL-PEG. The resonance peaks a (δ=4.05), b (δ=1.66), c (δ=1.41), and d (δ=2.31) were assigned to the protons of CH2 in the PCL (). In the 1H-NMR spectra of 6S-PCL-PEG, the high-intensity peak at 3.62 ppm () belongs to the protons in CH2 of PEG, whereas the other peaks assigned to 6S-PCL remained. All these results indicate the existence of PEG, which proves that 6S-PCL-PEG was synthesized successfully. We also calculated the molecular weight of the end product by comparing the integration areas of NMR spectra from PCL and PEG blocks (). The total molar ratio between caprolac-tone and ethylene glycol was about 276:552. Therefore, the molecular weight of the end product calculated by 1H-NMR was about 56,000.
Figure 2 Synthesis and characterization of 6S-PCL and 6S-PCL-PEG. (A) 1H-NMR spectrum of 6S-PCL. (B) 1H-NMR spectrum of 6S-PCL-PEG. (C) FTIR spectrometry of 6S-PCL and 6S-PCL-PEG. (D) Molecular weights of 6S-PCL and 6S-PCL-PEG investigated by GPC.
Notes: a, (δ=4.05 ppm); b, (δ=1.66 ppm); c, (δ=1.41 ppm); and d, (δ=2.31 ppm). Compared with the 6S-PCL spectrum, the high intensity of peak e (δ=3.62 ppm, −CH2 in PEG segments) in the 6S-PCL-PEG spectrum indicated the existence of PEG.
Abbreviations: 6S-PCL, six-arm poly(ε-caprolactone); 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); 1H-NMR, hydrogen-1 nuclear magnetic resonance; FTIR, Fourier-transform infrared; GPC, gel-permeation chromatography.
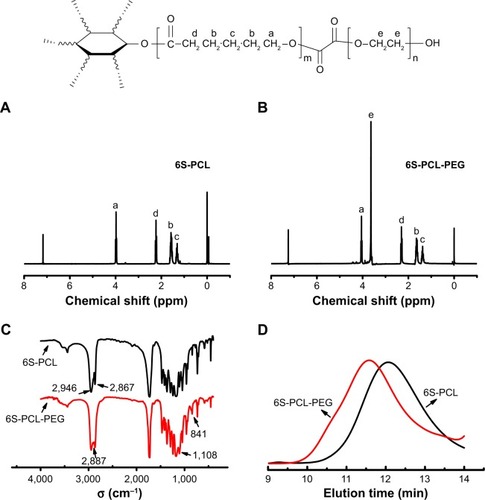
FTIR spectrometry was employed to identify the chemical structure of the copolymer. As shown in , the peaks of 2,946 cm−1 and 2,867 cm−1 were attribute to the saturated −CH bond and the peak at 1,725 cm−1 was a carbonyl group. However, after PEG modification, the absor-bance peaks 2,946 cm−1 and 2,867 cm−1 of the −CH bond in PCL were buried in the peak 2,887 cm−1 of −CH2 groups in PEG, and the new peaks of 1,108 cm−1 and 841 cm−1 of single-bond C−O were characteristic of PEG. All emerging peaks of 6S-PCL-PEG proved the existence of the PEG chain in the product.
The result of GPC is shown in . Compared to 6S-PCL, the reduction in the retention time in the GPC curve of 6S-PCL-PEG indicated an increase in molecular weight, demonstrating the attachment of PEG to 6S-PCL. The molecular weight of 6S-PCL was 38,211 and that of 6S-PCL-PEG was 59,953 L. The polydispersity of the copolymer was narrow (1.495 for 6S-PCL and 1.523 for 6S-PCL-PEG). The molecular weight of 6S-PCL-PEG measured by GPC was closed to the result calculated by 1H-NMR, which also indicates that 6S-PCL-PEG was synthesized successfully.
Biocompatibility of copolymer NPs
Drug carriers should not release toxic products or produce adverse reactions, and this can be evaluated through in vitro cytotoxicity tests. The cytotoxicity of blank NPs was assessed with a CCK-8 assay. NIH-3T3 cells and RAW-264.7 cells were incubated for 24 h with blank NPs, with concentrations ranging from 12.5 µg/mL to 100 µg/mL. shows that the blank 6S-PCL-PEG NPs did not exhibit obvious cytotoxicity compared with the control group, indicating the commendable biocompatibility of blank NPs.
Figure 3 Biocompatibility of 6S-PCL-PEG NPs. (A) NIH-3T3 and RAW-264.7 cell viability after co-culture with NPs for 24 h. (B) Cytokine release of RAW-264.7 cells after co-culture with NPs for 24 h. Bars are mean±SD (n=3). The statistical significance of differences were analyzed using Student’s t-test: **p<0.01.
Abbreviations: 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); NPs, nanoparticles; TNF-α, tumor necrosis factor-α.
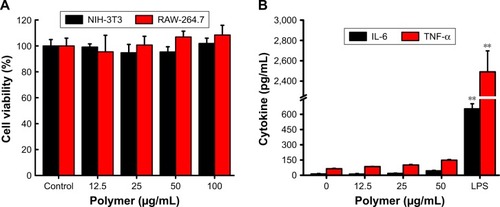
RAW-264.7 cells were also employed to evaluate the biocompatibility of copolymer NPs by detecting typical inflammatory cytokines such as IL-6 and TNF-α.Citation37 We measured the cytokines (IL-6 and TNF-α) released in culture supernatant after 24 h incubation with blank NPs. shows that there was no significant difference in cytokine secretion between the NP groups and the negative control group. However, much higher levels of cytokines were released in the positive group (LPS), which proved that the blank NPs would not elicit inflammatory responses in RAW-264.7 cells. This result was consistent with the above results, implying that the NPs have good biocompatibility.
Characterization of NPs
The NPs prepared by a double-emulsion (W1/O/W2) solvent evaporation method exhibited suitable particle size and negative charge. The particle size of 6S-PCL NPs was 190.7±1.665 nm and the polydispersity index (PDI) was 0.164±0.010 (). As shown in , OVA-loaded NPs had an average diameter of 233.4±0.565 nm, which is in the size range suitable for reducing uptake by the reticuloendothelial system and prolonging the circulation time in the blood.Citation38–Citation40 The 6S-PCL-PEG NPs would not easily be taken up by macrophages of the mononuclear–phagocyte system because macrophages readily phagocytose foreign matter in the range of 0.5–3 µm.Citation41,Citation42 Besides, the surface-grafted, flexible, and hydrophilic chains of PEG form conformational clouds, which also prevent protein (opsonin) adsorption to NPs and hence avoid capture by the mononuclear phagocyte system,Citation43,Citation44 which indicates that the NPs could have a long circulation period. The slightly negative charge of OVA-loaded 6S-PCL-PEG NPs (−10.4±0.9 mV) may decrease the undesirable clearance by the reticuloendothelial system, such as the liver, improving blood compatibility.Citation45 PEGylation of NPs could also decrease protein adsorption in the circulation, which would also benefit the circulation duration of NPs.Citation46
Figure 4 Characterization of 6S-PCL-PEG NPs in vitro. (A) Dynamic light scattering of 6S-PCL-PEG NPs. (B) The physical stability of 6S-PCL-PEG NPs in PBS was assessed by the change in NP size and PDI for 30 days. (C) TEM image of normal 6S-PCL-PEG NPs. Magnification is 5,000×. (D) OVA cumulative release from 6S-PCL-PEG NPs which were incubated in PBS (0.01 M, pH 7.4) for 28 days; the protein content was determined by a BCA assay. Bars are mean±SD (n=3).
Abbreviations: 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); NPs, nanoparticles; PDI, polydispersity index; TEM, transmission electron microscopy; OVA, ovalbumin; BCA, bicinchoninic acid.
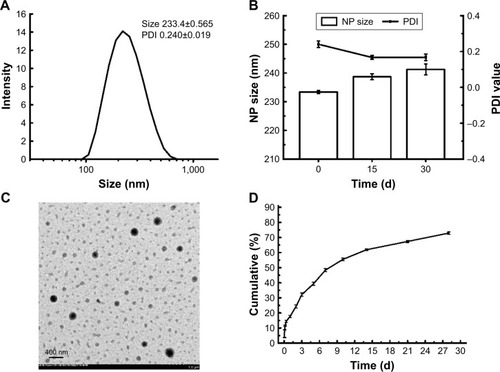
Stability is the “holy grail” for an excellent drug carrier. In this study, size change over 1 month was measured to evaluate the stability of NPs.Citation47 As shown in , there were no obvious size changes during 1 month at 4°C, indicating that the NPs have great stability.
The morphology of OVA-loaded NPs was observed by TEM. TEM images clearly showed NPs with a spherical shape and separated from each other (). Their diameter was consistent with the results of DLS. We further measured the protein EE and LC of 6S-PCL-PEG NPs. Significant increases in EE (85.8%±0.7%) and LC (28.5%±1.3%) can be ascribed to the PEGylation.Citation22
In vitro release of OVA-loaded NPs
In vitro release kinetics of OVA-loaded NPs in PBS (pH 7.4) during 28 days are shown in . The release curve of OVA from the 6S-PCL-PEG NPs exhibited a burst on the first day (which was less than 15% of the loaded OVA); however, a sustained-release profile was showed over 28 days and the cumulative release amount was about 70%. The fast release of OVA in the initial stage could be attributed to the release of OVA on the surface layers of the NPs.Citation48 The existence of poly(ε-caprolactone) and the structure of 6S-PCL-PEG copolymer in aqueous solution could form a more effective conformational cloud, which plays an important role in the sustained drug release.Citation25,Citation27,Citation49,Citation50 The time of sustainable protein release in this study was approximately 1 month, which was longer than in other reports.Citation8,Citation51,Citation52 The reason for this could be that PVA increases the viscosity of the external water phase and results in increased difficulty for the OVA aqueous solution to diffuse out.Citation53 Overall, the NPs were effective protein delivery carriers, with suitable size, loading capacity, stability, and sustained-release properties.
Stability and activity of OVA released from NPs
Far-ultraviolet CD (200–250 nm) was used to monitor the secondary structure of protein encapsulated into NPs. As shown in , the CD spectra of native OVA and the OVA released from 6S-PCL-PEG NPs were similar and each spectrum had two minima, at 208 nm and 222 nm, which indicated that OVA did not undergo any conformational changes during preparation and release from NPs.Citation54 shows that the molecular weight of OVA released from NPs was the same as with native OVA, which was consistent with the CD spectra results. The bioactivity of OVA released from NPs was examined by the MTT assay. The results in show that there was no significant difference in splenocyte proliferation between OVA and the OVA released from NPs, implying that the released OVA maintained its bioactivity.
Figure 5 Secondary structure and molecular weight of OVA encapsulated into 6S-PCL-PEG NPs. (A) CD spectra of OVA. (B) Electrophoretogram of OVA. (C) Proliferation of splenocytes stimulated by OVA. Released OVA means the OVA released from NPs. Bars are mean±SD (n=5). The statistical significance of differences were analyzed using Student’s t-test: ***p<0.001.
Abbreviations: OVA, ovalbumin; 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); CD, circular dichroism; NPs, nanoparticles.
Cellular uptake of OVA-loaded NPs
The cellular uptake efficiency of drug-loaded NPs affects the therapeutic effects.Citation50 Therefore, the cellular uptake efficiency of drug-loaded NPs was detected using NIH-3T3 cells as a model. shows the results of CLSM after 4 h incubation in NIH-3T3 cells with the pristine OVA. The 6S-PCL NPs and the 6S-PCL-PEG NPs had the same equivalent OVA concentration; however, the green fluorescence in NIH-3T3 cells treated with 6S-PCL-PEG NPs was significantly stronger than that in 6S-PCL NPs and native OVA, which indicated that 6S-PCL-PEG NPs showed enhanced cellular uptake compared with pristine OVA.
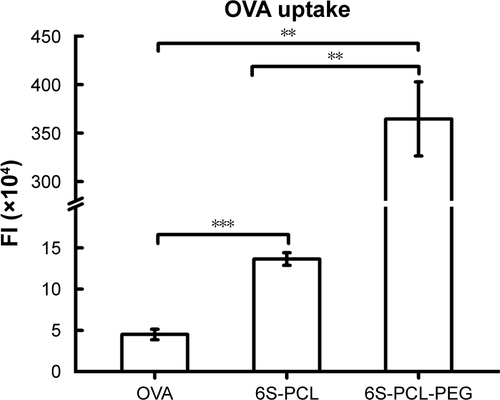
Figure 6 Cellular uptake of nanoparticles by NIH-3T3 cells. (A) NIH-3T3 cells were incubated with OVA-FITC (green) and OVA-FITC-loaded NPs for 3 h; the nucleus was labeled with DAPI (blue). The uptake of NPs was observed by confocal microscopy. (B) Percentage of OVA-FITC-positive cells. (C) Total FI of OVA-FITC; Total FI=% of positive cells × Mean fluorescence intensity. Bars are mean±SD (n=3). The statistical significance of differences were analyzed using Student’s t-test: *p<0.05, **p<0.01, ***p<0.001.
Abbreviations: 6S-PCL, six-arm poly(ε-caprolactone); 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); OVA, ovalbumin; DAPI, 4′,6-diamidino- 2-phenylindole; BF, bright field; FITC, fluorescein isothiocyanate; FI, fluorescence intensity.
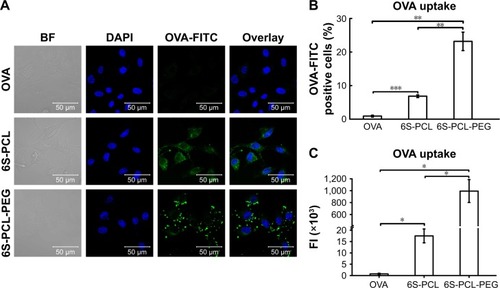
The cellular uptake of NPs was further investigated by C6 flow cytometry. and C shows the NIH-3T3 cellular uptake efficiency of the NPs compared with the native OVA after incubation for 4 h. The same equivalent OVA concentration in the culture medium was applied in the experiment. It is evident from that more 6S-PCL-PEG NPs were taken up by NIH-3T3 cells after 4 h incubation compared with the 6S-PCL NPs (3.4-fold) and pristine OVA-FITC (25.7-fold). The representative flow plot is presented in . A significant increase in the total fluorescence intensity of 6S-PCL-PEG was observed compared with soluble OVA and 6S-PCL NPs (). The mean fluorescence intensity plot is presented in . These data confirm the enhanced cellular uptake by NP formulations, which is consistent with the results obtained from CLSM.
All the results showed that 6S-PCL-PEG NPs have distinct properties as protein carriers, such as outstanding stability, high loading capability, and excellent biocompatibility, and enhance the cellular uptake to improve the effects of the protein.
Conclusion
In this study, the copolymer 6S-PCL-PEG, with excellent biocompatibility, was successfully synthesized in two steps: ring-opening polymerization then esterification by oxalyl chloride. After modification by PEG, 6S-PCL-PEG NPs had higher encapsulation efficiency and drug-loading capacity. Compared with 6S-PCL NPs and naked OVA, the uptake of 6S-PCL-PEG NPs by NIH-3T3 cells was significantly increased. In summary, a polymeric protein vehicle, 6S-PCL-PEG NPs, with robust efficacy in the delivery of proteins into living cells, was prepared. NPs have great potential application as protein sustained-release delivery systems.
Acknowledgments
The authors are grateful for the financial support from the National Natural Science Funds of China (31771097, 31600743), Tianjin Research Program of Application Foundation and Advanced Technology (17JCZDJC37400, 16JCQNJC13900), CAMS Innovation Fund for Medical Sciences (2017-I2M-1-016), and Tianjin Innovation and Promotion Plan Key Innovation Team of Immunoreactive Biomaterials.
Supplementary materials
Figure S1 Integration areas of NMR spectra of 6S-PCL-PEG.
Abbreviations: NMR, nuclear magnetic resonance; 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol).
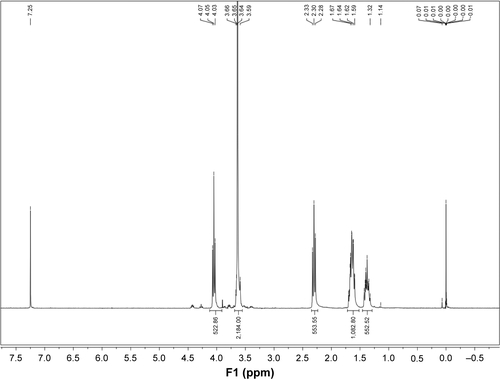
Figure S2 Size and PDI of 6S-PCL nanoparticles.
Abbreviations: PDI, polydispersity index; 6S-PCL, six-arm poly(ε-caprolactone).
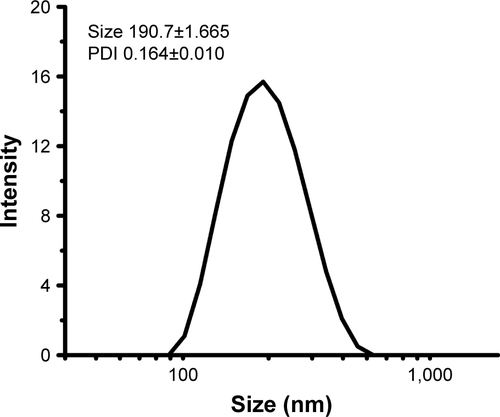
Figure S3 Representative flow plot of NPs taken up by NIH-3T3 cells.
Abbreviations: NPs, nanoparticles; OVA, ovalbumin; 6S-PCL, six-arm poly(ε-caprolactone); 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); FITC, fluor-escein isothiocyanate.
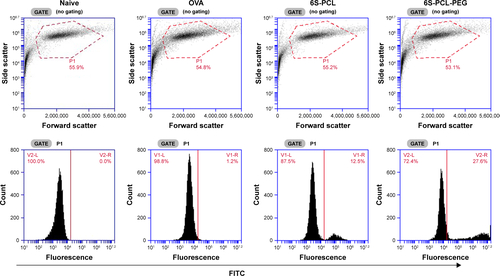
Figure S4 Mean fluorescence intensity plot of NPs taken up by NIH-3T3 cells. The statistical significance of differences were analyzed using Student’s t-test: **p<0.01, ***p<0.001.
Abbreviations: NPs, nanoparticles; OVA, ovalbumin; 6S-PCL, six-arm poly(ε-caprolactone); 6S-PCL-PEG, six-arm poly(ε-caprolactone)–poly(ethylene glycol); FI, fluorescence intensity.
Disclosure
The authors report no conflicts of interest in this work.
References
- HerreroEPAlonsoMJCsabaNPolymer-based oral peptide nano-medicinesTher Deliv20123565766822834409
- VilaASánchezATobioMCalvoPAlonsoMJDesign of biodegradable particles for protein deliveryJ Control Release2002781–3152411772445
- Van TommeSRHenninkWEBiodegradable dextran hydrogels for protein delivery applicationsExpert Rev Med Devices20074214716417359222
- LeeKYYukSHPolymeric protein delivery systemsProg Polymer Sci2007327669697
- van de WeertMHenninkWEJiskootWProtein instability in poly(lactic-co-glycolic acid) microparticlesPharm Res200017101159116711145219
- ZhangNLiJJiangWEffective protection and controlled release of insulin by cationic beta-cyclodextrin polymers from alginate/chitosan nanoparticlesInt J Pharm20103931–221221820394813
- Garcia-FuentesMPregoCTorresDAlonsoMJA comparative study of the potential of solid triglyceride nanostructures coated with chitosan or poly(ethylene glycol) as carriers for oral calcitonin deliveryEur J Pharm Sci200525113314315854809
- LiuGMaSSChengRMengFLiuHZhongZThe highly efficient delivery of exogenous proteins into cells mediated by biodegradable chimaeric polymersomesBiomaterials201031297575758520599266
- GutierroIHernándezRMIgartuaMGascónARPedrazJLSize dependent immune response after subcutaneous, oral and intranasal administration of BSA loaded nanospheresVaccine2002211677712443664
- SaitoNUsuiYAokiKCarbon nanotubes: biomaterial applicationsChem Soc Rev20093871897190319551170
- MalmstenMInorganic nanomaterials as delivery systems for proteins, peptides, DNA, and siRNACurr Opin Colloid Interface Sci2013185468480
- Al-RemawiMElsayedAMaghrabiIHamaidiMJaberNChitosan/lecithin liposomal nanovesicles as an oral insulin delivery systemPharm Dev Technol201722339039827470482
- PengQZhangZRGongTChenGQSunXA rapid-acting, long-acting insulin formulation based on a phospholipid complex loaded PHBHHx nanoparticlesBiomaterials20123351583158822112760
- XieSYWangSLZhaoBKHanCWangMZhouWZEffect of PLGA as a polymeric emulsifier on preparation of hydrophilic protein-loaded solid lipid nanoparticlesColloids Surf B Biointerfaces200867219920418829272
- FangFGongCYDongPWAcute toxicity evaluation of in situ gel-forming controlled drug delivery system based on biodegradable poly(epsilon-caprolactone)-poly(ethylene glycol)-poly(epsilon-caprolactone) copolymerBiomed Mater20094202500219208940
- GongCYShiSDongPWBiodegradable in situ gel-forming controlled drug delivery system based on thermosensitive PCL-PEG-PCL hydrogel: part 1 – Synthesis, characterization, and acute toxicity evaluationJ Pharm Sci200998124684469419367619
- KhodaverdiEAkbariATekieFSMohajeriSAZohuriGHadizadehFSustained delivery of amphotericin B and vancomycin hydrochloride by an injectable thermogelling tri-block copolymerPDA J Pharm Sci Technol201367213514523569074
- YooHSParkTGBiodegradable polymeric micelles composed of doxorubicin conjugated PLGA-PEG block copolymerJ Control Release2001701–2637011166408
- WeiXGongCGouMBiodegradable poly(epsilon-caprolactone)-poly(ethylene glycol) copolymers as drug delivery systemInt J Pharm2009381111819664700
- GouMZhengLPengXPoly(epsilon-caprolactone)-poly(ethylene glycol)-poly(epsilon-caprolactone) (PCL-PEG-PCL) nanoparticles for honokiol delivery in vitroInt J Pharm20093751–217017619427143
- ZhouSDengXYangHBiodegradable poly(ε-caprolactone)-poly(ethylene glycol) block copolymers: characterization and their use as drug carriers for a controlled delivery systemBiomaterials200324203563357012809785
- HarrisJMChessRBEffect of pegylation on pharmaceuticalsNat Rev Drug Discov20032321422112612647
- De KokerSCuiJVanparijsNEngineering Polymer Hydrogel Nanoparticles for Lymph Node-Targeted DeliveryAngew Chem Int Ed Engl20165541334133926666207
- PapiMCaputoDPalmieriVClinically approved PEGylated nanoparticles are covered by a protein corona that boosts the uptake by cancer cellsNanoscale2017929103271033428702661
- ChenYYangZLiuCSynthesis, characterization, and evaluation of paclitaxel loaded in six-arm star-shaped poly(lactic-co-glycolic acid)Int J Nanomedicine201384315432624235829
- ZengXTaoWMeiLHuangLTanCFengSSCholic acid-functionalized nanoparticles of star-shaped PLGA-vitamin E TPGS copolymer for docetaxel delivery to cervical cancerBiomaterials201334256058606723694904
- ZhuJZhouZYangCKongDWanYWangZFolate-conjugated amphiphilic star-shaped block copolymers as targeted nanocarriersJ Biomed Mater Res A201197449850821509931
- PoreeDEGilesMDLawsonLBHeJGraysonSMSynthesis of amphiphilic star block copolymers and their evaluation as transdermal carriersBiomacromolecules201112489890621341702
- LangMChuCCFunctionalized multiarm poly(∈-caprolactone)s: Synthesis, structure analysis, and network formationJ Appl Polym Sci201086922962306
- MaGZhangCZhangLDoxorubicin-loaded micelles based on multiarm star-shaped PLGA-PEG block copolymers: influence of arm numbers on drug deliveryJ Mater Sci Mater Med20162711726676863
- ZhuXLiuCDuanJSynthesis of three-arm block copolymer poly(lactic-co-glycolic acid)-poly(ethylene glycol) with oxalyl chloride and its application in hydrophobic drug deliveryInt J Nanomedicine2016116065607727895480
- YangJZengYZhangCThe prevention of restenosis in vivo with a VEGF gene and paclitaxel co-eluting stentBiomaterials20133461635164323199742
- AgrahariVAgrahariVHungWTChristensonLKMitraAKComposite Nanoformulation Therapeutics for Long-Term Ocular Delivery of MacromoleculesMol Pharm20161392912292226828415
- LiuQChenXJiaJpH-Responsive Poly(D,L-lactic-co-glycolic acid) Nanoparticles with Rapid Antigen Release Behavior Promote Immune ResponseACS Nano2015954925493825898266
- LuoZLiPDengJCationic polypeptide micelle-based antigen delivery system: a simple and robust adjuvant to improve vaccine efficacyJ Control Release2013170225926723742880
- YoonJJChungHJLeeHJParkTGHeparin-immobilized biodegradable scaffolds for local and sustained release of angiogenic growth factorJ Biomed Mater Res A200679493494216941589
- MirzaSHossainMMathewsCType 2-diabetes is associated with elevated levels of TNF-alpha, IL-6 and adiponectin and low levels of leptin in a population of Mexican Americans: a cross-sectional studyCytokine201257113614222035595
- HeCHuYYinLTangCYinCEffects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticlesBiomaterials201031133657366620138662
- AlexisFPridgenEMolnarLKFarokhzadOCFactors affecting the clearance and biodistribution of polymeric nanoparticlesMol Pharm20085450551518672949
- YuanFDellianMFukumuraDVascular permeability in a human tumor xenograft: molecular size dependence and cutoff sizeCancer Res19955517375237567641188
- LeeEKimSSeongKA biodegradable and biocompatible drug-delivery system based on polyoxalate microparticlesJ Biomat Sci Polym Ed2011221316831694
- WinKYFengSSEffects of particle size and surface coating on cellular uptake of polymeric nanoparticles for oral delivery of anticancer drugsBiomaterials200526152713272215585275
- MonfardiniCVeroneseFMStabilization of substances in circulationBioconjug Chem1998944184509667945
- LeeKDNirSPapahadjopoulosDQuantitative analysis of liposome-cell interactions in vitro: rate constants of binding and endocytosis with suspension and adherent J774 cells and human monocytesBiochemistry19933238898998422393
- XiaoKLiYLuoJThe effect of surface charge on in vivo biodistribution of PEG-oligocholic acid based micellar nanoparticlesBiomaterials201132133435344621295849
- AwasthiVDGarciaDGoinsBAPhillipsWTCirculation and biodistribution profiles of long-circulating PEG-liposomes of various sizes in rabbitsInt J Pharm20032531–212113212593943
- LinWJJuangLWLinCCStability and release performance of a series of pegylated copolymeric micellesPharm Res200320466867312739777
- LiRLiXXieLPreparation and evaluation of PEG-PCL nanoparticles for local tetradrine deliveryInt J Pharm2009379115816619524653
- ChenYLiYShenWControlled release of liraglutide using thermogelling polymers in treatment of diabetesSci Rep201663159327531588
- HuYXieJTongYWWangCHEffect of PEG conformation and particle size on the cellular uptake efficiency of nanoparticles with the HepG2 cellsJ Control Release2007118171717241684
- HsuYHLinCTYuYHChouYCLiuSJChanECDual delivery of active antibactericidal agents and bone morphogenetic protein at sustainable high concentrations using biodegradable sheath-core-structured drug-eluting nanofibersInt J Nanomedicine2016113927393727574423
- FeczkóTTóthJGyenisJComparison of the preparation of PLGA–BSA nano- and microparticles by PVA, poloxamer and PVPColloids Surf A Physicochem Eng Asp20083191–3188195
- YangYYChungTSNgNPMorphology, drug distribution, and in vitro release profiles of biodegradable polymeric microspheres containing protein fabricated by double-emulsion solvent extraction/evaporation methodBiomaterials200122323124111197498
- ZemserMFriedmanMKatzhendlerJGreeneLLMinskyAGorinsteinSRelationship between functional properties and structure of ovalbuminJ Protein Chem19941322612748060498
