Abstract
Efficient preparation and labeling of human induced pluripotent stem (iPS) cells is a great challenge in stem cell research and development. With the aim of investigating the feasibility of using nanotechnology to enhance the preparation efficiency of iPS cells and to label iPS cells for long-term tracing and imaging, in this paper, four transcription factor genes, ie, Oct4, Sox2, LIN28, and Nanog, and packaging plasmids such as PSPAX2 and PMD2.G were cotransfected into 293T cells using Generation 5.0 polyamidoamine dendrimer-modified magnetic nanoparticles (dMNPs) as a delivery system. The resultant supernatant liquids were incubated with human fibroblast cells at 37°C for 21 days, then the embryonic stem (ES) cell-like clones were screened, cultured, and identified. Finally, the prepared iPS cells were labeled with fluorescent magnetic nanoparticles (FMNPs). The results showed that dMNPs can efficiently deliver all vectors into 293T cells. The resultant lentiviruses’ titers were 10-fold more than those based on Lipofectamine™ 2000. Reverse transcription polymerase chain reaction analysis showed that four genes (Oct4, Sox2, LIN28, and Nanog) exhibited different expressions in iPS cells. Immunostaining analysis showed that specific surface markers of ES cells such as SSEA-3, SSEA-4, Tra-1-60, and Tra-1-81 were positive in iPS cells, and the terotomas were formed in NOD-SCID mice that were implanted with iPS cells. Red fluorescent signals could be observed in iPS cells labeled with FMNPs by fluorescent microscopy, and the magnetic signals were detected in labeled iPS cells by magnetic resonance imaging. In conclusion, human iPS cells can be efficiently generated using polyamidoamine dMNPs and lentivirus and labeled with FMNPs for long-term observation and tracking, which has great potential application in the research and development of stem cells in the near future.
Introduction
Since induced pluripotent stem (iPS) cells were first successfully generated from mouse embryonic fibroblasts (MEFs),Citation1 iPS cell research and development has become a hot topic. Mice have been cloned from iPS cells,Citation2 and stem cells have been labeled with fluorescent dyes such as NIR815 and ICG.Citation3,Citation4 Although iPS cells offer great progress for controllable manmade stem cells, several obstacles seriously affect the further research and development of iPS cells, including the development of advanced techniques to enhance the efficiency of iPS cell preparation and novel methods to label and track transplanted stem cells for long-term observation, as well as new nonvirus vectors to replace lentivirus vectors to enhance their safety. To date, the techniques of preparing iPS cells efficiently have not achieved a big breakthrough. How to fabricate many iPS cells within a limited timeframe has become a challenging problem. How to label iPS cells for long-term observation is another challenge.
In recent years, stem cell nanotechnology has emerged as a new, exciting field.Citation5 The importance of nanomaterials, nanostructures, and nanotechnology to the fundamental developments in stem cell-based therapies for injuries and degenerative diseases has been recognized.Citation6–Citation10 It is well known that nanomaterials have some unique effects. Ultimately, these effects can lead to new technological opportunities as well as new challenges.Citation11–Citation13 The application of nanotechnology in stem cell research and development has attractive technological prospects, which provide a new opportunity to solve current problems facing stem cell research and development. However, so far, few reports are closely associated with the use of nanotechnology to enhance the preparation efficiency of human iPS cells and labeling iPS cells for long-term tracking of their in vivo distribution and metabolism course. In our previous studies, we confirmed that polyamidoamine (PAMAM) dendrimer-modified magnetic nanopaticles (dMNPs) were one example of a highly efficient gene delivery system.Citation14–Citation16 Fluorescent magnetic nanoparticles (FMNPs) can be used as dual-modality imaging contrast reagents for long-term observation of tumor cells.Citation17–Citation19
In this study, we used PAMAM dMNPs and FMNPs, prepared human iPS cells by using Generation 5.0 (G5.0) PAMAM dMNPs and lentivirus vectors, and labeled iPS cells with FMNPs for long-term observation and tracking. Our aim was to investigate the feasibility of using PAMAM dMNPs to enhance the efficiency of preparing lentivirus with four genes and laying the foundations for further studies on iPS cell distribution, imaging, and tracking in vivo by using FMNPs. Our results show that PAMAM dMNPs could markedly increase the titer of lentivirus in supernatant liquids of 293T cells. The iPS cells can be successfully generated from human fibroblast cells transducted with prepared lentivirus and labeled with FMNPs, which has great potential application in long-term tracking and functional research into human iPS cells in vivo.
Materials and methods
Cell culture
Human dermal fibroblast (HDF) cells were primary cultured. 293T cells and MEF cells were provided by the Shanghai Institute of Digestive Diseases, Renji Hospital, Shanghai Second Medical University, Shanghai, People’s Republic of China. HDF, 293T, and MEF cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM, GIBCO®) containing 10% fetal bovine serum (FBS, GIBCO) and 1.0% penicillin and streptomycin (GIBCO). iPS cells were generated and maintained in a human embryonic stem (ES) cell medium, which was made up with F12/DMEM (GIBCO) supplemented with Knockout SR® (GIBCO), basic fibro-blast growth factor (Invitrogen), nonessential amino acids (GIBCO), L-glutamine (GIBCO), and β-mercaptoethanol (GIBCO). iPS cells were cultured on MEF feeder cell dishes, which were coated with BD Matrigel™ in advance according to the standard human ES cell culture protocol.Citation20
Plasmid construction
Packaging plasmid psPAX2, enveloping plasmid pMD2.G, and four transcription factors pSin-EF2-Oct4-Pur, pSin-EF2-Sox2-Pur, pSin-EF2-LIN28-Pur, and pSin-EF2-Nanog-Pur were purchased from Addgene Inc., Cambridge, MA, USA (Cat No. 12260, 12259, 16579, 16577, 16580, and 16578). We extracted the plasmids from the bacteria by QIAGEN® Maxi Kit according to the manufacturer’s protocol. These plasmids were identified by enzyme digestion. According to the plasmid maps, we chose EcoRV (TaKaRa) and SacI (TaKaRa) as restriction enzymes of psPAX2, NotI (TaKaRa) and HindIII (TaKaRa) as restriction enzymes of PMD2.G, and EcoRI (TaKaRa) and SpeI (TaKaRa) as restriction enzymes of Oct4, Sox2, Nanog, and LIN28.
Preparation and characterization of dMNPs
MNPs were prepared by coprecipitation of Fe2+ and Fe3+ in the presence of NaOH,Citation21,Citation22 and were dispersed in phosphate buffered saline (PBS) (pH 7.4). G5.0 PAMAM dMNPs were prepared according to our previous reports.Citation23,Citation24 Generation 0 (G0) dMNP represents the MNPs modified with only 3-aminopropyltrimethoxysilane (NH2(CH2)3-Si-(OCH3)3). Dendrimer generation was initiated with G0 methanol solution. Stepwise growth using methacrylate and ethylenediamine was repeated until the desired number of generations from 1.0 to 5.0 (G1.0–G5.0) was achieved. To characterize the samples, portions of the G5.0 PAMAM dMNP suspensions were dried at 80°C under vacuum. The average size was estimated using a transmission electron microscope (TEM, JEOL, JEM2010, Japan). Fourier transform infrared (FT-IR) spectra of suspensions were obtained using an FT-IR spectrophotometer (AVATAR 360, Nicolet, USA).
Transfection plasmid and lentivirus production
293T cells were plated in 100 mm dishes at 80% confluence and were transfected with plasmids using dMNPs. Respectively, different plasmids (Oct4, Sox2, LIN28, Nanog, PSPAX2, and PMD2.G) were mixed with dMNPs in DMEM without FBS and incubated for 30 minutes at room temperature under the condition of 10:1 of the charge ratio of dMNPs and plasmids complex (the mass ratio is 4:3:2 of transcription factor, PSPAX2, and PMD2.G). 293T cells were transfected with medium without serum by the addition of four kinds of dMNPs–plasmids complex (Oct4, Sox2, LIN28, Nanog), respectively. We changed the fresh medium after transfection for 4 hours. Then, the four kinds of supernatant of medium (Oct4, Sox2, LIN28, Nanog) were collected after transfection for 48 hours, respectively. Supernatants were filtered through 0.45 μm pore size cellulose acetate filters (Millipore, Billerica, MA, USA) and centrifuged at 23,000 rpm for 90 minutes at 4°C. The viral pellets were resuspended in DMEM to obtain a 10,000-fold concentration. Viral stocks were stored at −80°C until transduction. Viral titers were measured according to the protocol of the QuickTiter™ Lentivirus Quantitation Kit (Cell Biolabs, San Diego, CA, USA) and were determined by infection of 293T cells. Lipofectamine 2000 was used as a control transfection reagent.
Lentivirus transduction and reprogramming culture
HDF cells were plated in a 10 cm dish at 80% confluence before transduction. We prepared a medium with 10% FBS by the addition of virus solution, which mixed an equal volume of concentrated virus solution, such as Oct4, Sox2, LIN28, and Nanog (according to 10:1 of the number ratio of virus and cells), then we transducted the medium supplemented with 10 μg/mL polybrene into HDF cells and co-culture overnight at 37°C in a humidified 5% CO2 incubator. We used fresh medium every day for the first 3 days and a human ES cell medium every day for the following days. Twenty-one days after transduction, some colonies emerged. The colonies’ morphology was similar to that of human ES cells. We extracted colonies into an MEF feeder and cultured the iPS cells according to the standard human ES cell culture protocol.Citation20
Reverse transcription-polymerase chain reaction (RT-PCR)
PCR primers were designed and synthesized. The concrete sequences were as follows:
Oct4-forward 5′-CAGTGCCCGAAACCCACAC-3′
Oct4-reverse 5′-GGAGACCCAGCAGCCTCAAA-3′
Sox2-forward 5′-AGCTACAGCATGATGCAGGA-3′
Sox2-reverse 5′-GGTCATGGAGTTGTACTGCA-3′
Nanog-forward 5′-CAGAAGGCCTCAGCACCTAC-3′
Nanog-reverse 5′-ATTGTTCCAGGTCTGGTTGC-3′
LIN28-forward 5′-TGCGGGCATCTGTAAGTGG-3′
LIN28-reverse 5′-GGAACCCTTCCATGTGCAG-3′.
We isolated the total RNA from the iPS cells using a High Pure RNA Isolation Kit (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) according to the manufacturer’s protocol. First-strand cDNA were primed via random hexamers, which was carried out as described in the product protocol of the Transcriptor High Fidelity cDNA Synthesis Kit (Roche). PCR was then carried out with 1 μL cDNA for one cycle of 94°C for 2 minutes followed by 30 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 3 minutes using gene-specific primers and Taq polymerase. PCR productions were detected by 1% agarose gel electrophoresis with ethidium bromide.
Detection of human ES cell surface-specific biomarkers by immunofluorescence staining and flow cytometry analysis
The iPS cells were fixed with 4% paraformaldehyde (prepared freshly in PBS) for 20 minutes at room temperature and washed three times with PBS for 5 minutes each. Cells were blocked with 1% bovine serum albumin (BSA) and 10% normal donkey serum in PBS (blocking buffer) at room temperature for 45 minutes. After blocking, cells were probed with anti-SSEA-3 (Chemicon, Temecula, CA, USA), anti-SSEA-4 (Chemicon), anti-Tra-1-60 (Chemicon), and anti-Tra-1-81 (Chemicon) primary antibody. Then, cells were washed three times with 2 mL of 1% BSA PBS for 5 minutes each. After washing, cells were incubated with PE-goat-anti-rat (AbD serotec) and PE-goat-anti-mouse (Caltag) secondary antibody. Hoechst (Invitrogen) were used for counterstaining. Cells were then washed three times with PBS for 5 minutes each, and the plates were stored wrapped in aluminum foil at 4°C in 2 mL PBS until they could be visualized with a fluorescence microscope.
Adherent cells were individualized by trypsin treatment (0.05% trypsin/0.5 mM EDTA, Invitrogen) and were processed directly for antibody staining. The cells were filtered through a 40 μm mesh and resuspended in FACS buffer (PBS containing 2% FBS and 0.1% sodium azide). About 100 μL of cell suspension containing 5 × 105 cells was used in each labeling. Both primary and secondary antibody incubation was carried out at room temperature for 30 minutes. After washing, the cells were resuspended in 300–500 μL of the FACS buffer and proceeded for analysis on a BD FACSCalibur™ Flow Cytometer using CellQuest™ acquisition and analysis software. A total of 20,000 events were acquired.
Embryoid body formation and in vitro differentiation into three germ layers
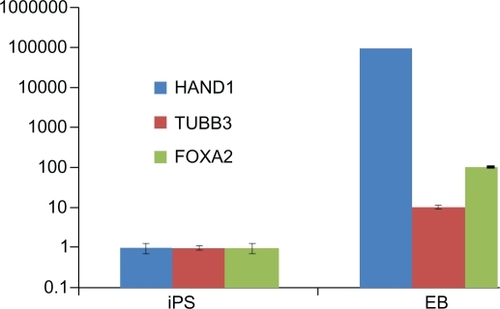
Expression of three germ layer differentiation markers was analyzed by quantitative RT-PCR. iPS cells were passaged five times, cell clumps were collected and washed four times with DMEM/F12, cell clumps were transferred to 6-well Low Attachment Culture Dishes (Corning®, NY, USA) with embryoid body (EB) medium change. EB formation was monitored by microscope. For quantitative gene expression analysis, EBs were collected at day 12, RNA was isolated using the High Pure RNA Isolation Kit (Roche), reverse transcription and real time PCR was performed using Transcriptor High Fidelity cDNA Synthesis Kit (Roche) and ExTaq™ (Takara Bio Inc., Shiga, Japan). β-actin was used as a normalization control. Expression of TUBB3 (ectoderm), HAND1 (mesoderm), and FOXA2 (endoderm) markers was quantified against undifferentiated control iPS cells.
Karyotype analysis
The iPS cells were passaged eight times and treated with 0.4 μg/mL colcemid for 2 hours at 37°C. The iPS cells were collected by 0.05% trypsin treatment and cocultured with the 0.5 mL 0.075 mol/L KCL in 37°C for 30 minutes, then swelled with glacial acetic acid and fixed with 2.5% glutaraldehyde and finally stained with Gemsa. Karyotype analysis was performed at the same time.
Terotoma formation and HE staining
The human iPS cells from the 100 mm dish grown on MEF feeder layers were harvested by collagenase IV treatment and collected into tubes and centrifuged, then suspended in 100 μL human ES cell medium and injected subcutaneously to the dorsal flank of 6-week-old NOD-SCID mice. HDF cells were also injected as control. One to 2 months postinjection, tumors were typically observed and surgically dissected from the mice and fixed with 4% paraformaldehyde. Tumor samples were embedded in paraffin, cut into 20 μm sections, stained with hematoxylin and eosin, and examined by light microscopy.
Labeling iPS cells with FMNPs
FMNPs were synthesized and saved by our laboratory. The preparation and characterization of FMNPs were reported in our previous papers.Citation25–Citation27 The iPS cells were seeded on the cover slips and treated with growth medium containing FMNPs (50 μg/mL) for 4 hours at 37°C with 5% CO2. Afterwards, the cells were rinsed with PBS and fixed with 2.5% glutaraldehyde for 30 minutes. Finally, the samples were attached to glass plates using mounting medium and were observed under fluorescent microscope. Labeled iPS cells were digested by trypsin treatment (0.05% trypsin/0.5 mM EDTA, Invitrogen) and proceeded for analysis on a BD FACSCalibur Flow Cytometer using CellQuest acquisition and analysis software. A total of 20,000 events were acquired. We also used the magnetic resonance imaging (MRI) system to observe iPS cells labeled with FMNPs. The iPS cells were treated with 50 μg/mL FMNPs for 4 hours and were harvested by magnetic iron. Then, the labeled iPS cells were fixed with 70% ethanol/PBS for 30 minutes on ice until imaging. MRI was performed using 3.0T field intensity by a GE HDX 3.0T MRI instrument equipped with GE Signa Excite 3.0T MRI software. The imaging protocol consisted of coronal and transverse T2-weighted spin echo (SE) pulse sequences. To produce T2 maps, the following imaging parameters were used: TR/TE = 1000/10, 20, 30, 40, 50, 60, 70, 80 ms; field of view = 8.0 cm; slice thickness = 2.0 mm; number of excitations = 2.
Statistical analysis
Each experiment was repeated three times in duplicate. The results were presented as mean ± standard deviation. Statistical differences were evaluated using the t-test and considered significant at P < 0.05.
Results
Identification of plasmids by enzyme digestion method
The pure vectors, including Oct4, Sox2, Nanog, LIN28, PMD2.G, and psPAX2, were extracted from the DH5α bacteria with lentivirus expression plasmids such as pSin4-EF2-Oct4-Pur, pSin4-EF2-Sox2-Pur, pSin4-EF2-Nanog-Pur, pSin4-EF2-LIN28-Pur, enveloping plasmids PMD2.G, and packaging plasmid psPAX2. As shown in (supporting data), the six plasmids’ single-site enzyme digestion results coincide with the dual-site enzyme digestion results, which show that six plasmids were successfully prepared.
Characterization of G5.0 dMNPs
The prepared dMNPs were characterized as shown in (supporting data). dMNPs were dispersed very well with an average diameter of 20–40 nm. The dendrimer modification process was proven by comparison of FT-IR spectra of the G5.0 dMNPs and MNPs. Compared with the MNP sample, the G5.0 dMNPs possess absorption bands at 2922 cm−1 and 2852 cm−1 due to the stretching vibration of the C–H bond, bands at 3422 cm−1 due to the bending vibration of the –NH2 group, and bands at 1719, 1637, 1560, and 1398 cm−1 due to the –CO–NH– group. All of those results proved the existence of dendrimer on the surface of MNPs.
Reprogramming HDF cells into iPS cells by dMNPs and lentivirus
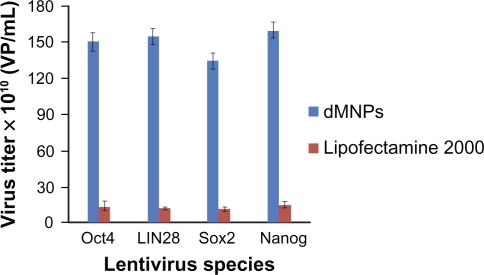
293T cells were cultured to prepare four kinds of lentivirus with Oct4, Sox2, Nanog, and LIN28 by using dMNPs as a transfection reagent. Compared with Lipofectamine 2000, as shown in , the titers of lentivruses based on dMNPs were 10-fold more than those based on Lipofectamine 2000. For example, the titers of Oct4 virus were, respectively, 1.5 × 1012 VP/mL for dMNPs, and 1.4 × 1011 VP/mL for Lipofectamine 2000. Similarly, the results of the other three kinds of virus titer were also similar to Oct4. For example, the titers of Sox2 virus were 1.35 × 1012 VP/mL and 1.2 × 1011 VP/mL, the titers of Nanog virus, respectively, were 1.6 × 1012 VP/mL and 1.5 × 1011 VP/mL, and the titers of LIN28 virus, respectively, were 1.55 × 1012 VP/mL and 1.3 × 1011 VP/mL based on dMNPs and Lipofectamine 2000 as transfection agents. Therefore, this proved that dMNPs could enhance the preparation efficiency of lentivirus with Oct4, Sox2, Nanog, or LIN28 genes. The transfection efficiency of dMNPs was 10-fold more than that of Lipofectamine 2000, which would provide more lentivirus for the following preparation of iPS cells.
Figure 1 Titering different kinds of lentivirus produced by different transfection reagents.
Abbreviation: dMNPs, dendrimer-modified magnetic nanoparticles.
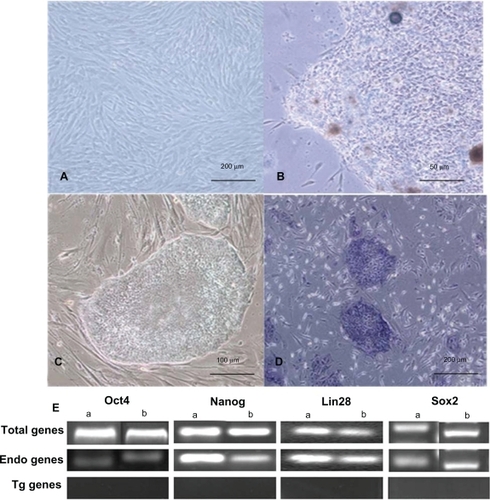
The 293T cells’ supernatants were collected and concentrated after 48 hours of transfection for transduction usage. shows the primary cultured HDF cells. At 21 days after transduction, we observed iPS cell clones that look like ES cell clones, as shown in . The cells had a clear boundary, brighter large nucleoli, and scant cytoplasm, similar to human ES cells, as shown in . The cells exhibited positive staining for alkaline phosphatase, which suggests that the clone cells should be iPS cells.
Figure 2 Generating iPS cells from HDF cells. A) The morphology of primary passage of human foreskin fibroblast (100X). B) Primary induced pluripotential stem cell colony (400X). C) iPS cells grown on irradiated MEFs (200X). D) Alkaline phosphatase staining of iPS cells (100X). E) The gene expression profiles of iPS cells by DNA electrophoresis.
Notes: aThe iPS cells were generated based on Lipofectamine 2000; bThe iPS cells were generated based on dMNPs.
Abbreviations: dMNPs, dendrimer-modified magnetic nanoparticles; HDF, human dermal fibroblast; iPS, induced pluripotent stem; MEF, mouse embryonic fibroblasts.
Identification of iPS cells
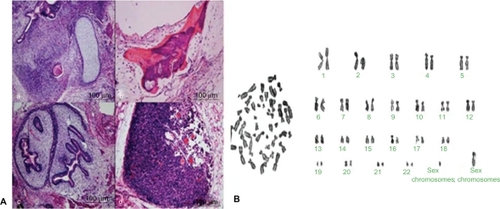
To identify the iPS cells at the RNA level, we examined the expression levels of four genes by RT-PCR. As shown in , endogenous Oct4, Sox2, LIN28, and Nanog were reactivated and the exogenous transgenes were silenced, indicating that the pluripotent state was not maintained by continuous expression of exogenous factors. To confirm the similarity of iPS cells and ES cells, we checked the expression levels of ES cell-specific biomarkers by fluorescent immunostaining and flow cytometry analysis. As shown in , the ES cell-specific surface markers, such as SSEA-3, SSEA-4, Tra-1-60, and Tra-1-81, exhibited positive expressions in the prepared iPS cells, but no positive staining was observed in the control HDF cells (data not shown). shows the flow cytometry expression analyses of human ES cell-specific markers in reprogrammed clones, which exhibited the prepared iPS cells highly expressed with SSEA-3, SSEA-4, Tra-1-60, and Tra-1-81. In addition, in order to demonstrate the pluripotency of iPS cells, we observed that the teratoma grew on the back of NOD-SCID mice, and HE staining results showed that the teratoma tissues were composed of three germ layers (endoderm, mesoderm, and ectoderm), as shown in , which fully demonstrates that the prepared iPS cells were able to differentiate into all three germ layers evidenced by the neural ganglia, supporting cartilage, bone and smooth muscle, submucosa glands, and neural epithelium. Simultaneously, we also carried out karyotype analysis of iPS cells. The iPS cells had a normal karyotype. G-banding chromosome analysis is represented in . To further determine the pluripotency of iPS cells, EBs were formed after iPS cells were cultured in suspension for 7 days. Quantitative RT-PCR analysis was performed on 12-day-old EBs with expression of HAND1 (heart and neural crest derivatives expressed 1-mesoderm), TUBB3 (tubulin beta 3-ectoderm), and FOX-A2 (forkhead box A2-endoderm), as shown in . All these data fully demonstrate that the iPS cells were successfully prepared.
Figure 3 A) Fluorescence microscopy images of the human induced pluripotent stem cells were stained with antibody recognizing SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81. B) Flow cytometry analysis of human embryonic stem cell-specific markers such as SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81 in induced pluripotent stem cells.
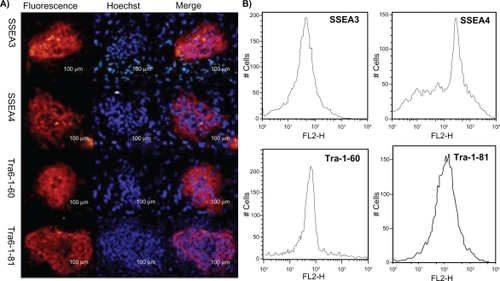
Figure 4 A) Histological analysis of teratomas formed from grafted colonies of induced pluripotent stem cells in severe combined immunodeficient mice: a) Neural ganglia and supporting cartilage; b) bone and smooth muscle; c) submucosa glands; d) neural epithelium. B) G-banding chromosome analysis of induced pluripotent stem cells.
Labeling iPS cells with FMNPs
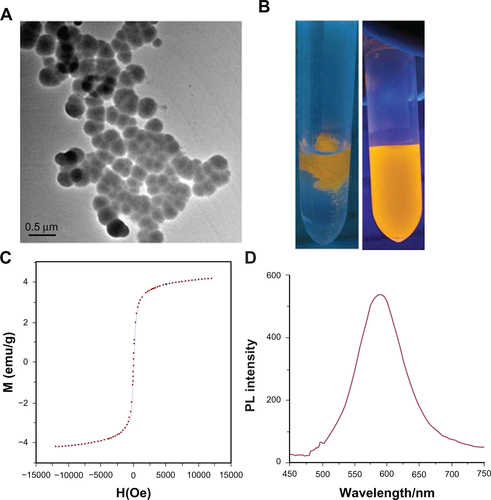
The prepared FMNPs were characterized as shown in (supporting data). TEM imaging exhibited that FMNPs were sphere with an average diameter of 100 nm. shows the fluorescence of FMNPs under ultraviolet radiation. FMNPs were assembled and the solution became transparent under the external magnetic field (left). After removal of the external magnetic field, the aggregations were rapidly redispersed evenly (right). shows a magnetic hysteresis curve of FMNPs. This clearly indicates that FMNPs maintained super-paramagnetic property at room temperature and reached a saturation magnetization (Ms) value of 4.0 emu g−1. The photoluminescence spectra of FMNPs are shown in . The emission peak of FMNPs remained symmetric.
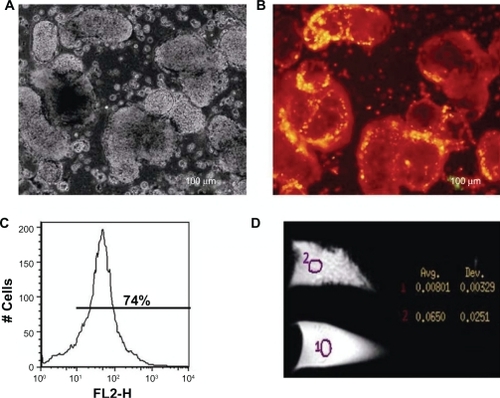
After FMNPs were incubated with iPS cells for 4 hours, we observed that FMNPs entered the iPS cells. shows the morphology of iPS cells under a bright field. shows iPS cells with a strong red fluorescent signal under fluorescent microscopy, which suggests that FMNPs are located inside iPS cells. shows the flow cytometry analysis result of intracellular FMNPs. The iPS cells labeled with 50 μg/mL FMNPs expressed 74% positive signals. In order to further confirm the iPS cells labeled with FMNPs, we used an MRI system to detect the magnetic signals of iPS cells. The labeled iPS cells and control iPS cells were, respectively, dispersed in 70% ethanol/PBS fixed buffer in eppendorf tubes. Cells underwent MRI under 3.0T field intensity. T2-weighted images were obtained by adding together the different T2-weighted data during raw data acquisition, as shown in . The average magnetic resonance intensity of iPS cells labeled with FMNPs was 0.0650T, and the average magnetic resonance intensity of iPS cells not labeled with FMNPs was 0.00801T. This exhibited that the labeled iPS cells had stronger magnetic signals than control iPS cells.
Figure 6 The images of induced pluripotent stem cells (iPS) labeled with fluorescent magnetic nanoparticles (FMNPs). A) The microscopy image of iPS cells labeled with FMNPs under bright light (200X). B) The fluorescent microscopy image of iPS cells labeled with FMNPs (200X). C) Flow cytometry analysis of iPS cells labeled with FMNPs. D) The magnetic resonance imaging of iPS cells with or without labeled FMNPs.
Discussion
Since the iPS cells were successfully produced, iPS cell research and development has become a hot topic. Although iPS cells exhibit an attractive application prospect in regenerative medicine, to date, how to obtain high-quality iPS cells is still a great challenge. So far, iPS cell in vivo distribution and development are still not clarified, which is due to the shortage of more efficient iPS cell-labeling technologies. However, using nanomaterials’ unique properties may help to solve current problems.
In this study, in order to obtain iPS cells efficiently, at first, we had to obtain enough lentivirus with Oct4, Sox2, LIN28, and Nanog. In our previous work, we found that G5.0 PAMAM dMNPs with good biosafety can take a lot of genes and highly efficiently deliver genes into different kinds of cells. Thus, we finally selected G5.0 PAMAM dMNPs as a delivery system for four transcription factor genes, ie, Oct4, Sox2, LIN28, and Nanog, and packaging plasmids such as PSPAX2 and PMD2.G to enter into 293T cells. The results showed that G5.0 PAMAM dMNPs successfully deliver many vectors with Oct4, Sox2, LIN28, and Nanog genes into 293T cells. The titers of Oct4, Sox2, LIN28, and Nanog viruses produced by dMNPs were 10 times more than those based on Lipofectamine 2000. Thus, we consider that G5.0 PAMMAM dMNPs could enhance the preparation efficiency of lentivirus. This step is very important in order to prepare iPS cells that can provide enough lentivirus for further preparation of iPS cells within a limited timeframe. In the course of follow-up experiments, we collected and concentrated supernatant liquids with lentivirus from 293T cells and coincubated it with human fibroblast cells at 37°C for 21 days. Then, we obtained ES-like cells. We confirmed those ES-like cells were iPS cells by using RT-PCR, immunostaining analysis, and the terotomas formation test. Although the preparation efficiency of iPS cells produced from HDF cells is only associated with integration efficiency of lentivirus into cell genome, not associated with lentivirus titer, high titer of lentivirus can transduct more HDF cells and produce more iPS cells, which is helpful and can save time. Therefore, we consider that preparation efficiency of iPS cells is indirectly enhanced.
Regarding potential mechanism, it is well known that dMNPs are one kind of nanocomposite with a positive charge that has excellent chemical structure and biocompatibility. As the generation of PAMAM dendrimers increased, the amount of plasmids absorbed by dMNPs also increased correspondingly. They were able to attach to the cell membrane surface via charge attraction and then induced nanoscale hole formation on the surface of cell membranes.Citation28,Citation29 They then took plasmids into cytoplasm highly efficiently. Because of the specific environment inside cells, exogenous vectors can be released from the dMNP–plasmid composites and enter the cell nucleus, producing a lot of virus plasmids, then interacting with the packaging plasmids, finally resulting in lentivirus virus formation and secretion into 293T cells’ supernatant. Because G5.0 PAMAM dMNPs can take more vectors into 293T cells than Lipofectamine 2000, the lentivirus titers based on dMNPs are higher than those based on Lipofectamine 2000.
In recent years, with the rapid progress of iPS cell research and development, how to prepare the labeled iPS cells for imaging and tracking in vivo has become an important question. Traditional labeling methods such as cytoplasm markers labeled with PKH26 and DIL, nucleic acid markers labeled with BrdU and DAPI, and gene markers labeled with LacZ and GFP have different limitations. The biggest problem is how to track stem cells in vivo.Citation30–Citation32 In this study, with the aim of investigating the feasibility of labeling iPS cells for long-term tracing and imaging, we successfully labeled iPS cells with FMNPs. Our prepared FMNPs are silica-coated quantum dots and FMNPs, which have both fluorescent signals and magnetic signals and have good biocompatibility. Fluorescent signals can be used for iPS cell tracking and imaging, and magnetic signals can be used for iPS cell isolation and MRI imaging. When prepared iPS cells are incubated with FMNPs for 4 hours or overnight, we observed FMNPs located inside the iPS cells. We continued to culture the iPS cells for 1 month and observed fluorescent signals existing in iPS cells, which highly suggests that FMNPs in iPS cells were not easily exited.
To date, there are some molecular imaging technologies that can be used to track stem cells in vivo, such as bioluminescence imaging (BLI), fluorescence imaging (FI), positron emission tomography (PET), single photon emission computed tomography (SPECT), and MRI.Citation33–Citation37 In this study, we used FI technology to observe iPS cells. The iPS cells exhibited red fluorescent signals. We also used MRI to image the collected iPS cells inside the eppendorf tubes. The labeled iPS cells exhibited higher magnetic intensity signals than the control iPS cells. Although the magnetic intensity difference between labeled iPS cells and control iPS cells was very small, the labeled iPS cells were attracted by an in vitro magnetic field and adhered to the tube wall (data not shown). Conversely, the control iPS cells could not stick to the tube wall. Therefore, we consider that the iPS cells were successfully labeled with FMNPs, which has laid the foundation for further tracking and imaging in the near future.
In conclusion, human iPS cells can be efficiently generated from HDF by using the dMNPs and a lentivirus system. dMNPs markedly enhanced the efficiency of lentivirus supernatants produced from 293T cells and indirectly enhanced the preparation efficiency of iPS cells. The prepared iPS cells were also successfully labeled with FMNPs, which lay the foundation for further isolating, in vivo imaging, and tracking of iPS cells in the near future. The established methods should be the first report. We believe that stem cell nanotechnology has great potential application in the research and development of stem cells in the near future.
Acknowledgements
This work is supported by the National Key Basic Research Program (973 Project) (2010CB933901), National 863 Hi-tech Project (2007AA022004), Important National Science and Technology Specific Project (2009ZX10004-311), National Natural Scientific Fund (No. 20771075 and No. 20803040), Special Project for Nano-technology from Shanghai (No. 1052nm04100), New Century Excellent Talent of Ministry of Education of China (NCET-08-0350, No. 20070248050), Shanghai Science and Technology Fund (10XD1406100), and Pu Jiang Project (09PJ1407300).
Supplemental information
Figure S1 The digestion maps of plasmid DNA. a) Single-site digestion map of plasmids DNA. b) Double-site digestion map of plasmid DNA.
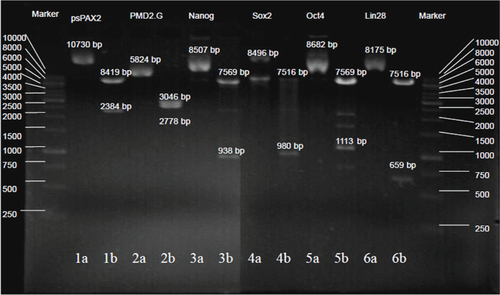
Figure S2 Characterizations of dendrimer-modified magnetic nanoparticles (dMNPs). A) Transmission electron microscope image of dMNPs. B) The Fourier transform infrared spectra of dMNPs: a) MNPs, b) dMNPs.
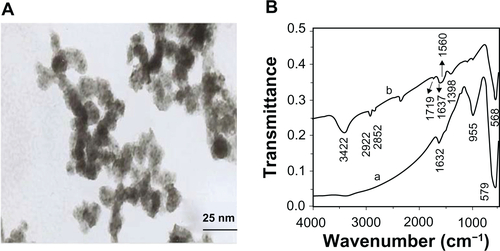
Figure S3 Characterizations of fluorescent magnetic nanoparticles (FMNPs). A) The transmission electron microscope image of FMNPs. B) The fluorescent image of FMNPs with/without external magnetic field. C) The field-dependent magnetization curve of FMNPs at room temperature. D) The photoluminescence spectra of FMNPs.
Disclosure
The authors report no conflicts of interest in this work.
References
- TakahashiKYamanakaSInduction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factorsCell200612666367616904174
- ZhaoXYLiWLvZiPS cells produce viable mice through tetraploid complementationNature20094617260869019672241
- LingCBrianGLiuXRStem cell tracking with membrane intercalating near infrared (NIR) fluorescent dyesJ Nucl Med201051Suppl 239120150248
- SophieBTobiasDHElizabethJSLabeling stem cells with fluorescent dyes for non-invasive detection with optical imagingJ Vis Exp20081468619066580
- WangZRuanJCuiDAdvances and prospect of nanotechnology in stem cellsNanoscale Res Lett2009459360520596412
- JiJRuanJCuiDAdvances of nanotechnology in the stem cells research and developmentNano Biomed Eng201026688
- AniruddhSJohnDKKi-BumLNanotechnology for regenerative medicine: nanomaterials for stem cell imagingNanomedicine20083456757818694318
- SvensonSTomaliaDADendrimers in biomedical applications: reflections on the fieldAdv Drug Deliv Rev2005572106212916305813
- DennigJDuncanEGene transfer into eukaryotic cells using activated polyamidoamine dendrimersJ Biotechnol20029033934712071232
- NageshaDLaevskyGSLamptonPIn vitro imaging of embryonic stem cells using multiphoton luminescence of gold nanoparticlesInt J Nanomedicine20072481381918203448
- CuiDZhangHWangZEffects of dendrimer-functionalized multi-walled carbon nanotubes on mice embryonic stem cellECS Transactions20081314111116
- JainKRole of nanotechnology in developing new therapies for diseases of the nervous systemNanomedicine2006191217716203
- JainKApplications of nanobiotechnology in clinical diagnosticsClin Chem200753200217890442
- SahooSParveenSPandaJThe present and future of nanotechnology in human health careNanomedicine20073203117379166
- PanBCuiDShengYDendrimer-modified magnetic nanoparticles enhance efficiency of gene delivery systemCancer Res200767815617804728
- ShakhbazauAShcharbinDSeviarynIUse of polyamidoamine dendrimers to engineer BDNF-producing human mesenchymal stem cellsMol Biol Rep2010372003200819649724
- KimMHKino-okaMSaitoAMyogenic induction of human mesenchymal stem cells by culture on dendrimer-immobilized surface with d-glucose displayJ Biosci Bioeng20101091556120129083
- YongKHuRRoyITumor targeting and imaging in live animals with functionalized semiconductor quantum rodsACS Appl Mater Interfaces2009171071920160901
- ShiDChoHChenYFluorescent polystyrene-Fe3O4 composite nanospheres for in vivo imaging and hyperthermiaAdv Mater20092121702173
- ThomsonJAItskovitz-EldorJShapiroSSEmbryonic stem cell lines derived from human blastocystsScience19982825391114511479804556
- PanBGaoFAoLInvestigation of interactions between dendrimer-coated magnetite nanoparticles and bovine serum albuminJ Magn Magn Mater2005293252258
- PanBGaoFGuHDendrimer modified magnetite nanoparticles for protein immobilizationJ Colloid Interface Sci20052841615752777
- PanBCuiDGaoFHeRGrowth of multi-amine terminated poly(amidoamine) dendrimerson the surface of carbon nanotubesNanotechnology2006172483248921727493
- PanBCuiDXuPSynthesis and characterization of polyamidoamine dendrimer-coated multi-walled carbon nanotubes and their application in gene delivery systemsNanotechnology20092012510119420458
- HeRYouXGShaoJCore/shell fluorescent magnetic silica-coated composite nanoparticles for bioconjugationNanotechnology20071831560117
- CuiDHanYLiZFluorescent magnetic nanoparticles for in vivo targeted imaging and hyperthermia therapy of prostate cancerNano Biomed Eng200916174
- YouXGHeRGaoFHydrophilic high-luminescent magnetic nanocompositesNanotechnology20071803570115
- WalkkerSSoflatMJKakarlatRCationic facial amphiphiles: a promising class of transfection agentsPNAS199693158515908643675
- PrabalKMTahirÇLinSTEffect of solvent and pH on the structure of PAMAM dendrimersMacromolecules20052383979991
- AliSMohammadsharifTHosseinBProgress and promise towards safe induced pluripotent stem cells for therapyStem Cell Rev20106229730620180049
- ChenLYLiuLCurrent progress and prospects of induced pluripotent stem cellsSci China Life Sci2009527622636
- MahmoodALuDChoppMMarrow stromal cell transplantation after traumatic brain injury promotes cellular proliferation within the brainNeurosurgery2004551185119315509325
- NtziaehristosVRipollJWangLVLooking and listening to light: the evolution of whole-body photonic imagingNat Biotech200523313320
- BindslevLHaaek-$orengenMBisgaardLabelling of human mesenchymal stem cells with indium-111 for SPECT imaging: effect on cell proliferation and differentiationEur J Nucl Med Mol Imaging2006331171117716763813
- WisenbergGLekxKZabelPCell tracking and therapy evaluation of bone marrow monocytes and stromal cells using SPECT and CMR in a canine model of myocardial infarctionCardiovasc Magn Reson200911116
- ChoiDKimJHLimMHepatocyte-like cells from human mesenchymal stem cells engrafted in regenerating rat liver tracked with in vivo magnetic resonance imagingTissue Eng Part C Methods200814152318454642
- RichardSRainerKJakubWTransferrin receptor upregulation: in vitro labeling of rat mesenchymal stem cells with superparamagnetic iron oxideRadiology200724451452317562811