Abstract
Background
Selenium nanoparticles (SeNPs) loaded with chemotherapeutic drugs provided a novel perspective for cancer therapy.
Materials and methods
Here, SeNPs were modified with cyclic peptide (Arg–Gly–Asp–d-Phe–Cys [RGDfC]) to fabricate tumor-targeting delivery carrier RGDfC-SeNPs and, then, doxorubicin (DOX) was loaded to the surface of RGDfC-SeNPs for improving the antitumor efficacy of DOX in non-small-cell lung carcinoma therapy.
Results
The chemical structure characterization of RGDfC-Se@DOX showed that DOX was successfully loaded to the surface of RGDfC-SeNPs to prepare functionalized antitumor drug delivery system RGDfC-Se@DOX. RGDfC-Se@DOX exhibited effective cellular uptake in A549 cells and entered A549 cells mainly by clathrin-mediated endocytosis pathway. Compared to free DOX or Se@DOX at the equivalent dose of DOX, RGDfC-Se@DOX showed greater activity to inhibit A549 cells’ proliferation and migration/invasion and induce A549 cells’ apoptosis. More importantly, compared with passive targeting delivery system Se@DOX, active targeting delivery system RGDfC-Se@DOX exhibited more significant antitumor efficacy in vivo.
Conclusion
Taken together, RGDfC-Se@DOX may be a novel promising drug candidate for the lung carcinoma therapy.
Introduction
Lung cancer that is known as pulmonary carcinoma or carcinoma of the lung can be categorized into non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC).Citation1–Citation3 The NSCLC makes up approximately 80% of all kinds of lung cancer, and the incidence and mortality ratio has been quickly rising worldwide.Citation4,Citation5 Chemotherapy is still an important treatment strategy for cancer therapy clinically. Doxorubicin (DOX) is a very common anticancer drug clinically.Citation6,Citation7 Nevertheless, the anticancer efficacy of DOX is not so ideal as expected partly because of its lack of targeted specificity, poor solubility, inadequate drug accumulation in the tumor, and serious side effects.Citation8,Citation9 Therefore, it is very crucial to improve the efficiency of chemotherapy by using new technologies.
Recently, nanotechnology application in the field of cancer therapy has provided a large number of advantages, such as early diagnosis, multifunctional therapy, and drug delivery systems.Citation10,Citation11 Many nanoscale drug delivery systems including polymers, mesoporous silica, liposomes, and nanoparticles were used to fabricate the nanoscale anticancer drug delivery systems with passive tumor-targeting property, which resulted from enhanced permeability and retention (EPR) effects.Citation12–Citation14 In these nanoscale drug delivery systems, selenium nanoparticles (SeNPs) have received a lot of attention as drug carriers.Citation15,Citation16 First, selenium (Se) as a trace element is very important to human biological process and involves many physiological functions.Citation17,Citation18 Second, Se plays a key role in cancer prevention and immune response.Citation19 Moreover, SeNPs showed some other advantages as drugs’ carrier, for example, the controlled size, potent drug loading capacity, improved antitumor effect, and low cytotoxicity.Citation20,Citation21 Thus, SeNPs gradually developed into a excellent anticancer drug carrier. However, there is still some deficiency, especially the lack of active tumor-targeted capacity still existed in such delivery carrier.Citation22 To obtain high targeting ability, several tumor-targeted molecules, such as arg-gly-asp (RGD) peptide, hyaluronic acid, and folate, were used for decorating the nanoparticles.Citation23–Citation26
The NSCLC A549 cell was a typical lung cancer cell; thus, A549 cells were used for lung cancer therapy researches in this study. In order to improve the anticancer capacity of DOX in NSCLC A549 cells, cyclic peptide (Arg–Gly–Asp–d-Phe–Cys [RGDfC]) was loaded to the surface of SeNPs. The RGDfC peptide is able to specifically bind with αvβ3 integrins, which are overexpressed on various cancer cells.Citation27 RGDfC was loaded to the surface of SeNPs to fabricate the tumor-targeted carrier RGDfC-SeNPs, and DOX was loaded to the surface of RGDfC-SeNPs to prepare functionalized antitumor nanomedicine RGDfC-Se@DOX, which was effectively capable of inhibiting the proliferation, migration, and invasion of the A549 cells and obtaining favorable in vivo antitumor efficacy.
Materials and methods
Materials
Cyclic peptide (RGDfC) was purchased from China Peptides Co., Ltd. (Shanghai, China) DOX hydrochloride (DOX⋅HCl), sodium selenite (Na2SeO3), ascorbic acid (vitamin C [Vc]), dimethyl sulfoxide (DMSO), Annexin V-fluorescein isothiocyanate (FITC)/PI kit, DAPI and MTT were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). The antibody was obtained from Cell Signaling Technology (Danvers, MA, USA). Penicillin–streptomycin, DMEM, and FBS were obtained from Thermo Fisher Scientific (Waltham, MA, USA).
Preparation and characterization of RGDfC-Se@DOX
SeNPs were prepared as previously reported with partial modification.Citation28 Briefly, 0.25 mL of Na2SeO3 (0.1 M) solution and 2 mL of Vc (0.5 mM) solution were slowly added into 22.75 mL of Milli-Q water in a 50 mL beaker. Then, the solution mixtures were magnetically stirred for 30 min at room temperature to prepare SeNPs. Then, RGDfC was added to SeNPs and was magnetically stirred for 2 h at room temperature to prepare RGDfC-modified SeNPs (RGDfC-SeNPs). After that, 2 mg of DOX HCl was dissolved in 5 µL of DMSO and, then, added into RGDfC-SeNPs, and the mixed solutions were magnetically stirred for another 8 h. At last, high-purity RGDfC-Se@DOX was obtained via dialyzing reaction solutions for 4 h. Morphology of nanoparticles was characterized by transmission electron microscopy (TEM). The average size of nanoparticles was characterized by dynamic light scattering (DLS) analysis. The elemental analysis of nanoparticles was characterized by energy-dispersive X-ray (EDX). The chemical structures were characterized by Fourier transform infrared (FTIR). The size of nanoparticles in the water solution was continuously observed for 16 days.
Cell culture
NSCLC A549 cells and human umbilical vein endothelial cells (HUVEC) were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA) and were cultivated in DMEM with 10% FBS at 37°C with 5% CO2.
Cellular uptake study
A549 cells at the density of 4×104 cells/well were incubated in a 24-well plate for 12 h. Then, A549 cells were exposed to Se@DOX or RGDfC-Se@DOX at the DOX dose of 4 µg/mL for 1, 2, and 4 h. Then, the cells were washed with PBS and stained with DAPI for 15 min. Subsequently, the cells were washed with PBS three times and the images of cells were observed under the fluorescence microscope (DMi8; Leica Microsystems, Wetzlar, Germany).
A549 cells were used to research the cellular uptake mechanism. Briefly, 0.5 mL of A549 cell suspension at a density of 2×105 cells/mL was seeded in a 12-well plate and incubated at 37°C for 24 h. After that, the cells were exposed to RGDfC-Se@DOX a the DOX dose of 4 µg/mL for 4 h in the absence of inhibitor at 4°C, or with 3 mg/mL of sodium azide (NaN3) +50 mM 2-deoxy-d-glucose (DOG) or various cellular uptake inhibitors, such as chlorpromazine (2 µg/mL), amiloride (5 µg/mL), and nystatin (4 µg/mL) at 37°C. Then, the cells were washed with PBS and gently collected. The collected cells were filtered using 22 µm nylon mesh and then measured by flow cytometer (BD Biosciences, San Jose, CA, USA).
In vitro release of DOX
The release profiles of DOX from RGDfC-Se@DOX were evaluated using the dialysis method. Briefly, 1 mg of RGDfC-Se@DOX was suspended in 1 mL of PBS and transferred into a dialysis tube. Afterward, the tubes were immersed in 49 mL of buffer solution at pH 5.4 or 7.4 under horizontal shaking (100 rpm). The released DOX was monitored for 30 h by measuring the fluorescence intensity of DOX at an excitation wavelength of 535 nm. At predetermined time points, aliquots of 1 mL were removed for fluorescence test and replenished with the equivalent volume of fresh PBS. The amount of payload released was determined by comparing with an experimentally determined standard curve. All the payload release assays were carried out in triplicate, and the results were expressed as the average data with SD.
MTT assay
The MTT assays were performed to test the cellular cytotoxicity of nanoparticles.Citation29 A549 cells at a density of 5×104 cells/mL were added to a 96-well culture plate and incubated for 24 h at 37°C. The cells were then incubated with free DOX, Se@DOX, and RGDfC-Se@DOX (various equivalent DOX concentrations) or with various concentrations of RGDfC-SeNPs at 37°C for 48 h, respectively. Then, the medium was taken away and 100 µL of medium containing 20 µL of MTT (0.5 mg/mL) was gently added to each well, followed by incubation for another 4 h. Then, the medium was taken away and 200 µL of DMSO was added to each well. The culture plate was incubated for another 0.5 h. The absorbance of each well was tested at 570 nm by a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA). The cytotoxicity of RGDfC-Se@DOX against HUVEC was also tested using the abovementioned method.
Wound healing assay
The cell migration was determined by wound healing assays.Citation30 In brief, A549 cells (5×104 cells/well) were seeded in a 24-well plate and cultured in a complete medium to reach full confluence. The monolayer cells were scratched using the sterile 10 µL pipette tip. Then, the medium was taken away, and a fresh DMEM containing 2% FBS was added to each well. Afterward, A549 cells were co-cultured with DOX, Se@DOX, and RGDfC-Se@DOX at an equivalent DOX concentration of 4 µg/mL and incubated at 37°C for 24 h. The scratch closure of scratched monolayer cells was observed and photographed at 0 and 12 h. Average scratched width between the sides of the wound was measured at three random areas. The migration rate was calculated according to the following equation: cell motilities (%) = [1− (distance of scratched area at 24 h/distance of scratched area at 0 h)] ×100%.
Transwell assay
The cell invasion was analyzed using transwell chambers (8 µm).Citation31 In brief, A549 cells (1×105 cells/mL) were seeded into the upper chambers of 24-well transwell plates. Then, the cells were exposed to free DOX, Se@DOX, and RGDfC-Se@DOX at an equivalent DOX concentration of 4 µg/mL for 24 h, respectively. After that, the cells at a density of 5×104 cells/well were resuspended in 100 µL of serum-free medium and then added to the upper chamber, and subsequently, 300 µL of complete medium was added to the bottom chamber. After 24 h, the filter was taken away from the plate and the cells remaining on the upper filter were wiped gently. The cells that migrated to the bottom chamber were fixed using methanol and stained with crystal violet for 2 min. The average migrating cells in six independent views were photographed using microscope.
Flow cytometry assay
The cell cycle distribution was tested using flow cytometry (BD Biosciences). Briefly, A549 cells were exposed to free DOX, Se@DOX, and RGDfC-Se@DOX at an equivalent DOX concentration of 4 µg/mL for 24 h and washed twice with cold PBS. The precooled 75% ethanol was added to the collected cells for fixation at −20°C overnight and then stained with propidium iodine (PI) in the dark place for 30 min. The DNA contents were analyzed by the Modfit software (Verity Software House, Brewer, ME, USA). Flow cytometry was also utilized to examine the effect of RGDfC-Se@DOX on the cellular apoptosis of A549 cells. Briefly, A549 cells were exposed to free DOX or Se@DOX or RGDfC-Se@DOX at an equivalent DOX concentration of 4 µg/mL and washed twice with PBS. Then, the cells were collected and stained with Annexin V-FITC/PI for 30 min in the dark. Finally, the stained cells were examined by flow cytometry and the data were analyzed by the FlowJo software (Treestar, Ashland, OR, USA).
Xenograft mouse model
All animal experiments were carried out according to the guideline of Experimental Animal Center of Guangzhou Medical University and approved by the ethics committee of Guangzhou Medical University. The BALB/c nude mice (about 5 weeks old) were used to study the in vivo antitumor efficacy of RGDfC-Se@DOX. A549 cells (1×107 cells/150 µL) were injected into the abdomens of mice subcutaneously. The mice were randomly categorized into four groups (n=6) after the volumes of tumors reached ~100 mm3. Subsequently, saline (control group), DOX, Se@DOX, and RGDfC-Se@DOX (at an equivalent DOX of 2 mg/kg) were intravenously injected to the tumor-bearing mice once every other day. The overall treatment time was 21 days. The tumor volumes were reckoned up using the following formula: the tumor volumes (mm3) =1/2 × length × widthCitation2.
Statistical analysis
All the data are represented as mean ± SD. The differences between two groups were compared using one-way ANOVA. The differences were considered to be significant and highly significant at P<0.05 and P<0.01, respectively.
Results and discussion
Preparation and characterizations of RGDfC-Se@DOX
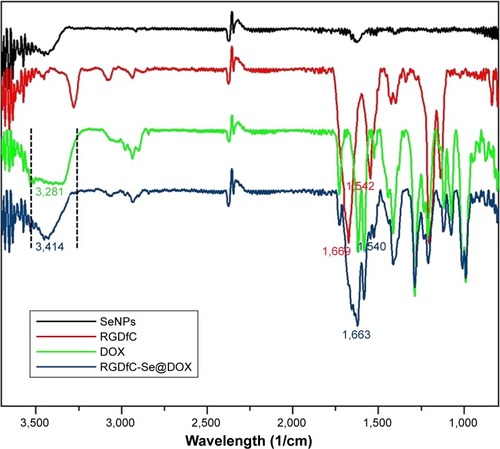
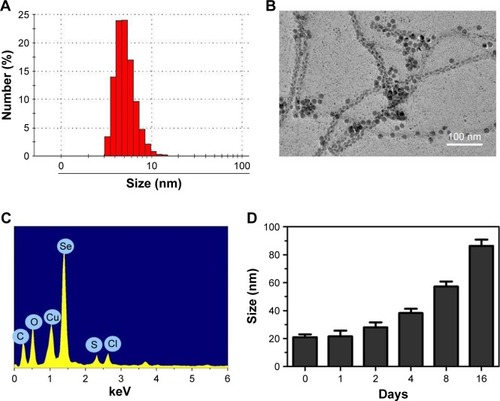
In this study, a novel tumor-targeting delivery system RGDfC-Se@DOX was prepared. The tumor-targeting molecular RGDfC was conjugated with SeNPs to fabricate tumor-targeting delivery carrier RGDfC-SeNPs, and then, the antitumor drug DOX was loaded to the surface of RGDfC-SeNPs to prepare the tumor-targeting delivery system RGDfC-Se@DOX. As shown in , the average size of RGDfC-Se@DOX was 18 nm. The morphology of nanoparticles in TEM image presented spherical particles with a size from about 7 to 12 nm (). A strong signal of Se atom, a typical S atom signal from RGDfC, and a Cl atom signal from DOX⋅HCl were the elemental compositions of RGDfC-Se@DOX (), indicating that RGDfC and DOX were successfully loaded to the surface of SeNPs. The size of RGDfC-Se@DOX in revealed that RGDfC-Se@DOX kept small sizes (<100 nm) for 16 days. It is well known that nanoparticles <200 nm possess passive targeting ability in solid tumors through the EPR effect. Thus, the relative small size in such nanoparticles can be beneficial to the delivery of drugs to the tumor sites. The FTIR spectra of RGDfC-Se@DOX, DOX, SeNPs, and RGDfC are shown in , and the typical peak of SeNPs also appeared in the spectrum of RGDfC-Se@ DOX. The peaks at 1,669 and 1,542/cm correspond to the amide band of RGDfC. After loading RGDfC to SeNPs, the typical amide bands (1,663 and 1,540 cm−1) appeared in the spectrum of RGDfC-Se@DOX, indicating the successful linking of SeNPs with RGDfC via amide bonds. The peak at ~3,281 cm−1 assigned to the characteristic hydroxyl group of DOX existed in the spectrum of RGDfC-Se@DOX, verifying the effective linking between DOX and RGDfC-SeNPs. The above data showed that RGDfC-Se@DOX was successfully synthesized and exhibited good stability in the water solution.
Figure 1 Characterization of RGDfC-Se@DOX.
Notes: (A) Particle size distribution of RGDfC-Se@DOX. (B) Representative TEM image featuring RGDfC-Se@DOX. (C) EDX analysis of RGDfC-Se@DOX. (D) Stability observation of RGDfC-Se@DOX in aqueous solution. RGDfC-Se@DOX, selenium nanoparticles conjugated with RGDfC and DOX.
Abbreviations: DOX, doxorubicin; EDX, energy-dispersive X-ray; TEM, transmission electron microscopy.
Cellular uptake studies
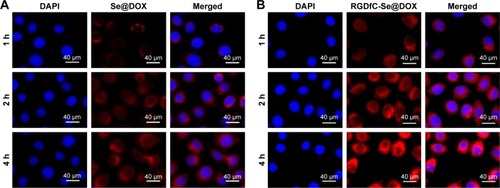
The drug delivery efficiency is closely related to the cellular uptake. High cellular uptake of drug can result in the effective treatment efficacy. Herein, the cellular uptake of passive tumor-targeting nanoparticles Se@DOX and active tumor-targeting nanoparticles RGDfC-Se@DOX was observed in A549 cells under the fluorescence microscope. The red fluorescence from DOX was observed in A549 cells exposed to Se@DOX or RGDfC-Se@DOX for 1 h (), suggesting that such nanoparticles could enter the A549 cells irrespective of the active targeting moiety. The intensity of fluorescence in both Se@DOX- and RGDfC-Se@DOX-treated groups increased as the incubation time prolonged to 4 h. Interestingly, the exposure of A549 cells to RGDfC-Se@ DOX resulted in greater cellular uptake compared with Se@DOX, presumably because of RGDfC-mediated specific endocytosis. After the exposure of RGDfC-Se@DOX to A549 cells for 4 h, a small part of RGDfC-Se@DOX obviously entered the nucleus, where RGDfC-Se@DOX exhibited antitumor activity. However, Se@DOX internalized into the cells and wrapped the cytoplasm at the same time of incubation.
Figure 3 Cellular uptake of Se@DOX (A) and RGDfC-Se@DOX (B) was visualized by red fluorescence from DOX in A549 cells.
Notes: RGDfC-Se@DOX, selenium nanoparticles conjugated with RGDfC and DOX; Se@DOX, selenium nanoparticles conjugated with DOX.
Abbreviations: DOX, doxorubicin; RGDfC, Arg–Gly–Asp–d-Phe–Cys.
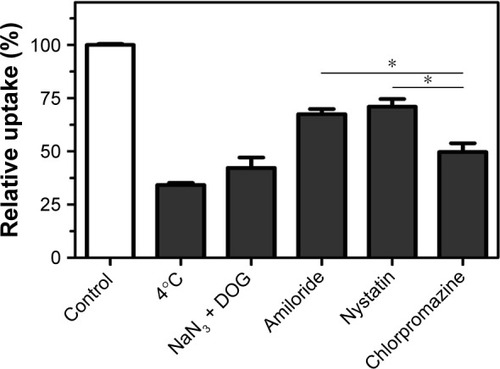
It has been reported that nanoparticles could enter the cells in an energy-dependent endocytosis way.Citation32 The incubation of A549 cells at 4°C or pretreated with NaN3/DOG markedly reduced the cellular uptake of nanoparticles (), indicating that the endocytosis of RGDfC-Se@DOX is an active energy-dependent process. The cells’ endocytosis mainly includes three pathways: clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis. To examine the endocytosis mechanism of RGDfC-Se@DOX in A549 cells, different endocytosis inhibitors were used to study the effect of RGDfC-Se@DOX on cellular uptake. Amiloride, nystatin, and chlorpromazine are usually used to inhibit micropinocytosis, caveolae-mediated endocytosis, and clathrin-associated endocytosis, respectively. After pretreating with amiloride or nystatin, the cellular uptake of RGDfC-Se@DOX was decreased by 32.4% or 28.3%, respectively. Nevertheless, pretreatment with chlorpromazine resulted in about 49.7% decrease in the cellular uptake of RGDfC-Se@DOX, suggesting that clathrin-associated endocytosis mainly contributed to the internalization of RGDfC-Se@DOX.
Figure 4 Effects of endocytosis inhibitors and temperature on the internalization of RGDfC-Se@DOX.
Notes: *P<0.05 vs chlorpromazine group. RGDfC-Se@DOX, selenium nanoparticles conjugated with RGDfC and DOX.
Abbreviations: DOG, 2-deoxy-d-glucose; DOX, doxorubicin; NaN3, sodium azide; RGDfC, Arg–Gly–Asp–d-Phe–Cys.
In vitro release of DOX
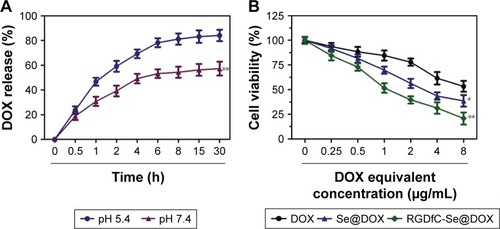
To evaluate the in vitro release behavior of RGDfC-Se@ DOX, we investigated the release profiles in two kinds of buffer solutions (pH 5.4 or 7.4) at 37°C using the dialysis method. The two different pH values were used to simulate the normal physiological environment and the endosomal/lysosomal microenvironments of cancer cells, which are more acidic (pH 4–6). The typical release profiles of DOX from RGDfC-Se@DOX are shown in , and the data revealed that its accumulative release behavior was acid dependent, and there was a significant initial burst payload release during the initial 4 h. The release rates of DOX significantly increased when the pH values changed from 7.4 to 5.4. As shown in , at pH 5.4, 84.2% of DOX was released in a sustained manner during 30 h. However, at the physiological pH value (pH 7.4), DOX was released at a slower rate (57.3%). The faster release in acidic environment may be due to the decrease in the surface negative charge of SeNPs in acidic pH, which weakens the electrostatic attraction of DOX and facilitates the release of DOX from SeNPs. This pH-dependent DOX release property of RGDfC-Se@DOX is quite beneficial for drug delivery systems in cancer therapy, because most tumors present a more acidic condition than normal tissues. Thus, RGDfC-SeNPs have great potential in carrying and releasing therapeutics for treating cancers.
Figure 5 (A) In vitro pH-triggered release of DOX from RGDfC-Se@DOX. **P<0.01 vs the pH 5.4 group. (B) In vitro cytotoxicity of DOX, Se@DOX, and RGDfC-Se@ DOX on A549 cells. *P<0.05 and **P<0.01 vs the free DOX group. RGDfC-Se@DOX, selenium nanoparticles conjugated with RGDfC and DOX; Se@DOX, selenium nanoparticles conjugated with DOX.
Abbreviations: DOX, doxorubicin; RGDfC, Arg–Gly–Asp–d-Phe–Cys.
In vitro cytotoxicity study
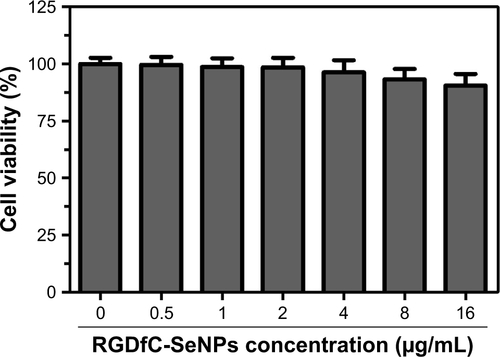
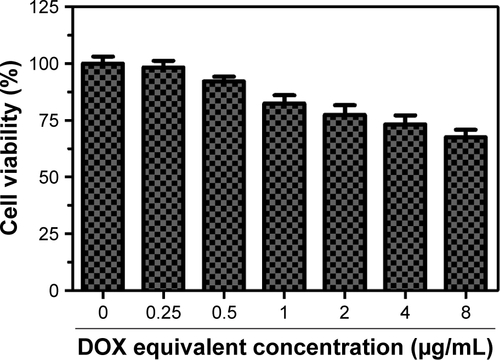
The MTT assay was used to investigate the cytotoxicity of different formulations of DOX against A549 cells in vitro. Free DOX and passive targeting nanoparticle Se@DOX were set as the negative control. As shown in , viability of A549 cells exposed to various formulations of DOX gradually declined with increasing DOX concentrations. Free DOX, Se@DOX, and RGDfC-Se@DOX at the DOX concentration of 8 µg/mL obviously suppressed A549 cell proliferation, and the cell viability rates were 53.3%, 38.6%, and 20.8%, respectively, suggesting that RGDfC-Se@DOX exhibited greater cytotoxicity in A549 cells compared with free DOX and Se@DOX, perhaps because of enhanced cellular uptake of RGDfC-Se@DOX in A549 cells. The cell viability of A549 cells was significantly <50% after treatment with RGDfC-Se@DOX at an equivalent DOX dose of 4 µg/mL; thus, such dose was used for further biological research. The MTT result indicated that the delivery of DOX using active tumor-targeted carrier RGDfC-SeNPs could effectively enhance the anticancer activity of DOX. The inhibition of proliferation of A549 cells treated with drug carrier RGDfC-SeNPs at the used dose was not obvious (), indicating the low cytotoxicity of RGDfC-SeNPs. HUVEC were used to evaluate the potential side effects of RGDfC-Se@DOX. As shown in , the cytotoxicity of RGDfC-Se@DOX against HUVEC was much lower in comparison with A549 cells, which verified the relatively low cytotoxicity of RGDfC-Se@DOX in human normal cell HUVEC.
RGDfC-Se@DOX suppresses the migration and invasion of A549 cells
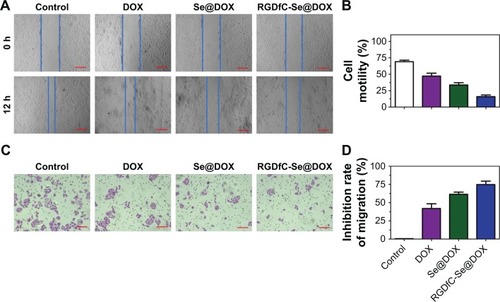
Tumor metastasis and invasion remain the leading causes of death in patients suffering from lung cancer. Effective inhibition of tumor metastasis and invasion is seen as a promising way of delaying tumor growth.Citation33 The wound healing assay of the cells was utilized to assess whether RGDfC-Se@ DOX could effectively inhibit the migration of cancer cells. As shown in , wound healing assay results showed that RGDfC-Se@DOX obviously decreased the migration of A549 cells during a 24 h interval. Meanwhile, the invasion of A549 cells was also strongly inhibited by RGDfC-Se@ DOX (). Furthermore, RGDfC-Se@DOX exhibited higher activity to inhibit A549 cells’ migration and invasion in comparison with the free DOX or Se@DOX (), indicating that RGDfC-Se@DOX is significantly superior to the free DOX and passive targeting delivery system Se@DOX to inhibit the motility and migration of A549 cells.
Figure 6 (A) The scratching width of A549 cells was observed at 12 h after the treatment with DOX, Se@DOX, and RGDfC-Se@DOX. The scale bars are 200 µm. (B) The cell motility of control, DOX, Se@DOX, and RGDfC-Se@DOX was quantitatively analyzed. (C) The effect of DOX, Se@DOX, and RGDfC-Se@DOX on the migration of A549 cells. The scale bar is 200 µm. (D) The inhibition rate of cell migration of DOX, Se@DOX, and RGDfC-Se@DOX was quantitatively analyzed. RGDfC-Se@DOX, selenium nanoparticles conjugated with RGDfC and DOX; Se@DOX, selenium nanoparticles conjugated with DOX.
Abbreviations: DOX, doxorubicin; RGDfC, Arg–Gly–Asp–d-Phe–Cys.
Flow cytometry studies
DOX is a very effective antitumor drug and can induce cancer cell apoptosis. Thus, flow cytometry was used to test whether RGDfC-Se@DOX exhibited greater activity to induce A549 cells’ apoptosis compared with free DOX or Se@DOX at the equivalent DOX dose. In this study, the apoptotic cells with DNA fragmentation were reflected as Sub-G1 peaks. shows that Sub-G1 apoptosis peak of cells in the RGDfC-Se@DOX-treated group was stronger (25.72%) than that in the DOX-treated (12.93%) and Se@DOX-treatment groups (17.98%), indicating that RGDfC-Se@DOX exhibited stronger capacity to induce A549 cells’ apoptosis at the equivalent DOX dose. However, there is no obvious difference in cell cycle distribution among the different treatment groups.
Figure 7 (A) Representative flow cytometry histograms of cell cycle analysis of A549 cells after incubation with various formulations of DOX for 24 h. (B) The percentages of apoptotic A549 cells after incubation with various formulations of DOX for 24 h. RGDfC-Se@DOX, selenium nanoparticles conjugated with RGDfC and DOX; Se@ DOX, selenium nanoparticles conjugated with DOX.
Abbreviations: DOX, doxorubicin; FITC, fluorescein isothiocyanate; PI, propidium iodine; RGDfC, Arg–Gly–Asp–d-Phe–Cys.
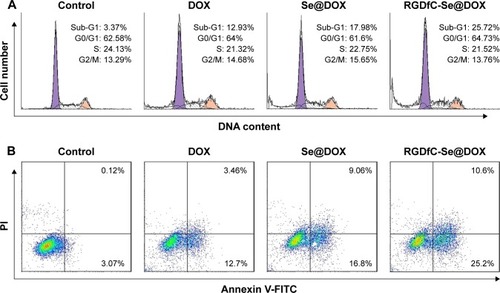
To further detect the apoptosis of A549 cells treated with various formulations of DOX, the cells were analyzed using Annexin V-FITC/PI dual staining. As shown in , RGDfC-Se@DOX treatment obviously induced A549 cells’ apoptosis and resulted in higher cell apoptosis rates (35.8%) compared with the cells treated with DOX (16.16%) or Se@DOX (25.86%) at the equivalent DOX dose. These results indicated that RGDfC-Se@DOX could enhance the anticancer activity of DOX to induce A549 cells’ apoptosis by loading the DOX onto the active tumor-targeting carrier RGDfC-SeNPs.
In vivo antitumor efficacy
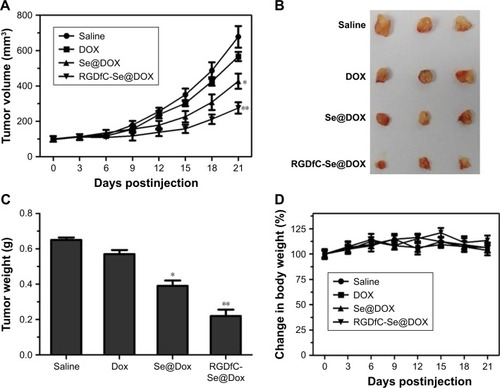
The A549 tumor xenografts were used to assess the antitumor efficacy of RGDfC-Se@DOX.Citation34 The mice were randomly assigned into four groups and then intravenously injected with RGDfC-Se@DOX, Se@DOX, free DOX, and saline, respectively. The tumor volumes and body weights of mice were tested every other day up to 21 days. As shown in , compared to the saline-treated control group, RGDfC-Se@DOX treatment obviously suppresses tumor growth during the treatment time. Moreover, RGDfC-Se@ DOX was more effective in suppressing tumor growth in comparison with free DOX or Se@DOX at the same dose of DOX, proving excellent antitumor efficacy of RGDfC-Se@ DOX. As shown in , tumor image and weight of RGDfC-Se@DOX-treated mice further verified the significant antitumor activity of RGDfC-Se@DOX. In addition, the body weight of mice kept relatively stable during treatment period, indicating that RGDfC-Se@DOX had no obvious side effects at the tested dose (). The in vivo antitumor efficacy showed that tumor-targeted delivery system RGDfC-Se@DOX exhibited a great potential for lung carcinoma therapy.
Figure 8 (A) Tumor growth curve of the xenograft nude mice bearing A549 cells after intravenous administration of saline and various formulations of DOX. (B) Morphology of all tumors stripped from the mice. (C) The tumor weight analysis of mice after 21 days of treatment. (D) The body weight change in mice during the treatment. RGDfC-Se@DOX, selenium nanoparticles conjugated with RGDfC and DOX; Se@DOX, selenium nanoparticles conjugated with DOX.
Abbreviations: DOX, doxorubicin; RGDfC, Arg–Gly–Asp–d-Phe–Cys.
Conclusion
A novel active tumor-targeting SeNP RGDfC-Se@DOX was successfully synthesized to effectively deliver DOX to treat NSCLC. The RGDfC-Se@DOX showed excellent cellular uptake in NSCLC A549 cells and resulted in significant anticancer efficacy. The RGDfC-Se@DOX was more superior to inhibit A549 cell proliferation and migration/invasion and to induce A549 cells’ apoptosis in vitro in comparison with free DOX and Se@DOX. Furthermore, RGDfC-Se@DOX exhibited stronger antitumor efficacy in vivo compared to free DOX and Se@DOX. Taken together, the tumor-targeting delivery system RGDfC-Se@DOX holds huge potential for NSCLC therapy.
Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (No 2017M612632), the Science and Technology Planning Project of Guangdong Province (No 2014A020212024), the Science and Technology Planning Project of Guangdong Province (No 2015A020211002), and the Technology Planning Project of Guangzhou City (No 201607010120).
Supplementary materials
Disclosure
The authors report no conflicts of interest in this work.
References
- WanXMinYBludauHDrug combination synergy in worm-like polymeric micelles improves treatment outcome for small cell and non-small cell lung cancerACS Nano20181232426243929533606
- YuanZQChenWLYouBGMultifunctional nanoparticles co-delivering EZH2 siRNA and etoposide for synergistic therapy of orthotopic non-small-cell lung tumorJ Control Release201726819821129061511
- XiongYZhaoYMiaoLLinCMHuangLCo-delivery of polymeric metformin and cisplatin by self-assembled core-membrane nanoparticles to treat non-small cell lung cancerJ Control Release2016244637327840166
- AuKMSatterleeAMinYFolate-targeted pH-responsive calcium zoledronate nanoscale metal-organic frameworks: turning a bone antiresorptive agent into an anticancer therapeuticBiomaterials20168217819326763733
- TartaroneARossiELeroseRPossible applications of circulating tumor cells in patients with non small cell lung cancerLung Cancer2017107596427339469
- SunHCaoDWuHLiuHKeXCiTDevelopment of low molecular weight heparin based nanoparticles for metastatic breast cancer therapyInt J Biol Macromol201811234335529409771
- WangZHeQZhaoWLuoJGaoWTumor-homing, pH- and ultrasound-responsive polypeptide-doxorubicin nanoconjugates overcome doxorubicin resistance in cancer therapyJ Control Release2017264667528837822
- HegerZPolanskaHKrizkovaSCo-delivery of VP-16 and Bcl-2-targeted antisense on PEG-grafted oMWCNTs for synergistic in vitro anti-cancer effects in non-small and small cell lung cancerColloids Surf B Biointerfaces201715013114027907860
- XiaFHouWZhangCpH-responsive gold nanoclusters-based nanoprobes for lung cancer targeted near-infrared fluorescence imaging and chemo-photodynamic therapyActa Biomater20186830831929292171
- MazumdarSItaliyaKSSharmaSChitkaraDMittalAEffective cellular internalization, cell cycle arrest and improved pharmacokinetics of Tamoxifen by cholesterol based lipopolymeric nanoparticlesInt J Pharm20185431–29610629578081
- SdJNamG-HKwakGYangYKwonICHarnessing designed nanoparticles: current strategies and future perspectives in cancer immunotherapyNano Today2017172337
- WuttkeSLismontMEscuderoARungtaweevoranitBParakWJPositioning metal-organic framework nanoparticles within the context of drug delivery – a comparison with mesoporous silica nanoparticles and dendrimersBiomaterials201712317218328182958
- FangYYangWChengLMengFZhangJZhongZEGFR-targeted multifunctional polymersomal doxorubicin induces selective and potent suppression of orthotopic human liver cancer in vivoActa Biomater20176432333329030307
- ZhouYQuanGWuQMesoporous silica nanoparticles for drug and gene deliveryActa Pharm Sin B20188216517729719777
- XiaYYouPXuFLiuJXingFNovel Functionalized Selenium Nanoparticles for Enhanced Anti-Hepatocarcinoma Activity In vitroNanoscale Res Lett2015101349
- XiaYLinZLiYTargeted delivery of siRNA using RGDfC-conjugated functionalized selenium nanoparticles for anticancer therapyJ Mater Chem B201753369416952
- LiYLinZZhaoMMultifunctional selenium nanoparticles as carriers of HSP70 siRNA to induce apoptosis of HepG2 cellsInt J Nanomedicine2016113065307627462151
- LiYGuoMLinZPolyethylenimine-functionalized silver nanoparticle-based co-delivery of paclitaxel to induce HepG2 cell apoptosisInt J Nanomedicine2016116693670227994465
- LiYLinZGuoMInhibitory activity of selenium nanoparticles functionalized with oseltamivir on H1N1 influenza virusInt J Nano-medicine20171257335743
- XiaYWangCXuTTargeted delivery of HES5-siRNA with novel polypeptide-modified nanoparticles for hepatocellular carcinoma therapyRSC Adv20188419171926
- XiaYXuTWangCNovel functionalized nanoparticles for tumor-targeting co-delivery of doxorubicin and siRNA to enhance cancer therapyInt J Nanomedicine20181314315929317822
- XiaYGuoMXuTsiRNA-loaded selenium nanoparticle modified with hyaluronic acid for enhanced hepatocellular carcinoma therapyInt J Nanomedicine2018131539155229588583
- LuesakulUPuthongSNeamatiNMuangsinNpH-responsive selenium nanoparticles stabilized by folate-chitosan delivering doxorubicin for overcoming drug-resistant cancer cellsCarbohydr Polym201818184185029254044
- LiWYiXLiuXZhangZFuYGongTHyaluronic acid ion-pairing nanoparticles for targeted tumor therapyJ Control Release201622517018226826304
- ZhangLLiGGaoMRGD-peptide conjugated inulin-ibuprofen nanoparticles for targeted delivery of EpirubicinColloids Surf B Biointerfaces2016144818927070055
- WangYWangXDengFThe effect of linkers on the self-assembling and anti-tumor efficacy of disulfide-linked doxorubicin drug-drug conjugate nanoparticlesJ Control Release201827913614629655991
- FangZSunYXiaoHTargeted osteosarcoma chemotherapy using RGD peptide-installed doxorubicin-loaded biodegradable polymeric micelleBiomed Pharmacother20178516016827930982
- LiYLinZGuoMInhibition of H1N1 influenza virus-induced apoptosis by functionalized selenium nanoparticles with amantadine through ROS-mediated AKT signaling pathwaysInt J Nanomedicine2018132005201629662313
- XiaYChenQQinXSunDZhangJLiuJStudies of ruthenium(ii)-2,2′-bisimidazole complexes on binding to G-quadruplex DNA and inducing apoptosis in HeLa cellsNew J Chem2013371137063715
- ZhangQHuangRTangQLeucine-rich alpha-2-glycoprotein-1 is up-regulated in colorectal cancer and is a tumor promoterOnco Targets Ther2018112745275229785123
- ZhangTLiuWMengWDownregulation of miR-542-3p promotes cancer metastasis through activating TGF-β/Smad signaling in hepatocellular carcinomaOnco Targets Ther2018111929193929670368
- BenferMKisselTCellular uptake mechanism and knockdown activity of siRNA-loaded biodegradable DEAPA-PVA-g-PLGA nanoparticlesEur J Pharm Biopharm201280224725622085653
- XiaYZhaoMChenYFolate-targeted selenium nanoparticles deliver therapeutic siRNA to improve hepatocellular carcinoma therapyRSC Adv20188462593225940
- TranTTTranPHAminHHLeeBJBiodistribution and in vivo performance of fattigation-platform theranostic nanoparticlesMater Sci Eng C Mater Biol Appl20177967167828629067