Abstract
A gene delivery concept based on ethylenediamine-functionalized single-walled carbon nanotubes (f-SWCNTs) using the oncogene suppressor p53 gene as a model gene was successfully tested in vitro in MCF-7 breast cancer cells. The f-SWCNTs-p53 complexes were introduced into the cell medium at a concentration of 20 μg mL−1 and cells were exposed for 24, 48, and 72 hours. Standard ethidium bromide and acridine orange assays were used to detect apoptotic cells and indicated that a significantly larger percentage of the cells (approx 40%) were dead after 72 hours of exposure to f-SWCNTs-p53 as compared to the control cells, which were exposed to only p53 or f-SWCNTs, respectively. To further support the uptake and expression of the genes within the cells, green fluorescent protein-tagged p53, attached to the f-SWCNTs was added to the medium and the complex was observed to be strongly expressed in the cells. Moreover, caspase 3 activity was found to be highly enhanced in cells incubated with the f-SWCNTs-p53 complex, indicating strongly induced apoptosis. This system could be the foundation for novel gene delivery platforms based on the unique structural and morphological properties of multi-functional nanomaterials.
Introduction
Nanomaterials, with their unique dimensional and structural properties, have been used in a number of nanomedicine-related applications ranging from cancer targeting,Citation1–Citation3 destruction,Citation4–Citation6 and visualizationCitation7 to tissue engineering and regeneration.Citation8,Citation9 In particular, carbon nanotubes with their 1D structures were reported to interact strongly with various biological systemsCitation10 and increase the activity of various drugs used in cancer treatment.Citation11 More recently, functionalized single-walled nanotubes (f-SWCNTs) have become one of the most frequently used nanodelivery vehicles and there is great interest in the use of these particles for multiple applications including diagnostics and drug delivery, as well as protein and gene delivery.Citation2,Citation6,Citation10,Citation12–Citation15 The generation of f-SWCNT-DNA complexes has been especially beneficial in two ways: 1) the presence of DNA in this complex highly stabilizes the carbon nanoparticle in aqueous suspensions and 2) the DNA molecule is protected from unwanted enzymatic cleavage and nucleic acid binding protein interference when attached to the carbon nanotubes.Citation15–Citation17 Another major advantage of using carbon nanotube-based materials as gene vectors is that they do not activate the naive primary B lymphocytes and are thus nontoxic.Citation18
One of the genes most frequently delivered to cancer cells is the tumor suppressor protein, p53, which in humans is encoded by the TP53 gene.Citation19–Citation21 Protein 53 plays a role in apoptosis, genetic stability, and inhibition of angiogenesis.Citation13 Normal cells contain two copies of functional p53 gene, and the protein produced by this gene is activated either to coordinate the DNA repair process or induce cell suicide if DNA damage proves to be irreparable.Citation19,Citation20,Citation22 Protein 53 can be activated in response to numerous stressors such as DNA damage, oxidative stress, osmotic shock, ribonucleotide depletion, or deregulated oncogene expression.Citation23 When the normal protective function of p53 is lost, the cells lose their potential to control growth, which results in rapid growth and a progression toward malignancy.Citation22 In many types of cancers, the loss of p53 activity may also result in resistance to radiotherapy and anticancer drugs.Citation24
Protein 53 genes are frequently delivered together with various chemotherapeutic agents, which dramatically increases the efficacy of those drugs.Citation24,Citation25 Several in vitro studies clearly suggest that delivery of this tumor suppressor, especially in synergistic combination with cancer drugs, is an efficient mode of cancer treatment.Citation26 Although numerous papers have emerged on the use of p53 as a model gene for testing various methods of gene delivery, such as targeted liposomesCitation27,Citation28 or polypeptide delivery vehicles,Citation29 to cancer cells, the use of f-SWCNTs has not been reported.
In this work, we have examined the possibility of delivering f-SWNT-DNA complexes containing p53 into MCF-7 breast cancer cells. The major goal of these studies has been to develop protocols for efficient DNA binding and delivery into cancer cells by using the well-characterized p53 gene as a model for cancer cell destruction. These studies have resulted in the successful generation of f-SWCNTs with circular plasmid DNA attached to the outer surface, which, when introduced in vitro to cancer cell cultures, were found to induce apoptosis in a significantly larger percentage of the cells as compared to the nanotubes without plasmid DNA attached to their outside surface. The methods presented here can now be applied as delivery platforms for other genes of interest for various biomedical applications. Using these particles as a foundation, further studies can be developed to generate more advanced particles that can be used for targeting molecules other than genes and/or powerful anti-cancer drugs.
Materials and methods
Unless otherwise mentioned, all chemicals were purchased from Sigma-Aldrich (St Louis, MO). The human breast cancer cell line MCF-7 and Dulbecco’s Modified Eagle Medium were obtained from American Type Cell Culture (Manassas, VA). The p53 plasmids were purchased from Addgene (Cambridge, MA). The caspase assay kit was purchased from BioVision Inc (Mountain View, CA), and the Gentamucin Reagent Solution was from MP Biomedicals, LLC (Illkirch, France).
Carbon nanotube synthesis
Single-walled carbon nanotubes were synthesized on a bimetallic Fe-Co/MgO (2.5:2.5:95 wt%) catalyst system, which was prepared as described previously.Citation30 After a thorough overnight drying, the catalyst system was calcinated at 500°C for a few hours and about 50 mg of the catalyst was placed on a graphite susceptor, which was inserted into a quartz tube. A radio frequency (RF) generator with a frequency of 350 kHz was used in the synthesis of the carbon nanotubes. Argon (Ar) was first introduced at 150 mL/minute into the quartz tube positioned inside the RF coil. After 10 minutes, the RF generator was turned on. Once the temperature of the graphite susceptor reached 800°C, methane was added at 40 mL/minute for 30 minutes. The carbon source and the RF generator were turned off and the sample was cooled to room temperature under Ar. The produced SWCNTs were mixed into a diluted HCl (1:1) solution and the mixture was stirred continuously for 24 hours. The SWCNT mixture was washed with deionized water to a pH of 7 and dried overnight at 100°C. The purified SWCNTs were oxidized at 430°C for 30 minutes to burn off amorphous carbon and to uncover the remaining catalyst nanoparticles. A second purification of SWCNTs was carried out with a nitric acid treatment to further remove any additional metal impurities. The final product contained SWCNTs functionalized with carboxylic groups with enhanced dispersion properties in various solutions.
Characterization techniques
A Mettler Toledo (Greifensee, Switzerland) TGA/SDTA 851e was used to carry out thermogravimetric analyses (TGA) with an airflow rate of 150 mL/minute. Approximately 3 mg of the sample was heated from 25 to 850°C at a rate of 5°C/minute. Transmission electron microscopy (TEM) images were collected on a field emission JEM-2100F TEM (JEOL Inc, Tokyo, Japan) equipped with a CCD camera and an acceleration voltage of 100 kV. For TEM analysis, SWCNTs were homogeneously dispersed in 2-propanol by ultrasonication for 30 minutes. A few drops of the suspension were deposited on the TEM grid and allowed to air-dry before analysis. A JEOL 7000F high-resolution scanning electron microscope was utilized to obtain scanning electron microscopy (SEM) images of the purified powdered samples. Before microscopy, the samples were mounted on aluminum pins with double-sided carbon tape.
The optical absorption spectra at UV-Vis-NIR range were recorded using a Shimadzu (Kyoto, Japan) UV-3600 double beam spectrophotometer with three detectors. For the optical measurements, SWCNTs were individually dispersed in an aqueous sodium cholate solution. To obtain a homogeneous solution, the mixture was first sonicated for 1 hour and then centrifuged for 2 hours at 15000 × g using a high speed centrifuge and the supernatant was utilized for optical analysis. Raman spectra of the SWCNTs were collected using a Horiba Jobin Yvon (Kyoto, Japan) Model LabRam HR800 system with a He-Ne laser (633 nm) as an excitation source. The Raman shifts were calibrated with a silicon wafer at a peak of 521 cm−1.
Synthesis of amine-functionalized SWCNTs (f-SWCNTs)
In a 100 mL round-bottomed flask SWCNTs (50 mg), dichloromethane (50 mL), and m-chloroperoxybenzoic acid (1 g) were mixed and the solution was refluxed for 48 hours. The solution was then filtered to collect the epoxidized SWCNTs and washed with dichloromethane (2 × 10 mL) and ethanol (2 × 10 mL). The epoxide-functionalized SWCNTs were mixed with lithium chloride (200 mg) and freshly distilled ethylenediamine (20 mL) and the mixture was refluxed for 18 hours under an inert atmosphere. After reflux, the reaction mixture was cooled and filtered to collect the product, which was then washed several times with ethanol (3 × 25 mL) to remove any excess ethylenediamine and lithium chloride. The resulting ethylenediamine f-SWCNTs were used for functionalization with the p53 genes.
Functionalization of f-SWCNT with p53 plasmid
Protein 53 plasmids (20 μg mL−1) were added to a suspension of f-SWCNTs in Dulbecco’s Modified Eagle Medium (DMEM) medium at a ratio (p53 plasmid:f-SWCNTs) of 1:7.4 (w/w). The solution was mixed thoroughly using a pipette to develop the complex of p53 and f-SWCNTs.
Cell culture
Human breast cancer cells (MCF-7) were grown in a 75 cm2 culture flask with DMEM, supplemented with 10% calf serum, 100 U mL−1 penicillin, 100 U mL−1 streptomycin, and 50 μg mL−1 gentamicin sulfate and incubated in a 5% CO2 atmosphere at 37°C. The cells were subcultured by trypsinization and maintained in aseptic conditions with medium changes every 2–3 days.
Cell treatment protocol
The cells were seeded at a density of 25 × 104 cells in each 35 mm cell culture dish and grown for 72 hours in normal growth medium (as detailed above) until they reached 60%–70% confluence. The medium was then removed and cells were supplied with medium containing 20 μg mL−1 of F-SWCNTs, 20 μg mL−1–2.7 μg mL−1 F-SWCNTs-p53. A negative control was prepared by incubating the cells only with normal growth medium. The cells were harvested after 24 hours, 48 hours, and 72 hours of incubation for further analysis. All experiments were carried out in triplicate.
Cell assessment with light microscopy
The cells were plated for microscopy as described above and were washed three times with 10× phosphate buffered saline (PBS) and examined with an Olympus BX51 light microscope.
Analysis of cell death with ethidium bromide and acridine orange staining
To determine the degree of cell death, cells were harvested by trypsinization after an appropriate incubation period, were washed twice with 1× PBS buffer and stained with 17 μL of a solution containing 100 mg mL−1 acridine orange and 100 mg mL−1 ethidium bromide in PBS. The harvested cells were placed on a slide and mounted with a cover slide. The stained cells were immediately visualized with UV light using an Olympus fluorescence microscope with a 10× objective equipped with a digital camera. Images were taken of random fields of view. The percentage of apoptosis was calculated by counting the number of live (green) and apoptotic (red) cells. Acridine orange stained the live cells, thus making them to appear green, whereas the apoptotic cells’ fragmented nuclear DNA was stained by ethidium bromide and appeared red when visualized under UV light using specific light filters. Each experiment was run in triplicate and the results expressed as mean ± standard deviation.
Caspase assay
Caspase assays were performed to assess the level of apoptosis using the Biovision CaspGLOW Red Active Caspase-3 staining kit. The cells were incubated following the previously mentioned treatment protocol. After incubation, both the control and treated cells were trypsinized and the cells were resuspended in 1 mL of normal growth medium and incubated with 1 μL of Red-DEVD-FMK for 1 hour at 37°C with 5% CO2 and then centrifuged for 5 minutes at 3000 rpm. The cells were resuspended and washed twice with wash buffer and the final pellet was resuspended in 100 μL of the wash buffer. A few drops of the cell suspension were transferred to a microscope slide, which was examined with a light microscope. The brightness level, which indicates the level of caspase activation in the cells, was analyzed for each sample.
Green fluorescent protein expression
The cells were incubated under the standard conditions using plasmids with green fluorescence protein (GFP) tagged p53. After incubation, both the control and treated cells were harvested, washed twice with 1× PBS buffer, and visualized under UV light by using an Olympus fluorescence microscope at 10× objective with an FTIC filter.
Statistical studies
The data were analyzed by one-way ANOVA test and expressed as mean ± SD among three separate samples for each condition, the P-values of 0.05 or less were considered to indicate significance.
Results and discussion
Synthesis and characterization of SWCNTS and f-SWCNTs and p53 complexed f-SWCNTs
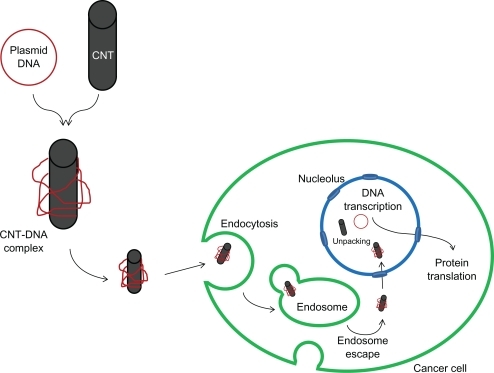
Our work explores the timely topic of nanodelivery of anticancer genes into cancer cells. It is postulated that the internalization of f-SWCNTs-p53 complex and gene expression of p53 plasmid is a multi-step process ().Citation15 First, the f-SWCNTs-p53 complex enters into the cytoplasm via endocytosis and then moves to the nucleus. There, the f-SWCNTs-p53 plasmid complex is hypothesized to dissociate, as the amine groups become deprotonated due to an increase in the pH inside the nucleus. The p53 plasmid then undergoes transcription followed by transport of the transcripts to the cytoplasm where the p53 RNA is translated. This p53 protein then activates the apoptosis pathways in the cytoplasm.
Figure 1 Schematic representation of f-SWCNT-p53 plasmid complex delivery into the cell and subsequent p53 gene expression.
Abbreviation: CNT, carbon nanotubes.
To produce the detailed experimental data needed to more fully understand each of these steps, fiber-like, single-walled carbon nanotubes (SWCNT) were synthesized and visualized by TEM (Inset in ). Since impurities such as catalyst nanoparticles or carbonaceous by-products strongly affect the electrical, mechanical, and optical properties of the carbon nanotubes, high purity samples with optimal performance are necessary for various applications. Thermogravimetric analysis is a useful technique for characterizing the purity of carbon nanotubes.Citation31 The weight loss profile of the SWCNTs (after purification and removal of amorphous carbon) is shown in and indicates that the purity of the SWCNTs utilized in this work was about 98.45%. Scanning electron microscopy (SEM) was used to analyze the morphology of the CNT products synthesized by RF catalytic chemical vapor deposition. and its inset show the SEM images of CNT bundles with an average length of several microns.
Figure 2 A) TGA analysis of the SWCNTs after purification and removal of amorphous carbon; B) SEM images of the SWCNT bundles synthesized on the bimetallic catalyst system, Inset; C) UV-Vis-NIR spectra of SWCNTs dispersed in aqueous solution. Inset: TEM images of the SWCNTs; D) Raman spectrum of the purified SWCNTs.
Abbreviations: SWCNTs, single-walled carbon nanotubes; SEM, scanning electron microscopy; TGA, thermogravimetric analyses.
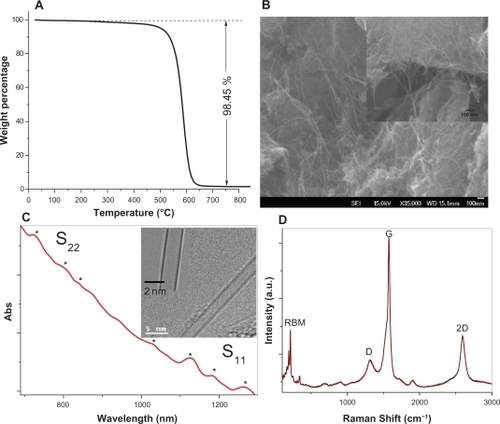
TEM and UV-Vis-NIR spectroscopy were utilized to analyze the diameter distribution of the SWCNTs. The inset in shows the TEM images of the SWCNTs with a diameter of 2 nm. The UV-Vis-NIR spectrum of the isolated SWCNTs () shows a few absorbance peaks (between 600 to 1300 nm wavelengths) in the first van Hove optical transitions (S11) and the second van Hove transition peaks (S22), which correspond to the absorption of the isolated SWCNTs with small diameters. TEM and optical analysis indicated that the SWCNTs utilized in this manuscript have diameters between 0.9 nm and 2.1 nm.Citation32 Raman spectroscopic analyses were also performed on the purified SWCNTs to further characterize their structural properties. The peaks present in the radial breathing mode (RBM) are often correlated with specific diameters of single-wall carbon nanotubes.Citation33 The Raman spectrum shown in indicated the presence of SWCNTs with high crystallinity and with diameters between 0.7–1.7 nm. These values, which were calculated based on the RBM peaks positions present in the Raman spectra of the SWCNTs utilized in this work, are in good agreement with the optical and microscopy findings.
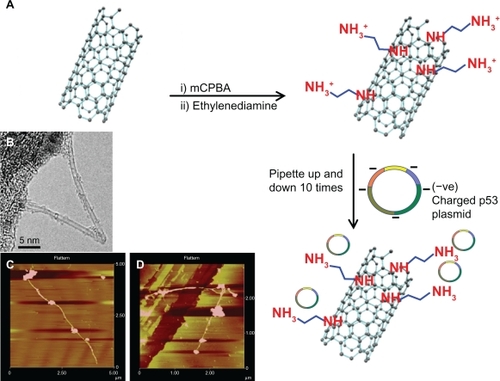
The purified SWCNTs were treated with chloroperbenzoic acid to form epoxidized SWCNTs. This type of functional group was chosen because epoxides are highly reactive, allowing further functionalization with ethylenediamine in the presence of lithium chloride to form amine f-SWCNTs. Due to the presence of free amine groups on the surface, f-SWCNTs remain protonated when exposed to cell culture medium. As a result, the negatively charged plasmid containing p53 DNA is introduced to the positively-charged f-SWCNTs in the medium and the two components form a stable complex due to their strong ionic attractions. A schematic representation of the sequence of the reaction and the steps involved in the production of the f-SWCNT-DNA complexes is shown in . Atomic force microscopy (AFM) images () show that the f-SWCNTs are individually dispersed and successfully complexed with the p53 plasmids, which are visible on the sides of the nanotubes. The majority of f-SWCNTs were found to be bound with p53 plasmid when analyzed by AFM. It is also evident from the images that the plasmid DNA is not bound over the entire surface of the f-SWCNTs but rather is localized in specific areas along their length. Most likely, docking of p53 plasmids on the f-SWCNTs occurred with the highest density of protonated amine groups.
Figure 3 A) Schematic representation of ethylenediamine functionalization of SWCNTs (f-SWCNTs) and formation of f-SWCNTs-p53 complex; B) TEM image of the SWCNTs used in this experiment; C and D) AFM images of f-SWCNTs-p53 complex formation. The presence of the p53 plasmids on the surface of the nanotubes can be clearly observed. In several cases, multiple p53 genes were found to be attached to a single nanotube.
Abbreviations: AFM, atomic force microscopy; SWCNTs, single-walled carbon nanotubes; SEM, scanning electron microscopy; TGA, thermogravimetric analyses.
Incorporation f-SWCNT-p53 complexes into MCF-7 cells: apoptosis assays and GFP expression
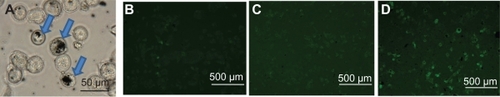
The uptake and internalization of the f-SWCNT-p53 complexes is shown by optical microscopy in , which indicates that the f-SWCNTs-p53 complexes are found adhered both on the membrane and inside the cells. The agglomeration seen here also supports the hypothesis that the f-SWCNTs-p53 complexes are internalized by the cell, most probably via endocytosis.
Figure 4 A) Optical image showing uptake of f-SWCNT-p53 complex by MCF-7 cells and agglomeration of f-SWCNT_p53 complex; B–D) Expression of GFP tagged p53 and f-SWCNT complex in MCF-7 cells after 24, 48, and 72 hours of incubation with 20 μg mL−1 of f-SWCNTs and 2.7 μg mL−1 of GFP tagged p53 at 37°C and 5% CO2 atmosphere. The arrows indicate the presence of nanotubes inside the cells.
Abbreviations: f-SWCNT, functionalized single-walled carbon nanotubes; GFP, green fluorescent protein.
In order to follow the events occurring after treatment of the cells with the f-SWCNTs-p53 complexes and to prove that the genes reach the nucleus of the cells, assays were also conducted with f-SWCNTs complexed with a plasmid coding for a p53-GFP fusion protein. Expression of the p53 protein was visualized by tracking the expression of the GFP-tag ().Citation35 Protein expression, as well as the number of cells expressing GFP, was found to increase with increasing incubation time, with the maximum signal seen after 72 hours.
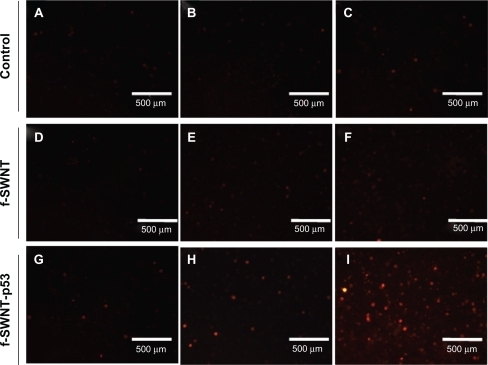
shows the result of incubation of cells with acridine orange and ethidium bromide to differentiate between viable and dead cells in the control and treatment groups.Citation36 shows the effect on MCF-7 cells in cultures with different incubation times (24, 48, and 72 hours; 5A) and treatment with f-SWCNTs (), p53 plasmid () and f-SWCNT-p53 () and the control group (). After 72 hours incubation, the percentage of apoptotic cell death increased dramatically in cells treated with f-SWCNT-p53 and was significantly higher than that seen at other incubation times. The effect of f-SWCNT-p53 in cell death remains very similar to the control groups until 48 hours of incubation, suggesting that cell death was due to initiation of the apoptosis pathway by the expression of p53 and not by any toxic effect of the over-expression of the p53 protein itself.
Figure 5 A) Percentage of dead MCF-7 cells in both control and treatment after incubation with normal growth medium and medium containing 20 μg mL−1 f-SWCNTs and f-SWCNTs-p53 plasmid (20 μg mL−1, 2.7 μg/mL respectively) complex for 24, 48, and 72 hours of incubation at 37°C temperature and 5% CO2 atmosphere; B) Illustration of the effect of F-SWCNTs delivered p53 on induction apoptosis by ethidium bromide/acridine orange assay in MCF-7 cell line, where live cells are green and dead cells appear red when exposed under UV light. A–C) show the control MCF-7 cells (without any treatment); D–F) represent incubation of MCF-7 cells with 20 μg/mL f-SWCNTs; G–I) MCF-7 cells were incubated with 2.7 μg mL−1 of p53 plasmid; J–l) show incubation of MCF-7 cells with 20 μg mL−1 of f-SWCNTs and 2.7 μg mL−1 of GFP tagged p53 plasmid complex at 37°C and 5% CO2 atmosphere.
Abbreviations: f-SWCNT, functionalized single-walled carbon nanotubes; GFP, green fluorescent protein.

One important fact is that, for the plasmid to be translocated to the nucleus, it must be separated from the f-SWCNT. This is probably achieved by a change in the pH in the nucleus compared to the cytoplasmic pH which facilitates the separation of the p53 plasmids from f-SWCNTs.Citation37 It should be noted that the percentage of dead cells in the f-SWCNT treated cells was slightly higher than in the negative control, due to the possible cytotoxic effects induced by the nanomaterials.Citation38 The positive control group, where cells were incubated with 2.7 μg mL−1 of p53 plasmid under identical experimental conditions, shows results very similar to the negative control group indicating no induction of apoptosis and proving that cells were undergoing natural apoptosis instead of induced apoptosis. This also supported the fact that f-SWCNTs indeed acted as very efficient carriers for the plasmid and incorporated them into the cells. In contrast, the p53 plasmids were unable to penetrate the cells when not attached to the f-SWCNTs, as no induced apoptosis was observed in this group. These data are also in very good agreement with GFP expression, which shows that the 72-hour incubation period provided adequate time for the plasmid to be expressed and activate the apoptotic pathways ().
Caspase assay and elucidation of a possible caspase activation pathway
Caspase assays were conducted to further confirm enhanced apoptosis and provide important information regarding the possible mechanism of apoptosis induced by the wild type p53 plasmid delivered by f-SWCNT.Citation39,Citation40 The negative control group showed negligible caspase activity (apoptosis) even after 72 hours () and the cells treated with f-SWCNT showed similar results (). The enhancement of the caspase signal after 72 hours was not significant when compared to the f-SWCNT-p53 treated group () although it has a higher signal than the negative control group. This could be explained by possible cytotoxic effects of the f-SWCNT also evident in the ethidium bromide/acridine orange assays.
Figure 6 Caspase-3 activity in MCF-7 cells. A–C) the caspase activity in control cells (without any treatment); D–F) caspase activity in MCF-7 cells incubated with 20 μg mL−1 f-SWCNTs; G–I) caspase activity in MCF-7 cells incubated with 20 μg mL−1 of f-SWCNTs and 2.7 μg mL−1 p53 plasmid complex for 24, 48, and 72 hours, respectively. The caspase-3 assay was performed following the manufacturer’s instructions for the Biovision CaspGLOW Red Active Caspase-3 staining kit; images were taken after exposing the cells under UV light with TRITC filter.
Abbreviation: f-SWCNT, functionalized single-walled carbon nanotubes.
The caspases are a group of proteins belonging to the cysteine protease family that plays a key role in the apoptosis process. Primarily these proteins remain in an inactivated zymogen form that, upon induction of apoptosis, becomes activated. There are various pathways through which the caspase response occurs during apoptosis. The initiator caspases are the first ones to be activated during apoptosis and are then involved in the activation of effector caspases. These effector caspases lead to the activation and cleavage of other caspases in the entire caspase cascade ultimately, leading to cell death by cleaving other cellular targets.
Based on our data, it is evident that the f-SWCNT-DNA complexes easily enter the cells and are translocated to the nucleus. This is followed by gene transcription and translation as is evident from the visualization of expressed GFP.
We have selected p53 as a model for our studies because it is one of the most significant genes in cancer proliferation. Identified in 1973, p53 was one of the first tumor suppressor genes to be identifiedCitation41 and is the most frequently mutated gene in many common human cancers, with mutations estimated to occur in 50% of all cancers. The major function of p53 is to repair cell damage or induce cell suicide, inhibiting the proliferation of abnormal cells and preventing neoplastic development. In normal, unstressed cells, the level of p53 protein is downregulated by the binding of proteins that promote the degradation of p53 via the ubiquitin/proteasome pathway. The majority of these proteins are also upregulated by p53 itself, which leads to a regulation loop that keeps p53 at very low levels in normal cells. Therefore, the p53 signaling pathway is not active under normal cellular conditions. Activation occurs in response to cellular stresses, such as DNA damage, oxidative stress, osmotic shock, ribonucleotide depletion, or deregulation of oncogene expression.Citation23 In these stressed cells, negative regulators are inhibited, allowing expressed p53 protein to accumulate, leading to over-translation of p53 RNA, and thus, p53 protein accumulation. Finally, a series of modulators (kinases, acetylases) activate p53 transcriptional activity.
Recently, knowledge of the upstream pathways regulating p53 activity has increased greatly and numerous transcriptional targets for p53 have been described. These data support an important role for p53 in mammary carcinogenesis. In breast cancer, p53 mutation is associated with more aggressive disease and worse overall survival rates.Citation39,Citation40 New data have also emerged that have resulted in the elucidation of other molecular mechanisms, in addition to mutation, by which p53 is disabled in breast cancer and have provided new insights into pathways of neoplasia in this tissue. Genetic and epigenetic changes induced by regulators specific to the activity and transcriptional targets of the p53 gene in breast cancers were recently identified.Citation42 A thorough understanding of the molecular pathology corresponding to the structure and expression of constituents of the p53 pathway is expected to have a major impact on the identification, precise diagnosis, and treatment of breast cancer. A number of innovative strategies have been proposed to restore p53 function to tumors.Citation43 Our study is among the most recent and we anticipate that ours and other novel therapeutic approaches targeted to the p53 pathway will significantly impact the clinical outcome for patients with breast cancer.
At present, we can only speculate as to the mechanism for the induction apoptotic events after f-SWCNT-p53 treatment. One pathway that could be involved is the upregulation of the BAX protein by expressed p53, with the help of cytochrome-C, which eventually activates the pro-apoptosome complex. The complex activates caspase-9 that eventually activates caspase-3, leading to the degradation and fragmentation of the chromosomal DNA of the apoptotic cell.Citation44,Citation45 However, other mechanisms of caspase activation may contribute to the death of cells and should be taken into consideration.Citation46,Citation47 The p53 gene over-expression upregulates the release and activation of one of the Bcl-2 procaspase family proteins. The BAX protein contributes to p53-dependent tumor suppression and apoptosis in vitro, allocated to the mitochondria, and eventually leads to the activation of cytochrome-C protein through the mitochondrial apoptosis pathway. The presence of the procaspase-9 and the Apaf-1/ATP cofactors will play an important role in apoptosome complex formation and the cleavage of active caspase-9 into the cytoplasm that has the potential for caspase-3 activation leading to the degradation and fragmentation of the chromosomal DNA of the apoptotic cell. Furthermore, our results demonstrate the potential of functionalized carbon nanotubes for increasing the induction of apoptosis by the p53 gene that will be implicated for targeted apoptosis-inducing gene therapy.
Conclusions
In this work, we have demonstrated the ability of f-SWCNT-p53 complexes to penetrate MCF-7 cells and facilitate the delivery of p53 plasmid DNA, resulting in the expression of this protein in these cells. The major purpose of this study was to optimize the methods for binding plasmid DNA to the f-SWCNTs, followed by detailed characterization of these complexes as gene delivery vehicles. It should be emphasized that the technique described in this work is designed to be a tool for the preliminary screening of different genes for their ability to affect cancer cell growth. At this time, the targeted delivery of the genes and the specific type of f-SWCNT used are not as important as their ability to deliver potential anticancer genes to these cells and assess their effects. Experiments using f-SWCNT-DNA complexes for use in selective delivery of specific antibodies to breast cancer cells, as well as f-SWCNTs that are easily biodegradable and nontoxic, are in progress. These studies will allow the techniques to be moved to in vivo models.
Acknowledgements
Financial support from the Arkansas Science and Technology Authority (ASTA) grant nr 08-CAT-03, the US Army Telemedicine, and Advanced Technology Research Center is highly appreciated. AR-P was supported by the NIH-GM075893 grant.
Disclosure
The authors declare no conflicts of interests in this work.
References
- OuZWuBXingDZhouFWangHTangYFunctional single-walled carbon nanotubes based on an integrin αvβ3 monoclonal antibody for highly efficient cancer cell targetingNanotechnology20092010510219417509
- XiaoYGaoXTaratulaOAnti-HER2 IgY antibody-functionalized single-walled carbon nanotubes for detection and selective destruction of breast cancer cellsBMC Cancer2009935119799784
- BhirdeAAPatelVGavardJTargeted killing of cancer cells in vivo and in vitro with EGF-directed carbon nanotube-based drug deliveryACS Nano2009330731619236065
- PanchapakesanBLuSSivakumarKTekerKCesaroneGWickstromESingle-wall carbon nanotube nanobomb agents for killing breast cancer cellsNano Biotechnology20051133140
- KamNWSConnellOMWisdomAJDaiHCarbon nanotubes as multifunctional biological transporters and near-infrared agents for selective cancer cell destructionProc Natl Acad Sci2005102116001160516087878
- DharSLiuZThomaleJDaiHLippardSJTargeted single-wall carbon nanotube-mediated Pt(IV) prodrug delivery using folate as a homing deviceJ Am Chem Soc2008130114671147618661990
- MalhotraRPatelVVaqueJPGutkindJSRuslingJFUltrasensitive electrochemical immunosensor for oral cancer biomarker IL-6 using carbon nanotube forest electrodes and multilabel amplificationAnal Chem2010823118312320192182
- ShiXSitharamanBPhamQPFabrication of porous ultra-short single-walled carbon nanotube nanocomposite scaffolds for bone tissue engineeringBiomaterials2007284078409017576009
- MacDonaldRALaurenziBFViswanathanGAjayanPMStegemannJPCollagen-carbon nanotube composite materials as scaffolds in tissue engineeringJ Biomed Mater Res, Part A200574A489496
- LadeiraMSAndradeVAGomesERMHighly efficient siRNA delivery system into human and murine cells using single-wall carbon nanotubesNanotechnology20102138510120798464
- MahmoodMKarmakarAFejlehASynergistic enhancement of cancer therapy using a combination of carbon nanotubes and anti-tumor drugNanomedicine2009488389319958225
- KamNWSDaiHCarbon nanotubes as intracellular protein transporters: generality and biological functionalityJ Am Chem Soc20051276021602615839702
- Brannon-PeppasLBlanchetteJONanoparticle and targeted systems for cancer therapyAdv Drug Delivery Rev20045616491659
- FerrariMCancer nanotechnology: opportunities and challengesNat Rev Cancer2005516117115738981
- SinghRPantarottoDMcCarthyDBinding and condensation of plasmid DNA onto functionalized carbon nanotubes: toward the construction of nanotube-based gene delivery vectorsJ Am Chem Soc20051274388439615783221
- WuYPhillipsJALiuHYangRTanWCarbon nanotubes protect DNA strands during cellular deliveryACS Nano200822023202819206447
- BellSKleinCMullerLHansenSBuchnerJp53 contains large unstructured regions in its native stateJ Mol Biol200232291792712367518
- CaiDDoughtyCAPotockyTBCarbon nanotube-mediated delivery of nucleic acids does not result in non-specific activation of B lymphocytesNanotechnology200718365101
- MatlashewskiGLambPPimDPeacockJCrawfordLBenchimolSIsolation and characterization of a human p53 cDNA clone: expression of the human p53 geneEMBO J19843325732626396087
- IsobeMEmanuelBSGivolDOrenMCroceCMLocalization of gene for human p53 tumor antigen to band 17p13Nature198632084853456488
- KernSEKinzlerKWBruskinAIdentification of p53 as a sequence-specific DNA-binding proteinScience1991252170817112047879
- StoreyAThomasMKalitaARole of a p53 polymorphism in the development of human papillomavirus-associated cancerNature19983932292349607760
- HanE-SMullerFLPerezVIThe in vivo gene expression signature of oxidative stressPhysiol Genomics20083411212618445702
- LiYXLinZBTanHRWild type p53 increased chemosensitivity of drug-resistant human hepatocellular carcinoma Bel7402/5-FU cellsActa Pharmacol Sin200425768214704126
- XuLTangW-HHuangC-CSystemic p53 gene therapy of cancer with immunolipoplexes targeted by anti-transferrin receptor scFvMol Med2001772373411713371
- GuntherMWagnerEOgrisMSpecific targets in tumor tissue for the delivery of therapeutic genesCurr Med Chem Anticancer Agents2005515717115777223
- SapraPTyagiPAllenTMLigand-targeted liposomes for cancer treatmentCurr Drug Delivery20052369381
- LedleyFDNonviral gene therapy: the promise of genes as pharmaceutical productsHum Gene Ther19956112911448527471
- MaeMLangelUCell-penetrating peptides as vectors for peptide, protein and oligonucleotide deliveryCurr Opin Pharmacol2006650951416860608
- DervishiELiZXuYThe influence of Fe-Co/MgO catalyst composition on the growth properties of carbon nanotubesPart Sci Technol200927222237
- ArepalliSNikolaevPGorelikOProtocol for the characterization of single-wall carbon nanotubes material qualityCarbon2004421783
- DervishiELiZWatanabeFThermally controlled synthesis of single-wall carbon nanotubes with selective diametersJ Mater Chem20091930043012
- DresselhausMSDresselhausGSaitoRJorioARaman spectroscopy of carbon nanotubesPhys Rep20054094799
- AlkhalafMMowafyE-AMOverexpression of wild-type p53 gene renders MCF-7 breast cancer cells more sensitive to the antiproliferative effect of progesteroneJ Endocrinol2003179556214529565
- SoboleskiMROaksJHalfordWPGreen fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cellsFASEB J20051944044215640280
- BabykuttySPadikkalaJSathiadevanPPApoptosis induction of centella asiatica on human breast cancer cellsAfr J Trad Cam20096916
- AltanNChenYSchindlerMSimonSMDefective acidification in human breast tumor cells and implications for chemotherapyJ Exp Med1998187158315989584137
- ZhangYAliSFDervishiECytotoxicity effects of graphene and single-wall carbon nanotubes in neural phaeochromocytoma-derived PC12 cellsACS Nano201043181318620481456
- AishaAFAAlrokayanSAAbu-SalahKMDarwisYMajid AbdulAMSIn vitro cytotoxic and apoptotic properties of the stem bark extract of sandoricum koetjape on breast cancer cellInt J Can Res20095123129
- BenjaminCWHiebschRRJonesDACaspase activation in MCF7 cells responding to etoposide treatmentMol Pharmacol1998534464509495810
- DeLeoABJayGAppellaEDuboisGCLawLWOldLJDetection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouseProc Natl Acad Sci U S A19797624202424221923
- GascoMShamiSCrookTThe p53 pathway in breast cancerBreast Cancer Res20024707611879567
- VogelsteinBKinzlerKWAchilles’ heel of cancer?Nature200141291492111528480
- SchulerMGreenDRMechanisms of p53-dependent apoptosisBiochem Soc Trans20012968468811709054
- GomyoYSasakiJ-IBranchCRothJAMukhopadhyayT5-aza-20-deoxycytidine upregulates caspase-9 expression cooperating with p53-induced apoptosis in human lung cancer cellsOncogene2004236779678715273730
- JanickeRUSprengartMLWatiMRPorterAGCaspase-3 is required for DNA fragmentation and morphological changes associated with apoptosisJ Biol Chem1998273935793609545256
- SteeleJAPrenticeGAHoffbrandVAp53-mediated apoptosis of CLL cells: evidence for a transcription-independent mechanismBlood20081123827383418682598