Abstract
Background
Plant defensins have a hallmark γ-core motif (GXCX3-9C) that is related to their antimicrobial properties. The aim of this work was to design synthetic peptides based on the region corresponding to the PvD1 defensin γ-core that are the smallest amino acid sequences that bear the strongest biological activity.
Methods
We made rational substitutions of negatively charged amino acid residues with positively charged ones, and the reduction in length in the selected PvD1 γ-core sequence to verify whether the increased net positive charges and shortened length are related to the increase in antifungal activity. Herein, we opted to evaluate the action mechanism of γ33-41PvD1++ peptide due to its significant inhibitory effect on tested yeasts. In addition, it is the smallest construct comprising only nine amino acid residues, giving it a better possibility to be a prototype for designing a new antifungal drug, with lower costs to the pharmaceutical industry while still maintaining the strongest antimicrobial properties.
Results
The γ33-41PvD1++ peptide caused the most toxic effects in the yeast Candida buinensis, leading to membrane permeabilization, viability loss, endogenous reactive oxygen species increase, the activation of metacaspase, and the loss of mitochondrial functionality, suggesting that this peptide triggers cell death via apoptosis.
Conclusion
We observed that the antifungal activity of PvD1 is not strictly localized in the structural domain, which comprises the γ-core region and that the increase in the net positive charge is directly related to the increase in antifungal activity.
Introduction
Plant defensins are peptides identified in different plant tissues and organs, and several studies demonstrated the important role they played in the innate immune system of plants.Citation1,Citation2 The participation of plant defensins in the defense responses is supported by reports of their strong antimicrobial activity in vitro, not only against plant pathogenic fungi, but also against human ones;Citation3,Citation4 the gene expression pattern in response to pathogen infection,Citation5 along with the demonstration that transformed plants engineering to constitutively express defensin genes, is resistant to fungal diseases.Citation6
Plant defensins present a diversity of primary structures coded in 45–54 amino acid residues. The amino acid composition of plant defensins make them highly basic, giving the molecules a net positive charge at neutral pH. Several amino acid residues are conserved among the plant defensin primary structures, such as the strictly conserved eight cysteine residues.Citation7,Citation8 The eight cysteine residues bond to each other in specific pairs, forming four disulfide bridges, which are responsible for the stabilization of their three-dimensional structure,Citation7,Citation8 allowing this family of plant peptides to resist harsh environmental conditions, such as protease degradation and pH and temperature extremes.Citation9 Despite the variation in the primary structure, a completely opposite effect is observed at the tertiary structure level, which is well defined and conserved, generally consisting of one α-helix and three antiparallel β sheets.Citation10 The diversity of plant defensins at their primary structure level is responsible for their different biological activities in vitro.Citation7,Citation11,Citation12 Among the activities already described, the best characterized is their growth inhibition of a large variety of filamentous fungi and yeasts, including those pathogenic to humans.Citation3,Citation7,Citation13–Citation17
The mechanism of action involved in the fungal growth inhibition by plant defensins has been investigated by several authors, and the evidence demonstrates several steps involving an extracellular mechanism acting on the cell wall and/or plasma membrane, further acting on intracellular targets.Citation7,Citation9,Citation15,Citation18 In addition to this whole membrane permeabilization process involving plant defensins, some defensins have been shown to induce apoptosis or programmed cell death in susceptible yeast and fungal species.Citation16,Citation17
In regard to the plant defensin structures, studies have demonstrated that two disulfide bridges, those formed between the Cys21 and Cys25 located in the α-helix and the Cys45 and Cys47 located in the last β-sheet, form a structural arrangement named the cysteine-stabilized αβ motif (CSαβ motif), characteristic of peptides with antimicrobial activity.Citation7,Citation19,Citation20 In addition to the CSαβ motif, plant defensins also have another conserved framework region characterized, called the γ-core, which has the conserved sequence (GXCX3-9C, where X may be any amino acid residue).Citation1,Citation21 Some studies have shown that the region responsible for the biological activity of plant defensins resides in the γ-core region. De Samblanx et alCitation22 showed that the substitution or removal of the amino acid residues located in this region, which encompasses β2 and β3 sheets, reduced the biological activity of RsAFP2 (a defensin from Raphanus sativus). Sagaram et alCitation23 demonstrated that the main determinants of the antifungal activity of MsDef1 and MtDef4 (defensins from Medicago sativa and Medicago truncatula, respectively) reside in their γ-core motifs. In another study, it was shown that the interaction of Psd1 defensin from Pisum sativum with the membrane is mediated, in part, by the amino acid residues that compose the γ-core region.Citation1
Knowing that the region comprising the plant defensin γ-core is related to the antimicrobial properties of these peptides, the principal aim of this work was to design synthetic peptides based on the region corresponding to the PvD1 defensin γ-core that are the smallest amino acid sequences that bear the strongest biological activity. PvD1 is an isolated defensin from Phaseolus vulgaris seeds with antifungal properties,Citation14,Citation15,Citation18 anti-Leishmania activity,Citation24 and anticancer activity.Citation25 Additionally, knowing that the charge, length, and hydrophobicity influence the activity of antimicrobial peptides,Citation26 we made rational substitutions of negatively charged amino acid residues with positively charged ones, as well as the reduction in length in the selected PvD1 defensin γ-core sequence to verify whether the increased net positive charge and the shortened length are related to the increase in antifungal activity. In addition, some tests were carried out to better understand the mechanism of action of the peptides derived from the γ-core region of PvD1 defensin.
Materials and methods
Microorganisms
The yeast species Candida albicans (CE022) and Candida buinensis (3982) were maintained in Sabouraud agar (1% peptone, 2% glucose, and 1.7% agar; Merck). The yeast cells were provided by Laboratório de Fisiologia e Bioquímica de Microrganismos, Centro de Biociências e Biotecnologia, Universidade Estadualdo Norte Fluminense Darcy Ribeiro, Campos dos Goytacazes, Rio de Janeiro, Brazil.
Analysis of amino acid sequence of PνD1 defensin
The amino acid sequence of the PvD1 defensin from P. vulgaris was obtained as previously described by Games et al.Citation14 In brief, the first 21 amino acid residues of PvD1 were obtained by Edman degradation of the purified peptide. Based on this 21 amino acid sequence, a degenerated primer was designed and used in an RT-PCR together with an oligo-dT primer. The amplicon was cloned into the pTZ57R/T vector (InsTAclone™ PCR Cloning Kit, Fermentas) and then submitted to sequencing by Big Dye1 Terminator v3.1 Kit (Applied Biosystems) in an ABI 3110 Genetic Analyzer (Applied Biosystems).
Synthetic peptide design
By comparison analysis of the primary structure of PvD1 defensin with other defensin primary structuresCitation7 and by superimposing onto these sequences the sequence of the γ-core, as defined by Yount and Yeaman,Citation27 we identified and selected the region corresponding to the PvD1 γ-core. This region encompasses the γ-core itself and amino acid residues that are part of the flanking β2 and β3 sheets. Based on these comparative analyses, two peptides with 15 amino acid residues and two peptides with 9 amino acid residues each were then chemically synthesized by the GenOne Soluções em Biotecnologia (Rio de Janeiro, Brazil). The purity level of the synthetic peptides was >95% as determined by high-performance liquid chromatography (4.6×250 mm PLRP-S 100A), using a solution A (0.1% trifluoroacetic acid in 100% acetonitrile) and a solution B (0.1% trifluoroacetic acid in 100% water) with the following gradient: 0%–15% of solution A from 0 to 25 minutes; 40%–100% of solution A from 25 to 30 minutes. The Cys (C) residues of these peptides were replaced by Ala (A) residues. Additionally, two Asp (D) residues were substituted by two Arg (R) residues in two peptides. All these peptides were resuspended in ultrapure water. Their molecular masses and isoelectric points were determined by the Expasy Compute pI/Mw tool.Citation28–Citation30
Effect of synthetic peptides on yeast growth
Yeast cell inocula from C. albicans and C. buinensis were removed from stock tubes containing Sabouraud agar and transferred to Petri dishes containing Sabouraud agar. Cells were grown at 30°C for 2 days. After this period, each cell aliquot was added to 10 mL sterile culture medium (Sabouraud broth, 1% peptone, 2% glucose, Merck). Yeast cells were quantified in a Neubauer chamber (LaborOptik). Initially, the C. albicans and C. buinensis yeast cells (1×104 cells mL−1) were incubated in 100 µL of Sabouraud broth containing the selected synthetic peptides at increasing concentrations from 18.35 to 293.6 µM.
The assay was performed on cell culture microplates (96 wells; Nunc) at 30°C for 24 hours. Optical readings at 620 nm (EZ Read 400, Biochrom) were collected 24 hours after the start of assay. Control cells were grown in the absence of synthetic peptides. The assay was done three times according to a methodology adapted from Broekaert et al.Citation31 The yeast growth inhibition data were evaluated by the one-way ANOVA, and the differences of the mean at P<0.05 were considered significant. All statistical analyses were performed using the GraphPad Prism software (version 6.0 for Windows).
Cell viability
To verify whether the growth inhibition of C. buinensis cells was caused by the fungicidal or fungistatic effect of the smallest active peptide, the control cells (without the peptide) were washed and diluted 1,000-fold. A 100 µL aliquot of the dilution was spread with a Drigalski spatula on the surface of a Petri dish containing Sabouraud agar and grown at 30°C for 48 hours. At the end of this period, the colony forming units were determined, and Petri dishes were photographed.Citation32 The same procedure was repeated with the yeasts treated with 36.7 and 73.4 µM of the smallest and the strongest active peptide for 24 hours. The experiments were performed in triplicate, and the results are shown assuming that the control represents 100% cell viability. These data were evaluated by one-way ANOVA, and the differences of the mean at P<0.05 were considered significant. All statistical analyses were performed using the GraphPad Prism software (version 6.0 for Windows).
Effect of a synthetic peptide on plasma membrane permeabilization
The plasma membrane permeabilization of C. buinensis cells treated with 25 µM of the smallest and the strongest active peptide was evaluated using Sytox green fluorescent dye (Invitrogen), according to the methodology described by Thevissen et al.Citation33 Sytox green is a dye that has high affinity for nucleic acids and penetrates the cell only when its membrane is compromised, emitting a strong green fluorescent.
After the growth inhibition assay, C. buinensis yeast cells grown in the absence (control) and in the presence of the smallest and the strongest active peptide were incubated with Sytox green at a final concentration of 0.2 µM (dissolved in dimethyl sulfoxide), according to instructions provided by the manufacturer. A positive control with 300 mM ethanol was done. After 15 minutes incubation at 25°C with constant agitation at 500 rpm, the cells were observed under an optical microscope (Axioplan.A2, Zeiss) coupled to an AxioCAM MRc5 (Zeiss) camera and the images were analyzed by the Axiovision software version 4.0 (Zeiss). The microscope was equipped with a set of fluorescent filters for fluorescein detection (excitation wavelength between 450 and 490 nm and emission of 500 nm). The results represent triplicate experiments.
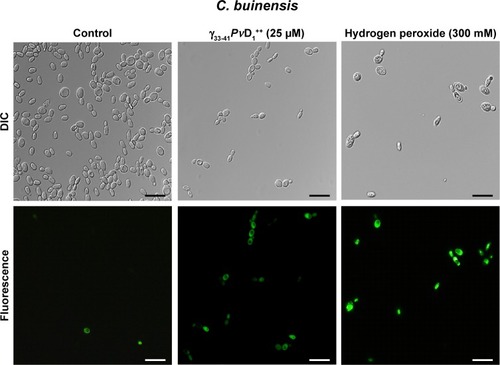
Determining the induction of intracellular oxygen reactive species
To evaluate whether the mechanism of action of the smallest and the strongest active peptide involves the induction of oxidative stress, the fluorescent probe 2′,7′-dichlorofluorescein diacetate (H2DCFDA) was used according to the methodology described by Mello et al.Citation15 This dye enters the cell passively, is deacetylated by intracellular esterases and after being oxidized by reactive oxygen species (ROS), becomes fluorescent. After 24 hours incubation with 25 µM of the smallest and the strongest active peptide, 50 µL of C. buinensis cells grown in the absence (control) and in the presence of the peptide were incubated with 20 µM of the H2DCFDA probe (Calbiochem), for 2 hours at 25°C with constant agitation at 500 rpm. A positive control was done with 300 mM hydrogen peroxide. After this period, the cells were observed under an optical microscope (Axioplan. A2, Zeiss) coupled to an AxioCAM MRc5 (Zeiss) camera, and the images were analyzed by the Axiovision software version 4.0 (Zeiss). The microscope was equipped with a set of fluorescent filters for fluorescein detection (excitation wavelength between 450 and 490 nm and emission of 500 nm). The results represent triplicate experiments.
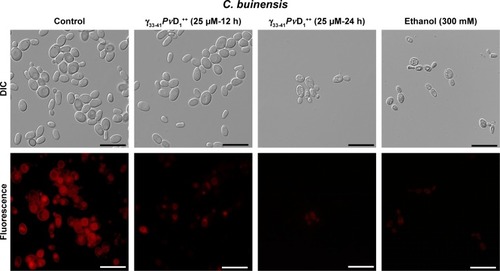
Analysis of mitochondrial functionality
Mitochondrial functionality was assessed by fluorescent dye Rhodamine 123 (Sigma). Rhodamine 123 is a cationic fluorescent dye that has a high affinity to the electrical potential of membranes; thus, it marks active mitochondria in living cells resulting in a bright red fluorescence. In contrast, the treated samples present a weak or absent fluorescent signal. This test was done as described in the section “Effect of synthetic peptides on yeast growth” with the following differences: after incubation with 25 µM of the smallest and the strongest active peptide for 12 and 24 hours, the C. buinensis yeast cells were resuspended, incubated with 10 µg mL−1 of Rhodamine 123 with constant agitation at 500 rpm for 2 hours and protected from light and then analyzed by differential interference contrast (DIC) under an optical microscope equipped with a fluorescence filter (with an excitation wavelength of 506 nm and emission wavelength of 530 nm). Control cells (in the absence of the peptide) had the same treatment as cells treated with the peptide and a positive control with cells treated with 300 mM ethanol for 30 minutes before the dye addition.
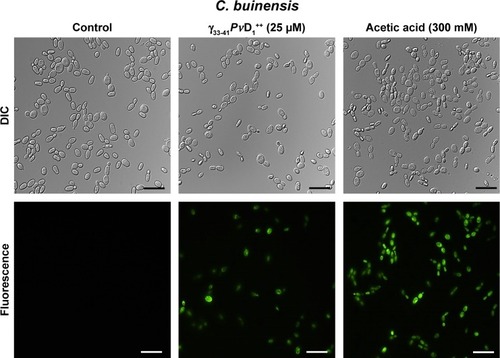
Detection of caspase activity induced by a synthetic active peptide
The detection of caspase activity was performed using the CaspACE FITC-VAD-FMK marker (Promega), as described by the manufacturer. The FITC-VAD-FMK marker is an analog of the Z-VAD-FMK caspase inhibitor (carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone) that enters the cell and irreversibly binds to activated caspases. This test was done as described in the section “Effect of synthetic peptides on yeast growth” with the following differences: after incubation with 25 µM of the smallest and the strongest active peptide for 24 hours, the C. buinensis cells were resuspended, washed once in 500 µL PBS (10 mM NaH2PO4, 0.15 M NaCl) with pH 7.4 and resuspended in 50 µL of the staining solution (supplied by the kit) containing 50 µM FITC-VAD-FMK marker. After incubation for 20 minutes at 30°C with constant agitation at 500 rpm, the cells were again washed in 500 µL PBS and resuspended in 20 µL PBS. Negative control (in the absence of the peptide) and positive control cells (incubated with 300 mM acetic acid) had the same treatment as cells treated with the peptide. The cells were analyzed by DIC on the optical microscope equipped with a fluorescence filter for fluorescein detection (excitation wavelength 450–490 nm and emission wavelength 500 nm).
Results
Synthetic peptide design
After the analysis of the primary structure of PvD1, previously obtained by Games et al,Citation14 and other defensin peptides,Citation7 the region corresponding to the PvD1 γ-core was selected and is highlighted in blue (). Based on the PvD1 γ-core sequence, together with parts of the β2 and β3 sheets (from Arg31 to Lys45), an amino acid stretch giving rise to the peptide of 15 residues with the following amino acid sequence was chosen: RSGRARDDFRAWATK, which was called γ31-45PvD1 (). Another peptide, based on the γ31-45PvD1 design, had its net positive charge increased with the replacement of the two Asp residues, at positions 37 and 38, by two Arg residues, giving rise to the following amino acid sequence: RSGRARRRFRAWATK, which was called γ31-45PvD1++ (). Two other peptides of nine residues each, smaller than γ31-45PvD1 and γ31-45PvD1++, and had the sequences comprising Gly33 to Cys41 (PvD1 γ-core itself) with the amino acid sequences GRARDDFRA and GRARRRFRA, which were called γ33-41PvD1 and γ33-41PvD1++, respectively, were also synthesized (). All the synthesized peptides had their Cys residues replaced by Ala residues (). These substitutions were made to prevent the formation of disulfide bridges. Similarly, Schaaper et alCitation34 replaced the Cys residues by α-aminobutyric acid, to avoid the formation of such undesired bridges. The biochemical characteristics of synthesized peptides are shown in .
Figure 1 Design, alignment and biochemical characteristics of the synthetic peptides.
Notes: (A) Alignment of the primary structures of PνD1 and the four synthetic peptides in amino acid one-letter code. Numbers above the PνD1 sequence indicate the peptide size in amino acids. The amino acid residues in blue represent the PνD1 γ-core region. The amino acid residues in red represent the replaced residues in the original PνD1 sequence as follows: C residues were replaced by A, and D residues were replaced by R. Amino acid residues in black are not part of the γ-core region and were not changed. Numbers in the synthetic peptide names stand for the amino acid position in the original PνD1. Double-plus (++) indicates the double replacement from the original D to R amino acid residues to increase the synthetic peptide positive net charge. (B) Biochemical characteristics of the synthetic peptides.

Growth inhibition assay
To analyze the effect of the synthetic peptides on the growth of yeasts, C. albicans and C. buinensis, γ31-45PvD1, γ31-45PvD1++, γ33-41PvD1, and γ33-41PvD1++ peptides were used at increasing concentrations from 18.35 to 293.6 µM ( and ).
Figure 2 Antifungal effects of γ31-45PνD1 and γ31-45PνD1++ incubated for 24 hours at different concentrations on Candida albicans and Candida buinensis. *Indicates significance by the one-way analysis of variance (ANOVA) (P<0.05).
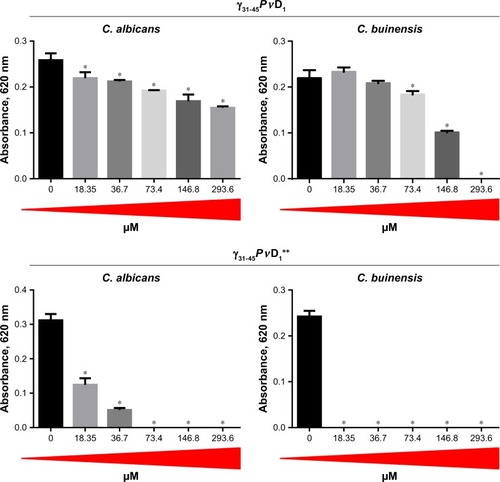
Figure 3 Antifungal effect of γ33-41PνD1 and γ33-41PνD1++ incubated for 24 hours at different concentrations on Candida albicans and Candida buinensis. *Indicates significance by the one-way analysis of variance (ANOVA) (P<0.05).
With the γ31-45PvD1 peptide, we observed that all used concentrations inhibited the growth of yeast C. albicans, reaching 40% inhibition at the concentration of 293.6 µM. The yeast C. buinensis had its growth inhibited starting from the 73.4 µM concentration, and it was possible to observe the 100% inhibition, when the peptide concentration was 293.6 µM (). When the positive net charge of this peptide was increased by the synthesis of γ31-45PvD1++, the peptide concentration of 18.35 µM inhibited 60% growth of the yeast C. albicans, and starting from the concentration of 73.4 µM, the yeast growth was completely inhibited (). For yeast C. buinensis, the lowest used peptide concentration of 18.35 µM completely inhibited yeast growth ().
When we used the smallest peptides, we observed that γ33-41PvD1 was not able to inhibit the growth of C. albicans at any concentrations tested (). For C. buinensis, this peptide inhibits growth by 15% and 17% at the two highest used concentrations, respectively (). However, the peptide γ33-41PvD1++ that had its positive net charge increased showed a greater inhibition potential, being able to inhibit the growth of C. albicans at all concentrations tested and reaching 63% inhibition at the concentration of 293.6 µM (). For C. buinensis, this peptide had an even greater inhibitory potential, being able to inhibit 100% of its growth starting from the concentration of 36.7 µM ().
Based on the results from and , the designed smallest peptide that had the strongest antimicrobial activity was γ33-41PvD1++ when incubated with C. buinensis. Because of this outcome, this peptide and this yeast species were chosen for the next experiments.
Cell viability
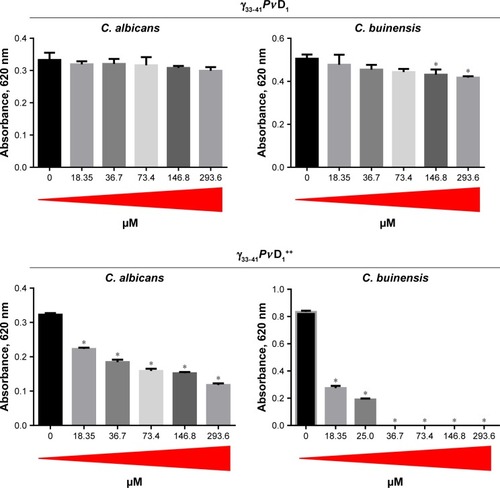
Based on the results obtained in the growth inhibition assay, we found that the minimal inhibitory concentration (MIC) of γ33-41PvD1++ peptide was 36.7 µM for C. buinensis cells (). The γ33-41PvD1++ peptide at 36.7 and 73.4 µM induced the viability loss in C. buinensis cells. The cells treated with 36.7 µM of this peptide had an 84% loss of viability and the cells treated with 73.4 µM had a 100% loss of viability, showing that the effect of this peptide is fungicidal to C. buinensis (). Starting from these experiments, all following assays were performed with yeast C. buinensis with the γ33-41PvD1++ at 25 µM, because at this concentration, below the determined MIC, we were able to obtain reasonable amounts of cells for visualization by optical microscopy.
Figure 4 MIC determination and cell viability loss.
Notes: (A) Images of the plate wells at the end of the growth inhibition assay (at 24 hours) showing the growth pattern of Candida buinensis cells at the bottom of the wells in the absence (control) and in the presence of different γ33-41PνD1++ concentrations. (B) Table showing the number of CFU and the percentage of viability loss of C. buinensis cells after treatment with 36.7 and 73.4 µM of γ33-41PνD1++ for 24 hours, respectively. *Indicates significance by the one-way analysis of variance (ANOVA) (P<0.05). (C) Images of Petri dishes showing the CFU as described in (B). The experiments were carried out in triplicate.
Abbreviations: CFU, colony forming units; MIC, minimal inhibitory concentration.
Effect of γ33-41PνD1++ on plasma membrane permeabilization
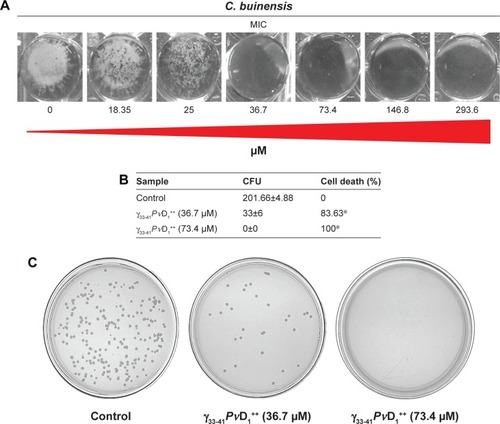
The ability of the γ33-41PvD1++ to permeabilize the C. buinensis plasma membrane was analyzed using Sytox green. The analysis by fluorescence microscopy revealed that C. buinensis cells were labeled by the dye, when treated with γ33-41PvD1++ at 25 µM; thus, these data suggest that γ33-41PvD1++ acts on the plasma membrane of C. buinensis, structurally compromising it and allowing its permeabilization for the labeling dye. In the control (in the absence of peptide), no fluorescence was observed and in the positive control (300 mM ethanol), fluorescent cells were observed ().
Figure 5 Images of membrane permeabilization assay of Candida buinensis cells after treatment with γ33-41PνD1++ (25 µM) for 24 hours. Control cells were treated only with the Sytox green probe and positive control cells were treated with 300 mM ethanol. Bars =20 µm.
Abbreviation: DIC, differential interference contrast.
Reactive oxygen species assay detection
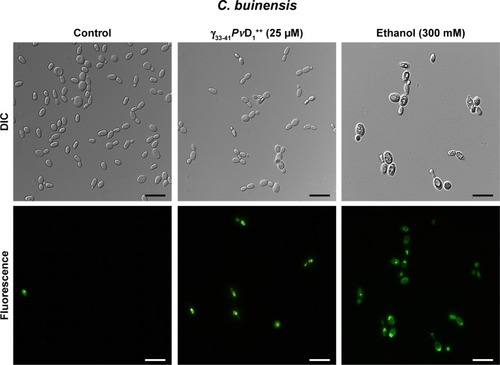
After the growth inhibition assay, C. buinensis cells were incubated with the H2DCFDA probe for the detection of endogenous ROS production. When incubated in the presence of 25 µM of γ33-41PvD1++, the cells showed ROS labeling (), suggesting that a γ33-41PvD1++-induced increase in oxidative stress may underlie the growth inhibitory effect on this yeast. In the control (in the absence of peptide), no fluorescence was observed and in the positive control (300 mM hydrogen peroxide), fluorescent cells were observed ().
Figure 6 Images of reactive oxygen species assay detection in Candida buinensis cells after treatment with γ33-41PνD1++ (25 µM) for 24 hours. Control cells were treated only with the 2′,7′-dichlorofluoresceindiacetate probe and positive control cells were treated with 300 mM hydrogen peroxide. Bars =20 µm.
Abbreviation: DIC, differential interference contrast.
Analysis of mitochondrial functionality
In C. buinensis cells treated with γ33-41PvD1++ at 25 µM for 12 hours is observed a diminished mitochondrial activity as indicated by the weaker red fluorescent signal of Rhodamine 123 () in comparison to the untreated cells, and after additional 24 hours in the presence of γ33-41PvD1++ it is also observed a shrinkage and condensation of cell cytoplasm. A similar weak fluorescent signal was observed for the 300 mM ethanol-treated samples (positive control). Control cells with functional mitochondria evinced a strong signal of Rhodamine 123 fluorescence (). This result indicated that after treatment with γ33-41PvD1++, the C. buinensis cells lost their electrical mitochondrial membrane potential, causing the dysfunction of their mitochondria.
Figure 7 Images of mitochondrial functionality assay of Candida buinensis cells after treatment with γ33-41PνD1++ (25 µM) for 12 and 24 hours. Control cells were treated only with Rhodamine 123 probe and positive control cells were treated with 300 mM ethanol. Bars =20 µm.
Abbreviation: DIC, differential interference contrast.
Detection of caspase activity induced by γ33-41PνD1++
The detection of caspase activity was performed using the CaspACE FITC-VAD-FMK in situ marker. The C. buinensis yeast cells grown in the absence (control) and in the presence of 25 µM of γ33-41PvD1++ were subsequently incubated with the FITC-VAD-FMK marker. It can be observed that the cell treatment with γ33-41PvD1++ resulted in the activation of metacaspases, making it possible to observe a strong labeling of these cells. Positive control cells, treated with 300 mM acetic acid, a known apoptosis inducer in yeast,Citation35 also showed labeling. However, the untreated cells (negative control) did not show metacaspase activity (). These data suggest a programmed cell death that occurs by an apoptotic pathway.
Figure 8 Images of detection of metacaspase activity assay of Candida buinensis cells after treatment with γ33-41PνD1++ (25 µM) for 24 hours. Control cells and cells treated with γ33-41PvD1++ were incubated with CaspACE FITC-VAD-FMK probe. Positive control cells were treated with 300 mM acetic acid and analyzed by fluorescence microscopy. Bars =20 µm.
Abbreviation: DIC, differential interference contrast.
Discussion
Plant defensins are peptides whose activity and mechanisms of action are primarily described against a wide variety of filamentous fungi and yeasts; however, their mechanism of action has not yet been fully elucidated.Citation3,Citation7 The defensins have a structure composed of an α-helix and two β-sheets connected by four disulfide bridges. Yount and YeamanCitation27 showed that the peptides stabilized by disulfide bridges possess amino acid residues that are important for antimicrobial activity in the γ-core region that encompasses β2 to β3 sheets. Many studies have already shown that the activity of plant defensins is related to this structural domain.Citation22,Citation23,Citation36
In the previous work performed by our group, the PvD1defensin isolated from P. vulgaris seeds inhibited growth of various yeasts.Citation14,Citation15,Citation18 In this study, we designed synthetic peptides based on the PvD1 γ-core. Initially, we designed two peptides of 15 residues that comprised the region from Arg31 to Lys45, called γ31-45PvD1++ and γ31-45PvD1, the latter having its positive net charge increased (). Some studies have identified charge, length, and hydrophobicity as the parameters that influence the activity of antimicrobial peptides.Citation26,Citation27 Regarding the length, it has already been reported that larger peptides that cover the entire β2 and β3 sheets, where the γ-core region is inserted, have higher activity.Citation34 Sagaram et alCitation23,Citation37also demonstrated that synthesis of peptides extending beyond the γ-core region improves antifungal activity and that the synthetic peptide (GMA-4C) derived from the γ-core region also contained the C-terminus of MtDef4 defensin with cationic and hydrophobic amino acids that were important for its antifungal activity. Regarding the charge, Lacerda et alCitation10 demonstrated that the positively charged amino acids located in the γ-core were essential for the antifungal activity of peptides, since the substitution of neutral residues by positively charged amino acid residues in the γ-core region increased their inhibitory activity against pathogenic fungi. The mechanism behind this interconnection between positive charge increasing and antimicrobial inhibition increasing is based mainly in the opposite charge attraction.Citation38 Fungal cells have negatively charged structures in their cell walls, such as phosphomanolipids,Citation39 that may serve as primary anchor sites for the positively charged peptides. Then the peptides became able to interact with the membrane. The interaction with the membrane is possible because of the amphipathic character of antimicrobial peptides which turning them able to interact with the negative charge of phospholipids hear groups and also with the hydrophobic core of the membrane.Citation11,Citation33 This interaction explains the ability of antimicrobial peptide to cause membrane permeabilization. Similarly, the His and Arg residues present in the γ-core were important for the oligomerization and antifungal activity of the peptide derived from MtDef5 defensin against the filamentous fungi Neurospora crassa and Fusarium graminearum.Citation36 Thus, we designed the peptide γ31-45PvD1, which had additional amino acids of the flanking β2 and β3 sheets, beside the amino acids that compose the γ-core and increased the charge of γ31-45PvD1++ by replacing two Asp37-38 by two Arg. When we tested the antifungal activity of γ31-45PvD1, it inhibited the growth of C. albicans at all concentrations tested. For C. buinensis, we observed inhibition only starting from the concentration of 73.4 µM, with MIC being 293.6 µM concentration. When we increased the net positive charge of γ31-45PvD1++, it exhibited higher antifungal activity. All tested concentrations of this peptide inhibited the C. albicans growth, with MIC being 73.4 µM. For yeast C. buinensis, MIC was determined at the lowest concentration used, at 18.35 µM ().
Although some previous works have shown that the peptide length influenced its antifungal activity, if a smaller molecule obtained from the native sequence of a peptide still had biological activity, it would be more commercially interesting due to the low cost of its synthesis and could be seen as a promising molecule for the control of fungal diseases.Citation34 To find an even smaller molecule with antifungal potential that could be used as a novel drug, we designed two peptides of nine amino acid residues each that comprised the γ-core region from Gly33 to Cys41, called γ33-41PvD1++ and γ33-41PvD1, where the latter also had its positive net charge increased as discussed above (). Our choice of the smallest sequence with the strongest biological activity is based on requirements of desired characteristics for the pharmaceutical industry to alleviate problems like toxicity, drug stability, specificity, and less immunogenicity.Citation40 For these reasons, we chose to work with the γ33-41PvD1++ peptide.
When we tested the activity of these peptides, the γ33-41PvD1 peptide at different concentrations did not inhibit the growth of C. albicans cells, whereas for yeast C. buinensis, it showed low activity at high concentrations (). These results are consistent with the works of Schaaper et alCitation34 and Sagaram et alCitation37 in the sense that the smallest peptides, only encompassing the γ-core, as in our study, did not bear strong biological activity, at least for C. albicans and C. buinensis. Additionally, as in another example of Sagaram et al,Citation23 the γ-core alone may or may not present biological activity similar to the entire defensin. These authors tested the activity of two synthetic peptides called GMA1 and GMA4 derived, respectively, from the γ-core of the defensins MsDef1 and MtDef4, against the filamentous fungus F. graminearum. The peptide GMA1, even at the concentration of 96 µM, had no antifungal activity. In contrast, the peptide GMA4 exhibited antifungal activity at 6 µM, and when used at 12 µM, was able to completely inhibit the conidial germination of the tested fungus. These results make it evident that only the γ-core region of the MtDef4 defensin is sufficient for antifungal activity; however, the γ-core of the MsDef1 defensin by itself is not sufficient for antifungal activity, similar to the γ-core (γ33-41PvD1) of the PvD1 defensin.
On the other hand, when we tested the antifungal activity of γ33-41PvD1++, with the positive net charge increased by replacing two Asp37-38 residues by two Arg residues, we observed that it inhibited the growth of C. albicans and C. buinensis, with the MIC for C. buinensis of 36.7 µM. Previous studies showed that higher the net positive charge, the higher will be the antifungal activity of peptides, and the positively charged amino acids, such as Arg and Lys, were very important for antifungal activity. The synthetic peptide MBGo1 derived from RsAFP2 defensin, which had its net positive charge increased due to the substitution of some amino acid residues by Arg residues, showed higher activity against the filamentous fungus Fusarium culmorum.Citation34 Sagaram et alCitation23 showed that MtDef4 defensin had five basic amino acids in its γ-core, and MsDef1 defensin had two basic amino acids and two acidic amino acids. After an exchange of the γ-core region between these two defensins, MsDef1 had a two fold higher antifungal activity, thus showing the importance of positively charged amino acid residues for the higher antifungal activity of this peptide.
In the growth inhibition assays, γ33-41PvD1++ peptide showed higher activity against C. buinensis cells, and this inhibitory effect on yeast growth was fungicidal (). Taveira et alCitation41,Citation42 found that another cationic peptide isolated from Capsicum annuum fruits, belonging to thionin family, called CaThi, had a fungicidal effect against six species of Candida genus and Fusarium solani. It is worth emphasizing that the fungicidal characteristic is very important for the development of new therapeutic drugs, since substances that present fungistatic effect can contribute to the development of resistant microorganisms.Citation43,Citation44
Considering the results obtained, we began to investigate the mechanism of action of γ33-41PvD1++, responsible for the inhibition of C. buinensis growth. One characteristic of the cationic antimicrobial peptides is their ability to permeabilize the plasma membrane of microorganisms.Citation15,Citation36 Initially, we used Sytox green, a dye that has high affinity for nucleic acids and penetrates cells only when their plasma membrane is compromised. We observed that the C. buinensis cells, when treated with the γ33-41PvD1++ peptide, were marked by the dye (). Sagaram et alCitation23 showed that MsDef1γ4 peptide was also able to permeabilize the membrane; however, the tested fungus was F. graminearum. In another work, Islam et alCitation36 found that the cationic amino acid residues, present in the γ-core of MtDef5 defensin, were responsible for its antifungal activity and that at micromolar concentrations, MtDef5 caused the membrane permeability of the filamentous fungi F. graminearum and N. crassa. These same authors also observed that there was an increase in the production of intracellular ROS in F. graminearum and N. crassa hyphae after these cells were treated with MtDef5.
ROS have been considered as primary regulators of cell death and are linked to many crucial apoptotic pathways in yeasts.Citation45 Studies have shown that an increase of ROS in the medium can be toxic to the organisms, leading to the destruction of several cell types through apoptotic pathways.Citation1,Citation17 Previously, our research group showed that PvD1 induced ROS in C. albicans and Fusarium oxysporum cells.Citation15 In this study, we observed that C. buinensis cells were labeled by the 2′,7′-dichlorofluorescein diacetate probe when they were treated with γ33-41PvD1++, thus indicating that it caused an increase in endogenous ROS production in these cells (). Due to the increased ROS production in the presence of γ33-41PvD1++, our next objective was to verify if the apoptotic process could be taking place when the C. buinensis cells were treated with γ33-41PvD1++. For this process, we verified the presence of active metacaspasesCitation46 and the dissipation of mitochondrial membrane potential in these cells. Caspases play a central role in signaling death by apoptosis. These are described as specific cysteine-containing aspartate proteases that are among the key markers of the apoptotic pathway, including yeasts.Citation46,Citation47 In these two assays, it was possible to observe that γ33-41PvD1++ activated the metacaspases in C. buinensis cells and caused a collapse of mitochondrial membrane potential in these cells ( and ). Taveira et alCitation42 found that CaThi caused an activation of metacaspase in F. solani, indicating that programmed cell death could be triggered by CaThi in this fungus. In another study, Soares et alCitation17 showed that ApDef1 defensin, isolated from the seeds of Adenanthera pavonina, caused an increase in the ROS production and accumulation that led to permeabilization of the plasma membrane and, consequently, the death of Saccharomyces cerevisiae cells through a metacaspase-dependent apoptotic process. It has also been observed by Vieira et alCitation48 that C. albicans cells lost mitochondrial functionality when treated with the Lp-Def1 defensin, isolated from Lecythis pisonis seeds.
Conclusion
In conclusion, taken together, our results suggest that the antifungal activity of PvD1 is not strictly localized in the structural domain, which comprises the γ-core region; however, we can state that the addition of amino acid residues beyond the γ-core region, which comprises parts of the β2 and β3 sheets, is important for antifungal activity. Additionally, we observed that the increase in the net positive charge is directly related to the increase in antifungal activity. In this work, we opted to evaluate the mechanism of action of the γ33-41PvD1++ peptide due to its significant inhibitory effect on tested yeast. In addition, γ33-41PvD1++ is the smallest construct comprising only nine amino acid residues, which gives it a better possibility to be the basis for a design of a new anti- fungal drug, with lower costs to the pharmaceutical industry, but still maintaining its antimicrobial properties. In relation to the antifungal activity of γ33-41PvD1++, it was fungicidal, and its effects on the growth inhibition of C. buinensis are related to membrane permeabilization, an endogenous increase in ROS, the loss of mitochondrial functionality and caspase activation, suggesting that γ33-41PvD1++ triggers C. buinensis cell death via apoptosis, as demonstrated by the key markers of this pathway.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Acknowledgments
This work was performed at the Universidade Estadual do Norte Fluminense Darcy Ribeiro. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil(CAPES) – Finance Code 001. We acknowledge the financial support from the Brazilian agencies CNPq(305766/2013-9) FAPERJ (E-26/203090/2016; E-26/202.132/2015; E-26/202.735/2016). We are grateful to Souza LCD and Kokis VM for general laboratory technical support.
Disclosure
The authors report no conflicts of interest in this work.
References
- de ConinckBCammueBPAThevissenKModes of antifungal action and in planta functions of plant defensins and defensin-like peptidesFungal Biol Rev2013264109120
- CoolsTLVriensKStruyfsCThe antifungal plant defensin HsAFP1 is a phosphatidic acid-interacting peptide inducing membrane permeabilizationFront Microbiol20178229529209301
- CarvalhoAOGomesVMPlant defensins – prospects for the biological functions and biotechnological propertiesPeptides20093051007102019428780
- JarczakJKościuczukEMLisowskiPDefensins: natural component of human innate immunityHum Immunol20137491069107923756165
- JhaSChattooBBExpression of a plant defensin in rice confers resistance to fungal phytopathogensTransgenic Res201019337338419690975
- SarkarPJanaKSikdarSROverexpression of biologically safe Rorippa indica defensin enhances aphid tolerance in Brassica junceaPlanta201724651029104428770337
- CarvalhoAOGomesVMPlant defensins and defensin-like peptides – biological activities and biotechnological applicationsCurr Pharm Des201117384270429322204427
- WongJHIpDCNgTBChanYSFangFPanWLA defensin-like peptide from Phaseolus vulgaris cv. “King Pole Bean”Food Chem2012135240841422868107
- ParisiKShafeeTMAQuimbarPvan der WeerdenNLBleackleyMRAndersonMAThe evolution, function and mechanisms of action for plant defensinsSemin Cell Dev Biol Epub2018223
- LacerdaAFVasconcelosEAPelegriniPBGrossi de SaMFAnti-fungal defensins and their role in plant defenseFront Microbiol2014545911024478763
- ThevissenKFerketKKFrançoisIECammueBPInteractions of antifungal plant defensins with fungal membrane componentsPeptides200324111705171215019201
- HegedüsNMarxFAntifungal proteins: more than antimicrobials?Fungal Biol Rev201326413214523412850
- BroekaertWFCammueBPAde BolleMFCAntimicrobial peptides from plantsCrit Rev Plant Sci1997163297323
- GamesPDdos SantosISMelloEOIsolation, characterization and cloning of a cDNA encoding a new antifungal defensin from Phaseolus vulgaris L. seedsPeptides200829122090210018786582
- MelloEORibeiroSFCarvalhoAOThe antifungal activity of PvD1, a plant seed defensin of Phaseolus vulgaris, involves plasma membrane permeabilization, inhibition of medium acidification and induction of reactive oxygen species in yeast cellsCurr Microbiol2011621209121721170711
- ThevissenKde Mello TavaresPXuDThe plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicansMol Microbiol201284116618022384976
- SoaresJRJosé Tenório de MeloEda CunhaMInteraction between the plant ApDef1 defensin and Saccharomyces cerevisiae results in yeast death through a cell cycle- and caspase-dependent process occurring via uncontrolled oxidative stressBiochim Biophys Acta Gen Subj201718611 Pt A3429344327614033
- de O MelloÉdos SantosISde O CarvalhoAFunctional expression and activity of the recombinant antifungal defensin Pv D1r from Phaseolus vulgaris L. (common bean) seedsBMC Biochem2014151724690228
- CornetBBonmatinJMHetruCHoffmannJAPtakMVovelleFRefined three-dimensional solution structure of insect defensin AStructure1995354354487663941
- ThommaBPCammueBPThevissenKDefensinsPPlant defensinsPlanta2002216219320212447532
- MuñozAChuMMarrisPISpecific domains of plant defensins differentially disrupt colony initiation, cell fusion and calcium homeostasis in Neurospora crassaMol Microbiol20149261357137424773060
- de SamblanxGWGoderisIJThevissenKMutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activityJ Biol Chem19972722117111798995418
- SagaramUSPandurangiRKaurJSmithTJShahDMStructure-activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearumPLoS One201164e1855021533249
- do NascimentoVVMello ÉdeOCarvalhoLPPvD1 defensin, a plant antimicrobial peptide with inhibitory activity against Leishmania amazonensisBiosci Rep2015355e0024826285803
- FigueiraTNOliveiraFDAlmeidaIChallenging metastatic breast cancer with the natural defensin PvD1Nanoscale2017943168871689929076508
- SchmidtchenAPasupuletiMMalmstenMEffect of hydrophobic modifications in antimicrobial peptidesAdv Colloid Interface Sci201420526527423910480
- YountNYYeamanMRMultidimensional signatures in antimicrobial peptidesProc Natl Acad Sci U S A2004101197363736815118082
- BjellqvistBHughesGJPasqualiCThe focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequencesElectrophoresis19931410102310318125050
- BjellqvistBBasseBOlsenECelisJEReference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositionsElectrophoresis1994153–45295398055880
- GasteigerEHooglandCGattikerAProtein identification and analysis tools on the ExPASy ServerWalkerJMThe Proteomics Protocols HandbookNew YorkHumana Press2005571607
- BroekaertWFTerrasFRGCammueBPAVanderleydenJAn automated quantitative assay for fungal growth inhibitionFEMS Microbiol Lett1990691–25559
- VermelhoABPereiraAFCoelhoRRRSouto-PadrónTPráticas de MicrobiologiaRio de JaneiroGuanabara Koogan2006239
- ThevissenKTerrasFRBroekaertWFPermeabilization of fungal membranes by plant defensins inhibits fungal growthAppl Environ Microbiol199965125451545810584003
- SchaaperWMPosthumaGAPlasmanHHSynthetic peptides derived from the beta2-beta3 loop of Raphanus sativus antifungal protein 2 that mimic the active siteJ Pept Res200157540941811350601
- AertsAMCarmona-GutierrezDLefevreSThe antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicansFEBS Lett2009583152513251619596007
- IslamKTVelivelliSLSBergRHOakleyBShahDMA novel bi-domain plant defensin MtDef5 with potent broad spectrum antifungal activity binds to multiple phospholipids and forms oligomersBiosci Rep2017716157
- SagaramUSEl-MounadiKBuchkoGWStructural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: identification of an RGFRRR motif governing fungal cell entryPLoS One2013812e8248524324798
- FindlayBZhanelGGSchweizerFCationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffoldAntimicrob Agents Chemother201054104049405820696877
- ChaffinWLCandida albicans cell wall proteinsMicrobiol Mol Biol Rev200872349554418772287
- RameshSGovenderTKrugerHGde La TorreBGAlbericioFShort AntiMicrobial Peptides (SAMPs) as a class of extraordinary promising therapeutic agentsJ Pept Sci201622743845127352996
- TaveiraGBCarvalhoAORodriguesRTrindadeFGda CunhaMGomesVMThionin-like peptide from Capsicum annuum fruits: mechanism of action and synergism with fluconazole against Candida speciesBMC Microbiol20161611226819228
- TaveiraGBMello ÉricaOCarvalhoAOAntimicrobial activity and mechanism of action of a thionin-like peptide from Capsicum annuum fruits and combinatorial treatment with fluconazole against Fusarium solaniBiopolymers20171083e23008
- XieDYaoCWangLAn albumin-conjugated peptide exhibits potent anti-HIV activity and long in vivo half-lifeAntimicrob Agents Chemother201054119119619858258
- KołaczkowskaAKołaczkowskiMDrug resistance mechanisms and their regulation in non-albicans Candida speciesJ Antimicrob Chemother20167161438145026801081
- Carmona-GutierrezDEisenbergTBüttnerSMeisingerCKroemerGMadeoFApoptosis in yeast: triggers, pathways, subroutinesCell Death Differ201017576377320075938
- MadeoFCarmona-GutierrezDRingJBüttnerSEisenbergTKroemerGCaspase-dependent and caspase-independent cell death pathways in yeastBiochem Biophys Res Commun2009382222723119250922
- ZivnaLKrocovaZHärtlovaAActivation of B cell apoptotic pathways in the course of Francisella tularensis infectionMicrob Pathog201049522623620600796
- VieiraMEVasconcelosIMMachadoOLGomesVMCarvalhoAOIsolation, characterization and mechanism of action of an antimicrobial peptide from Lecythis pisonis seeds with inhibitory activity against Candida albicansActa Biochim Biophys Sin201547971672926245301
