Abstract
Purpose
The primary goal of the present study was to develop the nano-drug consisting of doxorubicin and exosome derived from mesenchymal stem cells, and to explore its effect on osteosarcoma in vitro.
Methods
The exosomes were isolated from bone marrow MSCs (BM-MSCs) by an Exosome Isolation Kit. The exosome-loaded doxorubicin (Exo-Dox) was prepared by mixing exosome with Dox-HCl, desalinizing with triethylamine and then dialyzing against PBS overnight. The nanoparticle tracking analysis (NTA) and transmission electron microscope (TEM) were used to characterize of the exosome and Exo-Dox. The cytotoxicity of Exo-Dox was determined by CCK-8 assay. Further, the cellular uptake of different drugs was analyzed using inverted fluorescence microscope and flow cytometry.
Results
The typical exosome structures can be observed by TEM. After loading with doxorubicin, its size is larger than free exosome. Compared with the free Dox, the prepared Exo-Dox showed enhanced cellular uptake efficiency and anti-tumor effect in osteosarcoma MG63 cell line but low cytotoxicity in myocardial H9C2 cell line.
Conclusion
The prepared Exo-Dox could be used as an excellent chemotherapeutic drug for treatment of osteosarcoma in vitro. Considering the tumor-homing feature of BM-MSCs, the Exo-Dox may be a good candidate for targeted osteosarcoma treatment in future study.
Introduction
The rapid advance of nanocarriers for drug delivery has led to the emergence of many targeted drugs for cancer treatment.Citation1,Citation2 These nanocarriers have some advantages such as specific tumor targeting and accumulation, stabilized drug and easy release in tumor microenvironment.Citation3,Citation4 Many materials have been used for preparing nanocarriers, such as nanogold particles,Citation5 carbon nanotubes,Citation6 micelles and liposomes,Citation7,Citation8 polymeric nanoparticlesCitation9 and polymer conjugates.Citation10 Among these materials, lipid-based nanocarriers have attracted increasing attentions due to their good biocompatibility and biodegradability, the ease of breaching the plasma membrane, easy isolation of drugs from the encircling environment, and the ability of entrapping both hydrophilic and hydrophobic capsule.Citation11–Citation13 For example, Feng et al.Citation14 fabricated cisplatin-prodrug-constructed liposomes with the advantages of precisely defined/easily tunable drug compositions, stealth-like pharmacokinetics, efficient tumor intake, and loading with various types of imaging or therapeutic agents simultaneously. Fang et al.Citation15 developed a PEGylated liposomal system loading with SN38 prodrug, which effectively improved its systemic circulation and preferential accumulation in tumors. Despite the great successes in efficacy and targeting of these lipid-based nanocarriers, there is still attention needed to be paid in immune rejection by host immune system.Citation16 Thus, the natural particulates which have a natural tropism toward target cells with high selectivity and efficiency have been exploited as drug delivery vehicles. These natural particulates include bacteria, viruses, red blood cells, macrophages, lymphocytes, stem cells and exosome, among which exosome is a more promising natural particulate for its wide distribution in biological fluids and intrinsic homing capability.Citation17–Citation19
Exosome, as a membrane vesicle with a diameter of 30–100 nm, has a bi-lipid membrane with the same orientation as plasma membrane and contains proteins and genetic materials which play an important role in intercellular communication.Citation20 Due to its many excellent attributes including wide distribution in biological fluids, intrinsic homing capability and penetrable to blood–brain barrier, exosome undoubtedly could be an ideal drug delivery vehicle.Citation21–Citation23 Many types of cells such as epithelial cells,Citation24 hematopoietic cells,Citation25 tumor cellsCitation26 and mesenchymal stem cells (MSCs)Citation27 are known to secrete exosome. Among these cells, MSCs are recognized as an ideal cell candidate for the mass production of exosomes because of its good performances such as the ease of isolation from ethically noncontroversial human materials, the large ex vivo expansion capacity, the tropism toward tumor tissue, low immunogenicity and side effect.Citation28–Citation31
Motivated by the excellent performances of MSCs and exosome, in this work, the exosomes from bone marrow MSCs (BM-MSCs) were used as nanocarrier to load chemotherapeutic drug doxorubicin (Dox) for osteosarcoma treatment. Compared with the free Dox, the exosome-loading Dox exhibited better cellular uptake and antitumor effects for osteosarcoma. Also, these exosomes from BM-MSCs had been demonstrated no cytotoxicity for osteosarcoma cell line, which may make it a good tumor-targeted drug nanocarrier in further study.
Materials And Methods
Cell Lines And Materials
The human osteosarcoma cell line MG63 and myocardial H9C2 cell line were purchased from the American Type Culture Collection (ATCC). The mouse bone marrow MSC with stable transfection of GFP was purchased from Cyagen Biosciences Inc (Guangzhou, China), the quality inspection report was done by the company. Phosphate-buffered saline (PBS) was purchased from HyClone Laboratories (Logan, Utah, USA). Trypsin-EDTA (0.25%), penicillin–streptomycin, Dulbecco’s Modified Eagle Medium (DMEM) and Minimum Essential Medium (MEM) were all obtained from Gibco Life Technologies (Grand Island, NY). Mesenchymal stem cell medium-serum free (MSCM-sf) was from ScienCell (San Diego, California, USA). Triethylamine was purchased from Aladdin Industrial Corporation (Shanghai, China). Doxorubicin (Dox) hydrochloride was purchased from Ourchem (Shanghai, China). D36mm Dialysis belt (8000–14,000 wt) was from Beijing DingGuo Changsheng Biotechnology (cat. AT-C-026, Beijing, China). Exosome Isolation Kit was purchased from Umibio (UR52121, Shanghai, China). Bicinchonininc acid (BCA) protein assay kit was purchased from Beyotime Biotechnology (Jiangsu, China). Cell lysis buffer (10X), phenylmethanesulfonyl fluoride (PMSF) and Protease Inhibitor Cocktail (100X) were all purchased from Cell Signaling Technology (Danvers, MA, USA). Cell counting kit-8 (CCK-8) was obtained from Dojindo Molecular Technologies (Kumamoto, Japan). Cell Culture Plates were from Fisher Scientific (cat. 310109008–96 Wells, Flat Bottom).
Cell Culture
Osteosarcoma MG63 cell line and myocardial H9C2 cell line were propagated in a DMEM medium supplemented with 10% fetal bovine serum (FBS, Hyclone Thermo Scientific, USA) and 1% penicillin–streptomycin. MSCs were cultured in MEM medium supplemented with 10% fetal bovine serum (FBS, ScienCell, San Diego, California, USA) and 1% antibiotics. All cells were maintained at an atmosphere containing 5% CO2 and maintained at 37°C in a humidified chamber (Thermo Fischer Scientific, Waltham, MA, USA), and the medium was routinely replaced every 2–3 days.
Isolation Of Exosomes
After growing to 70–80% confluency, MSCs were cultured in MSCM-sf medium supplemented with 1% MSCGS-sf (ScienCell, cat. 7562) and 1% P/S Solution (ScienCell, cat. 0503) for 48 hrs. Then, the conditioned medium was collected and exosomes were isolated with Exosome Isolation Kit according to the manufacturer’s instructions. Briefly, the conditioned medium was centrifuged at 3000g for 10 min to dislodge cells and debris. The Kit reagents and the conditioned medium were mixed in proportion to 1:4. Then, the mixture was vortexed for 1 min and incubated for 2 hrs at 4°C before centrifugation at 10,000g for 60 min to separate exosomes. After that, 1× PBS was used to resuspend exosome-containing pellet, and then centrifuged at 3000g for 10 min in Exosome Purification Filter. All the centrifugation steps were carried out at 4°C to maintain the activity of exosomes. The weight of exosome was measured by BCA kit according to the manufacturer’s instructions. The isolated exosomes were immediately stored at −80°C until further experiments.
Preparation Of Dox-Loaded Exosome (Exo-Dox) And Drug Release
Doxorubicin (Dox) hydrochloride was appropriately diluted by sterile water for injection (SWFI). The 70 μL of Dox HCl (1 mg mL−1) was mixed with 930 μL exosome solution (1 mg mL−1) for 30 min and desalinized with triethylamine for 1 hr at room temperature (RT). The mix solution was then added to dialysis tube prepared by dialysis belt and centrifuge tube and dialyzed against PBS overnight at 4°C to prepare the Exo-Dox complex. The amount of DOX loaded into exosomes was calculated from a standard curve obtained from BioTek ELX800 (Gene Company Limited, USA) by detecting the absorbance value at the wavelength of 490 nm.
In order to measure the in-vitro drug release profile, 3 mL of Exo-Dox solution was transferred into dialysis tubes immersed in PBS at pH 7.4 or pH 4.5. The release of Dox was performed at 37°C and different time intervals. At 1, 2, 4, 8, 12, 24 and 36 h, the concentrations of Dox were determined based on calibration curve by Bioequivalence ELx800 automated enzyme immunoassay analyzer.
Characterization Of The Exosome And Exo-Dox
After purification of exosomes and preparation of Exo-Dox, the hydrodynamic size and concentration of samples were measured using nanoparticle tracking analysis (NTA) with Zeta View PMX 110 (Particle Metrix, Meerbusch, Germany) and corresponding software Zeta View 8.04.02.
The sample solutions were fixed on Formvar-carbon copper grids and were negatively stained for 2 min and air-dried for morphologic visualization using a transmission electron microscope (TEM, HT7700, Hitachi, Japan).
Cytotoxicity Of Exo-Dox In Vitro
The cytotoxicity of Exo-Dox was determined by CCK-8 assay. MG-63 cells and H9C2 cells were seeded into 96-well plates at a density of 3000 or 5000 cells/well and were cultured at 37°C for 24 hrs. Then, the cells were treated with exosomes (0, 5, 10, 20, 100 and 200 μg mL−1) and Dox formulations (Dox and Exo-Dox) with the equivalent Dox concentrations of 0, 0.25, 0.5, 1, 5, 10, 20, 50, 100 μg mL−1for 24 hrs. After that, the cells were incubated with CCK-8 solution for 2 hrs, then the optical density (OD) at 450 nm was detected.
Cell Uptake And Localization
In order to detect the cellular uptake of different drugs, MG-63 cell and H9C2 cell were incubated with Dox and Exo-Dox for 1 hr, 4 hrs and 24 hrs. Cells without any treatment were used as control. After incubation, cells were washed three times with PBS and fixed in 4% paraformaldehyde for 15 min, the cell nuclei were labelled by using DAPI or anti-fluorescence quenching seal solution (containing DAPI). Finally, the localization of Exo-Dox and Dox in cells was visualized by inverted fluorescence microscope (Carl Zeiss Microscope Systems, Jena, Germany). To further investigate the cellular uptake process of Dox and Exo-Dox. MG63 cell and H9C2 cell were seeded into 12-well plates at a density of 1× 105 cells/well and were cultured for 24 h. The cells were also incubated with Dox and Exo-Dox for 1, 4 and 24 hrs. Then, the cells were washed twice with PBS, and cell pellets were collected by centrifugation at 1000rpm for 5 min. The fluorescence signals in cells were analyzed using flow cytometry (BD Accuri C6, USA).
Statistical Analysis
Statistical analysis of data was performed using Student’s t-test and one-way analysis of variance (ANOVA). Probability values lower than 0.05 were considered statistically significant.
Results And Discussion
Characterization Of Exosome Derived From BM-MSCs And Dox-Loaded Exosome
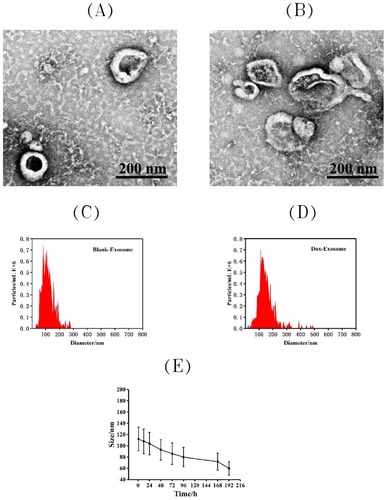
The morphological investigation of exosomes before and after loading with Dox was performed by transmission electron microscope (TEM). As shown in , the typical exosome structures can be observed. Nanoparticle tracking analysis (NTA) () indicated that the exosomes had a relatively narrow size distribution, and the peak diameters were at 112.4 nm for free exosome. After loading with Dox, the peak diameters increased to 152.7 nm. The stability of these exosomes over time was also investigated by NTA (), and the results showed that the size did not change significantly within 7 days at −20°C.
Figure 1 Characterization of exosomes: the representative TEM image of blank exosome (A) and exosome-doxorubicin (B). Size distributions of blank exosome (C) and exosome-doxorubicin (D) based on NTA measurements. The peak diameters were at 112.4 nm for free exosome and 152.7 nm for exosome-doxorubicin. (E) Particle size by nanoparticle tracking analysis for MSCs-derived exosomes during storage at −20 °C.
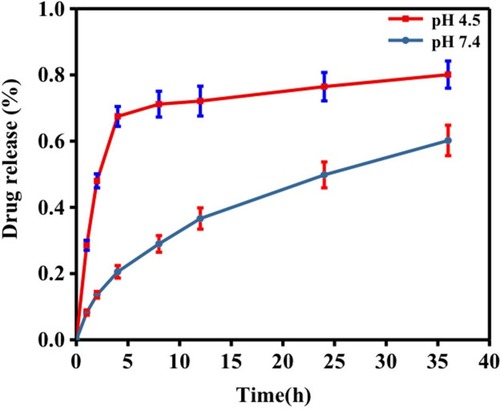
Drug Encapsulation Efficiency And Release Profile
The drug encapsulation efficiency is an important parameter to evaluate the drug delivery potential of a drug delivery carrier, so it is needed to test the drug encapsulation efficiency of Exo-drug. After calculation, the drug encapsulation efficiency was about 12% when the mass ratio of Exo/Dox was 93:7.
To investigate in-vitro drug release profile of Exo-Dox, we determined the release of Dox when putting the Exo-Dox into PBS dialysate with pH of 7.4 or 4.5 () which mimicked the physiological environment and endolysosomal compartment interior environment.Citation32 In PBS dialysate with pH 7.4, the release of Exo-Dox was about 60% after 36 hrs, while in PBS dialysate with pH 4.5, the release rate of Exo-Dox boosted rapidly and achieved about 80%, suggesting that the decrease of the pH could result in the protonation of DOX and accelerate the release of Dox. The drug release profile of Exo-Dox indicated that the acidic environment, such as late endosomes and lysosomes of cancer cells, can accelerate the Dox release of Exo-Dox, this characteristic may make it a good targeted drug in cancer treatment in further study.
The Cellular Uptake Of Exo-Dox In Vitro
The cellular uptake profile of Exo-Dox was evaluated. Because the MSCs were stably transfected by GFP, the exosomes from the MSCs also had the GFP. After incubating MG 63 cells with free Dox and Exo-Dox for 1 hr, the fluorescence microscope observation indicated that all the Dox and Exo-Dox entered into cell (), and the exosome (green) and Dox (red) were mostly co-localized (), suggesting that the exosome and Dox could simultaneously enter into the MG63 cells and the exosome can retain the intact structure after entering into cells. After incubation with MG 63 cells for 4 hrs, it was observed more Exo-Dox entered into nucleus comparing with free Dox (), showing that the cellular uptake efficiency of Exo-Dox was higher than that of free Dox at the incubation time of 4 hrs, which could be attributed to the easy binding of MSCs-derived exosome with MG63 cell. After incubation with MG 63 cells for 24 hrs, almost all the free Dox and Exo-Dox entered into the nucleus.
Figure 3 Intracellular distribution of Dox (A) and Exo-DOX (B) after 1 hrs, 4 hrs and 24 hrs incubation with MG63 cells, bar was 50μm. The histogram (C) and Quantification (D) of internalized Dox, Exo-Dox using flow cytometry after incubation with MG63cells for 1 h,4 h and 24 h. All of the data are presented as mean±s.d. (n=3) and analyzed with two-way ANOVA (****P < 0.0001).
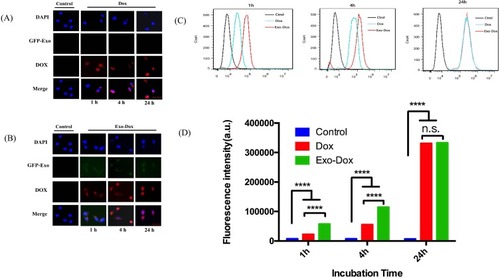
The FACS of cellular uptake of Exo-Dox and free Dox by MG63 cell were also investigated as shown in . From these two figures, we can see that fluorescence intensity of Exo-Dox is much stronger than that of free Dox, showing that more Exo-Dox entered into cell comparing with free Dox. These results were consistent with the results of fluorescence microscope observation.
Because the major side effect induced by doxorubicin is cardiac toxicity, the uptake of Exo-Dox and free Dox by myocardial H9C2 cell was also investigated using inverted fluorescence microscope and flow cytometry. The results showed that fluorescence of Exo-Dox is much weaker than that of free Dox, indicating that less Exo-Dox entering into cell comparing with free Dox (Figure S1). The opposite results indicated that the uptake of Exo-Dox is easier by MG63 than H9C2 cell.
The Cytotoxicity And Antitumor Effects Of Exo-Dox In Vitro
The cytotoxic effects of Exo-Dox on osteosarcoma MG63 cell and myocardial H9C2 cell were examined by Cell Counting Kit-8 (CCK-8) cell viability assay (). As shown in , the cell viability was maintained as high as 92% after incubating with free exosome at the concentration of 200 μg mL−1 for 24 hrs, demonstrating that the exosome had an excellent cytocompatibility.
Figure 4 Cell viability of MG63 cells exposed to different concentrations of free doxorubicin, exosome-doxorubicin (A) and blank exosome (B). The IC50 of free doxorubicin and exosome-doxorubicin (C), where the value was calculated according to Figure A.
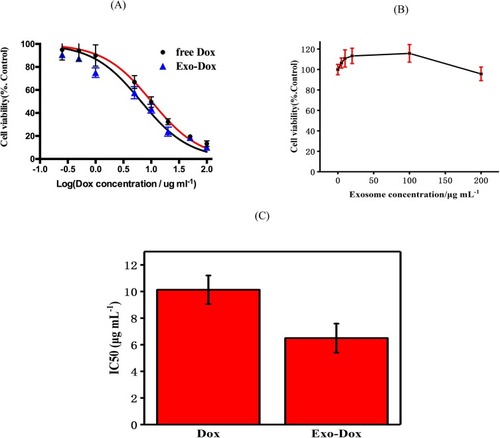
To further investigate the antitumor effect of Exo-Dox in vitro, MG63 cells were treated with free Dox and Exo-Dox with the same concentration of Dox for 24 h followed by CCK-8 assay (). The half-maximal inhibitory concentrations (IC50) of Dox, Exo-Dox were calculated to be 10.13 and 6.48 μg mL−1, respectively (). The tumor cell inhibition was in accordance with the cellular uptake efficiency at 4 hrs.
Moreover, we then further investigated the cytotoxicity of Dox and Exo-Dox on myocardial H9C2 cell line. As shown in Figure S2, the IC50 value of Exo-Dox is much higher than that of free Dox (29 μg mL-1 for Exo-Dox and 7.312 μg mL-1 for Dox, respectively), indicating that the cytotoxicity of Exo-Dox is much weaker for the myocardial cell comparing with free Dox, which is the main priority to use exosome as nanocarrier.
Together, all the results mentioned above, we can see that the Exo-Dox showed selectively anti-tumor effect while had low cytotoxicity for myocardial cells, this may be attributed to the interaction of the membrane proteins in the surface of MSCs-derived exosomes and MG63 cells.
Conclusion
In summary, this work reported that the exosome isolated from MSCs could be used as a drug nanocarrier to load chemotherapeutic drug Dox. The exosome showed low cytotoxicity, and the Exo-Dox could be taken up by MG63 cells and induce cell death. Compared with free Dox, the Exo-Dox can kill the osteosarcoma cells more effectively and show low cytotoxicity in myocardial cells. Considering the tropism toward tumor tissue of MSCs, the exosome derived from BM-MSCs may provide a promising delivery vehicle for tumor-targeted treatment in further study.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
The authors gratefully acknowledge the financial support of Joint Funds for the Innovation of Science and Technology, Fujian Province (2016Y9019), the Outstanding Youth Fund of Fujian Province (2017J06017), the Special Fund of Youth Top-Notch Innovative Talents of Fujian Province (SQNBJ201601) and Trauma Medical Center of Fujian Province.
References
- Ji T, Zhao Y, Ding Y, Nie G. Using functional nanomaterials to target and regulate the tumor microenvironment: diagnostic and therapeutic applications. Adv Mater. 2013;25:3508–3525. doi:10.1002/adma.20130029923703805
- Ding Y, Li S, Nie G. Nanotechnological strategies for therapeutic targeting of tumor vasculature. Nanomedicine. 2013;8:1209–1222. doi:10.2217/nnm.13.10623837858
- Ji T, Li S, Zhang Y, et al. An MMP-2 responsive liposome integrating antifibrosis and chemotherapeutic drugs for enhanced drug perfusion and efficacy in pancreatic cancer. ACS Appl Mater Interfaces. 2016;8:3438–3445. doi:10.1021/acsami.5b1161926759926
- Li S, Zhang Y, Wang J, et al. Nanoparticle-mediated local depletion of tumour-associated platelets disrupts vascular barriers and augments drug accumulation in tumours. Nat Biomed Eng. 2017;1:667–679. doi:10.1038/s41551-017-0115-831015598
- Ali I, Mukhtar SD, Lone MN, Ali HS, Aboul-Enein HY. Recent advances in mesoporous silica and gold based nanovectors in anticancer drug delivery system. Curr Org Chem. 2017;21(23):2400–2415. doi:10.2174/1385272820666161025161853
- Son KH, Hong JH, Lee JW. Carbon nanotubes as cancer therapeutic carriers and mediators. Int J Nanomedicine. 2016;11:5163–5185. doi:10.2147/IJN.S11266027785021
- Cagel M, Tesan FC, Bernabeu E, et al. Polymeric mixed micelles as nanomedicines: achievements and perspectives. Eur J Pharm Biopharm. 2017;113:211–228. doi:10.1016/j.ejpb.2016.12.01928087380
- Andriyanov AV, Portnoy E, Koren E, et al. Therapeutic efficacy of combined PEGylated liposomal doxorubicin and radiofrequency ablation: comparing single and combined therapy in young and old mice. J Controlled Release. 2017;257:2–9. doi:10.1016/j.jconrel.2017.02.018
- Banik BL, Fattahi P, Brown JL. Polymeric nanoparticles: the future of nanomedicine. WIREs Nanomed Nanobiotechnol. 2016;8(2):271–299. doi:10.1002/wnan.1364
- Cai H, Wang XJ, Zhang H, et al. Enzyme-sensitive biodegradable and multifunctional polymeric conjugate as theranostic nanomedicine. Appl Mater. 2018;11:207–218. doi:10.1016/j.apmt.2018.02.003
- Esfanjani AF, Assadpour E, Jafari SM. Improving the bioavailability of phenolic compounds by loading them within lipid-based nanocarriers. Trends Food Sci Technol. 2018;76:56–66. doi:10.1016/j.tifs.2018.04.002
- Kim CH, Lee SG, Kang MJ, Lee S, Choi YW. Surface modification of lipid-based nanocarriers for cancer cell-specific drug targeting. J Pharm Invest. 2017;47(3):203–227. doi:10.1007/s40005-017-0329-5
- Dolatabadi JEN, Omidi Y. Solid lipid-based nanocarriers as efficient targeted drug and gene delivery systems. TrAC Trends Anal Chem. 2016;77:100–108. doi:10.1016/j.trac.2015.12.016
- Feng LZ, Gao M, Tao DL, et al. Cisplatin-prodrug-constructed liposomes as a versatile theranostic nanoplatform for bimodal imaging guided combination cancer therapy. Adv Funct Mater. 2016;26:2207–2217. doi:10.1002/adfm.201504899
- Fang T, Dong YH, Zhang XM, Xie K, Lin L, Wang HX. Integrating a novel SN38 prodrug into the PEGylated liposomal system as a robust platform for efficient cancer therapy in solid tumors. Int J Polym Mater Polym Biomater. 2018;67(6):39–48.
- Yoo JW, Irvine DJ, Discher DE, Mitragotri S. Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov. 2011;10:521–535. doi:10.1038/nrd349921720407
- Simons M, Raposo G. Exosomes- vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi:10.1016/j.ceb.2009.03.00719442504
- van den Boorn JG, Schlee M, Coch C, Hartmann G. SiRNA delivery with exosome nanoparticles. Nat Biotechnol. 2011;29:325–326. doi:10.1038/nbt.183021478846
- Lakhal S, Wood MJA. Exosome nanotechnology: an emerging paradigm shift in drug delivery. Bioessays. 2011;33:737–741. doi:10.1002/bies.20110007621932222
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi:10.1038/nri85512154376
- Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi:10.1002/pmic.20080010918780348
- Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi:10.1158/0008-5472.CAN-10-445521478294
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi:10.1038/nbt.180721423189
- Richesa A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells - a new regulatory pathway. Eur J Cancer. 2014;50(5):1025–1034. doi:10.1016/j.ejca.2013.12.01924462375
- Hornick NI, Doron B, Abdelhamed S, et al. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci Signal. 2016;9(44):ra88. doi:10.1126/scisignal.aaf279727601730
- Hoshino A, Costa-Silva B, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi:10.1038/nature1575626524530
- Liang XL, Zhang LN, Wang SH, Han Q, Zhao RC. Exosomes secreted by mesenchymal stem cells promote endothelial cell angiogenesis by transferring miR-125a. J Cell Sci. 2016;129:2182–2189. doi:10.1242/jcs.17037327252357
- Lai RC, Arslan F, Tan SS, et al. Derivation and characterization of human fetal MSCs: an alternative cell source for large-scale production of cardioprotective microparticles. J Mol Cell Cardiol. 2010;48:1215–1224. doi:10.1016/j.yjmcc.2009.12.02120064522
- Yeo RWY, Lai RC, Zhang B, et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–341. doi:10.1016/j.addr.2012.07.00122780955
- Kidd S, Spaeth E, Dembinski JL, et al. Direct evidence of mesenchymal stem cell tropism for tumor and wounding microenvironments using in vivo bioluminescent imaging. Stem Cells. 2009;27:2614–2623. doi:10.1002/stem.v27:1019650040
- Rezaie Z, Ardeshirylajimi A, Ashkezari MD. Improved anticancer properties of stem cells derived exosomes by prolonged release from PCL nanofibrous structure. Gene. 2018;665:105–110. doi:10.1016/j.gene.2018.05.00329729379
- Mindell JA. Lysosomal acidification mechanisms. Annu Rev Physiol. 2012;74:69–86. doi:10.1146/annurev-physiol-012110-14231722335796