Abstract
Background
Tuberculosis is a public health problem worldwide, and new easy to perform diagnostic methods with high accuracy are necessary for optimal control of the disease. Recently, fluorescent silica nanoparticles (FSNP) has attracted immense interest for the detection of pathogenic microorganisms. The aim of this study was to detect Mycobacterium tuberculosis in clinical samples using bioconjugated FSNP compared with microscopic examination, polymerase chain reaction (PCR), nested PCR, and culture as the gold standard.
Methods
In total, 152 sputum specimens were obtained from patients who were suspected to have pulmonary tuberculosis. All samples were examined by the four techniques described.
Results
The assay showed 97.1% sensitivity (95% confidence interval [CI] 91–99.2) and 91.35% specificity (CI 78.3–97.1). Furthermore, assays using variable bacterial concentrations indicated that 100 colony forming units/mL of M. tuberculosis could be detected. There were no differences between the results obtained from two types of mouse monoclonal antibody against Hsp-65 and 16 KDa antigens.
Conclusion
We performed this assay in a large number of clinical samples to confirm the diagnostic specificity and sensitivity of the test and can recommend its application for diagnosis of M. tuberculosis. We believe that this method is more convenient for routine diagnosis of M. tuberculosis in sputum and will be more easily applicable in the field, and with sufficient sensitivity.
Introduction
Tuberculosis is a public health problem of global importance. Mycobacterium tuberculosis is the main etiology of the disease. According to the World Health Organization, in 2009 there were an estimated 9.4 million incident cases of tuberculosis globally (equivalent to 137 cases per 100,000 population), and approximately 1.7 million people died of tuberculosis in that year. The absolute number of cases continues to increase slightly every year. Most of the cases in 2009 (55%) occurred in Asia.Citation1,Citation2 Multidrug- resistant and extensively drug-resistant strains of M. tuberculosis are spreading rapidly, so this bacterium poses a serious dilemma for public health. There were an estimated 440,000 cases of multidrug-resistant tuberculosis in 2008. Drug-resistant isolates of M. tuberculosis have been found in all countries. The four countries that had the largest number of estimated cases of multidrug-resistant tuberculosis in 2008 were China, India, the Russian Federation, and South Africa.Citation3 Rapid detection of tuberculosis is extremely important for early therapy and designing control measures for prevention of the spread of bacteria in the human population and inhibiting emergence of multidrug-resistant and/or extensively drug-resistant isolates. The gold standard for diagnosis of tuberculosis is the cultivation of M. tuberculosis. Cultivation can detect 100 bacilli/mL of sputum in comparison with the 5,000–10,000 bacilli/mL needed for microscopy.Citation4 Commercial biochemical kits are available for the detection and phenotypic characterization of M. tuberculosis, but molecular methods developed in recent years are more rapid and accurate for this purpose.Citation5 Generally, molecular methods offer several advantages over conventional techniques for rapid detection and identification of mycobacterium. However, molecular methods are expensive and require more skilled personnel, so that implementation of these methods is difficult, especially in regions where M. tuberculosis infection is more common.Citation6 Rapid and accurate detection of trace amounts of organisms, such as pathogenic bacteria, is important for clinical diagnosis and prevention of accidental outbreaks. Recently, nanoparticles have been successfully used for detection of micro-organisms. Unlike traditional methods for the detection of bacteria, which require amplification or enrichment of the target bacteria in samples, fewer numbers of bacteria are needed for detection by the nanoparticle method in a shorter time.Citation7 Some of the different nanomaterials developed can be used as a convenient surface for the attachment of biomolecules. They can be used as a convenient surface for molecular assembly and may be composed of inorganic or polymeric materials.Citation8 Stöber et al first reported a method for synthesis of a dye-doped nanoparticle which encapsulates many thousands of dye molecules inside silica nanoparticles.Citation9,Citation10 There are several advantages to using dye-doped silica nanoparticles, including their small size (20–70 nm in diameter) brightness, photostability, and ease of functionalization. Using surface modification, it is possible to conjugate a dye-doped silica nanoparticle to a particle, so that thousands of dye molecules can be attached to a biomolecule. Consequently, a significantly greater signal is emitted from signal binding events, which leads to increased sensitivity in comparison with a single fluorophore-labeled assay.Citation11 Recently, a study was reported for detection of M. tuberculosis using a reference strain of M. tuberculosis.Citation12 The aim of the present study was to evaluate the validity of the FSNP assay in comparison with culture as the gold standard, acid-fast staining, and polymerase chain reaction (PCR) in order to develop a simple, rapid, and sensitive method for detection of M. tuberculosis in sputum specimens.
Materials and methods
Tris (2,2′-bipyridyl) dichlororuthenium(II) hexahydrate(RuBpy), Triton X-100, protein A from Staphylococcus aureus, acetonitrile, glycine, N acetyl-L-cysteine (NALC), cyanogen bromide, tetraethyl orthosilicate (TEOS), n-hexanol, cyclohexane, acetone, ammonium hydroxide (NH4OH, 28–30 wt%) were purchased from Sigma-Aldrich (St Louis, MO). All other chemicals were of analytical reagent grade. Mouse IgG (IgG2a) monoclonal antibodies to M. tuberculosis were purchased from AbCam (Cambridge, UK) against Hsp-65 antigen and AbD Serotec (Oxford, UK) against 16 KDa antigen. Lowenstein- Jensen medium (Merck, Darmstadt, Germany) was used for culture of M. tuberculosis. Primer oligonucleotides for conventional and nested PCR were purchased from TaG Co (Copenhagen, Denmark). Distilled deionized water was used in the preparation of all aqueous solutions. A standard strain of reference M. tuberculosis (H37Rv) and Escherichia coli (American Type Culture Collection 2532) were obtained from the Pasteur Institute in Iran.
Instrumentation
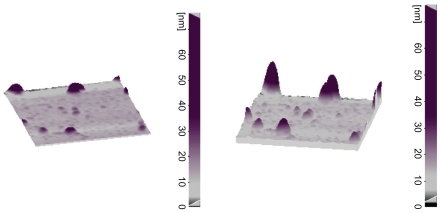
An atomic force microscope (SPM probe scanner DS 95–50 E, Danish Microengineering A/S, Herlev, Denmark) and particle size analyzers (Analystte 22-Nano Tec, Fritsch, Germany) were used for visualization of the morphology and estimation of the size of the silica nanoparticles. Protein concentration was evaluated using a ultraviolet-visible spectrophotometer (Biphotometer Plus, Eppendorf, Germany). A spectrofluorometer (RF-5301 PC, Shimadzu, Japan), and an ultraviolet-visible spectrophotometer (Varian Carry-100, Australia) was used for evolution of emission and absorbance of nanoparticles after synthesis. The presence of an amide group after conjugation of protein A was confirmed using an infrared spectrometer (Bomem MB 155S FT-IR spectrometer, ABB Bomem Inc, Canada). A Techne TC thermal cycler (Techne Co Ltd, Leicester, UK) for amplification of extracted DNA, and a fluorescent microscope (Olympus BX51 and DP70 digital camera system, Olympus America Inc, Center Valley, PA) was used for visualization of microscopic slides and their imaging.
Sputum collection and processing
One hundred and fifty-three sputum samples were obtained from the reference mycobacteriology laboratory at Ahvaz Jundishapur University of Medical Sciences from March 2009 to April 2010. All specimens were collected from patients who were suspected to have pulmonary tuberculosis. Specimens were examined by four techniques, ie, microscopic examination, culture, PCR, and FSNP assay. Specimens were processed immediately according to standard routine diagnostic procedures using the NALC method.Citation13 Mycobacteria were concentrated for culture and smear preparation, and 250 μL of each concentrated sample were inoculated into Lowenstein–Jensen media. All cultures were incubated at 37°C with 5% CO2 and humidity. After 8 weeks of incubation, cultures showing no growth were reported as negative and discarded. For microscopic examination, using a disposable Pasteur pipette, one drop of the sediment of the sputum specimen was spread onto a clean microscopic slide and, after air-drying and fixation by heat, the smears were stained using the Ziehl–Neelsen method.Citation14
DNA extraction and PCR assay
Chromosomal DNA was extracted using proteinase K and phenol:chloroform, then precipitated by ethanol and isopropanol according to a method described elsewhere.Citation15 Purified DNA was amplified with a pair of primers specific to the M. tuberculosis complex strains (IS6110) and two specific pairs of external and internal primers for this bacterium (ie, nested PCR). Conventional and nested PCR for detection of M. tuberculosis were performed as described by Eisenach et alCitation16 and Wilson et al, respectivelyCitation17 ( and ).
Table 1 Characteristics of DNA oligonucleotide primers used for IS6110-based Mycobacterium tuberculosis complex polymerase chain reaction and nested polymerase chain reaction for detection of M. tuberculosis
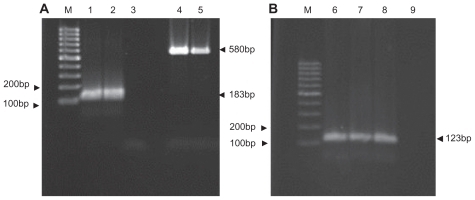
Figure 1 Conventional and nested polymerase chain reaction products of Mycobacterium tuberculosis IS6110 fragment by agarose gel electrophoresis (2%). (A) Nested polymerase chain reaction, M 100 bp DNA size marker, (second round) (1) positive control, (2) clinical positive sample, (3) negative control (first round), (4) positive control, (5) clinical positive sample. (B) Conventional polymerase chain reaction, (6) positive control, (7, 8) positive clinical samples, (9) negative control.
Synthesis of dye-doped silica nanoparticles
RuBpy-doped silica nanoparticles were synthesized using a microemulsion method as described by Stöber et al.Citation9,Citation18 In brief, a microemulsion was prepared by addition of 20 mM RuBpy dye to water (0.48 mL), TEOS 100 μL, cyclohexane 7.5 mL, n-hexanol 1.8 mL, and Triton X-100 1.77 mL. After mixing for 20 minutes, 60 μL of NH4OH was added to initiate polymerization. The reaction was allowed to continue for 24 hours. The nanoparticles were isolated by destabilizing the water-in-oil microemulsion system using acetone, followed by centrifuging and washing with ethanol and water several times to remove any surfactant molecules. The fluorescent intensity, size, and shape of the synthesized nanoparticles was evaluated using a spectrofluorometer, atomic force microscope, and particle size analyzer, as described previously. The shape and size of the silica nanoparticles are shown in and .
Surface modification of FSNP and covalent immobilization of protein A
There are several methods available to biofunctionalize silica nanoparticles, including amino group cross-linkage, avidin-biotin linkage bridge, disulfide coupling, chemical binding, and cyanogen bromide modification. In this assay, we used cyanogen bromide for surface modification of silica nanoparticles in order to immobilize protein A covalently on FSNP. Protein A is a highly stable cell surface receptor produced by several strains of Staphylococcus aureus. This protein is capable of binding to the Fc portion of immunoglobulins, especially IgGs, from a large number of species. One molecule of this protein is capable of binding to at least two molecules of IgG simultaneously.Citation19 Using this technique, FSNP with a natural surface sianol group is activated by cyanogen bromide reagent, forming a reactive –OCN derivative of FSNP. Briefly, a solution of cyanogen bromide in acetonitrile is added to a suspension of silica nanoparticles to yield an –OCN group on the particle surface.Citation20 The particles are then available for bioconjugation to the protein A biomolecules containing free amino groups. To block the protein A-immobilized nanoparticles, 6 mL of 0.3 M glycine solution (pH 8.0) is added and incubated for 16 hours at 4°C. For quality control, the amount of immobilized protein A on the nanoparticles is measured using the Bradford method.Citation21 The presence of an amide group (–NHCO–) is confirmed by infrared spectra assay.
Bioconjugation of a specific monoclonal antibody to M. tuberculosis
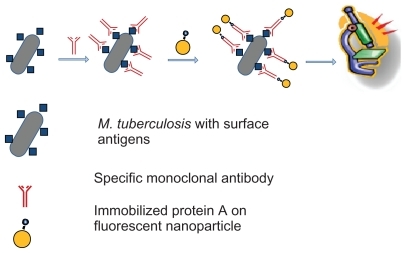
In the next step, mouse monoclonal anti-M. tuberculosis antibody was conjugated to the agents in the sputum specimen. Briefly, mouse monoclonal antibody was added to 500 μL of a sediment suspension of sputum in phosphate-buffered saline following processing (final antibody concentration 5 μg/mL), and after 1 hour of incubation at 37°C, the suspension was washed twice with phosphate-buffered saline. Afterwards, 0.1 mg/mL of protein A-conjugated nanoparticles were added, and the suspension was incubated for 1 hour at 37°C. Free FSNP protein A-conjugates which did not bind to bacteria were removed by centrifugation (8000 rpm per 2 minutes). The primary amine group of the M. tuberculosis monoclonal antibody was reacted with the activated FSNP, forming antibody-conjugated FSNP.
Microscopic examination
The fluorescent microscope used in this project was equipped with a 450–490 nm band pass excitation and a 515 nm long pass emission filter. The pellet was spread on a glass slide and observed by fluorescent microscopy. is a schematic representation of this method for detecting M. tuberculosis. To remove the fluorescence background during microscopic examination, quartz microscope slides (UQG Optics, Cambridge, UK) were used ().
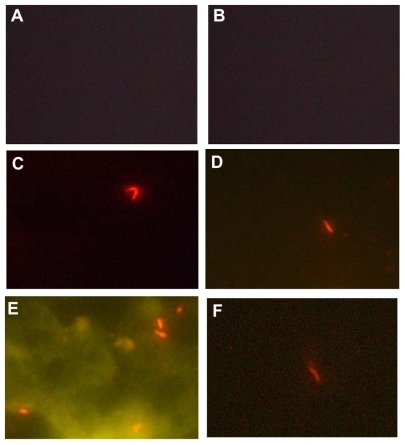
Figure 4 Fluorescence images (100× oil). (A) negative control, phosphate-buffered saline in place of the specific monoclonal antibody, (B) negative control, Escherichia coli in place of the Mycobacterium tuberculosis. (C, D and F) Specific interaction of bioconjugated nanoparticles with M. tuberculosis; (E) the nonspecific interaction (autofluorescence) despite displaying M. tuberculosis with a bright fluorescence.
Determination of limit of bacterial detection by FSNP assay
In this step, the ability of culture and FSNP assay for detection of different M. tuberculosis concentrations was evaluated. A single cell suspension was prepared by harvesting M. tuberculosis (H37Rv strain) in phosphate-buffered saline (pH 7.4) using a previously described method.Citation22 The bacterial suspension was counted in a Petroff-Hausser chamber. Different concentrations of bacteria (10,000, 5000, 1000, 500, 100, 50, and 10 colony forming units/mL) were prepared. Distilled water was used as a negative control in the process, and a suspension of M. tuberculosis adjusted to 0.5 McFarland turbidity (about 109 colony forming units/mL) was used as a positive control. All experiments were performed in duplicate.
Data analysis
Analysis of the data was performed using the SPSS 15 (SPSS Inc, Chicago, IL), and diagnostic sensitivity and specificity and other statistical parameters were calculated using Stats-Direct statistical software (Version 2.7.2). The investigators were blinded to the clinical data and samples used during the experiment for detection of M. tuberculosis.
Results
Smear of sputum, culture, PCR, and FSNP assays were performed for all the 152 specimens taken from patients. Reference strains were treated similarly to the field samples. By acid-fast staining, 63.2% (96/152) of the specimens were positive for M. tuberculosis, while 69.7% (106/152) were culture-positive. Of 152 samples, 86 (56.6%) were positive by PCR and 100 (65.8%) were positive by nested PCR for M. tuberculosis.
Of the 152 specimens, 107 (70.3%) were positive by FSNP assay. Of 107 specimens, 103 (96.2%) were culture-positive and 88 (82%) were positive by both acid-fast staining and culture. Thus, four samples that were positive by FSNP assay were reported negative by the culture method. Furthermore, of these 107 samples, 78 (72.8%) were positive by both PCR and nested PCR techniques, and 10 samples were negative by both techniques ().
Table 2 Comparison of four techniques versus culture method for diagnosis of Mycobacterium tuberculosis
Comparison of limit of bacterial detection by FSNP and culture methods
The limits of the FSNP assay against culture for the bacterial agent was calculated in this section. As described previously, all bacterial solutions were tested in duplicate for confirmation. Limits of detection for both methods were 100 colony forming units/mL, ie, there was no difference between the two methods in ability to detect bacteria in solution.
Analytical specificity and sensitivity
In the present study, the overall diagnostic sensitivity, specificity, efficiency, predictive value for a positive test, and predictive value for a negative test using the FSNP assay versus culture as a gold standard method were 97.1% (95% CI 91.3–99.2), 91.3% (95% CI 78.3–97.1), 95.5% (efficiency), 96.2% (95% CI 90.1–98.7), and 93.3% (95% CI 80.6–98.2), respectively. The sensitivity and specificity of the other techniques performed in this study are shown in . As previously described, two types of mouse monoclonal antibody against Hsp-65 and 16 kDa antigen from the M. tuberculosis surface antigen group were used for confirmation of the FSNP method. No differences were found in the results obtained with the two different monoclonal antibodies for detection of M. tuberculosis.
Discussion
In spite of considerable advances in the diagnosis of M. tuberculosis, the gold standard method for laboratory diagnosis is culture. Initiation of appropriate therapy is the main reason for developing a rapid test for diagnosis of pulmonary tuberculosis. In this era of molecular biology, efforts are focused on development of more rapid and sensitive tests. Recent advances in the field of nanoscience have provided new opportunities for rapid laboratory diagnosis of tuberculosis. The specificity, sensitivity, and rapidity of this method encouraged us to trial this technique for detection of M. tuberculosis in sputum. In this paper, we have reported the feasibility of the FSNP assay for detection of M. tuberculosis and compared this assay with other detection methods.
Recently, two studies were performed to detect bacteria using bioconjugated silica nanoparticles. Qin et al showed the ability of bioconjugated RuBpy-doped nanoparticles to detect M. tuberculosis in pure and mixed bacterial samples in comparison with commercial FITC-conjugated antirabbit M. tuberculosis antibody.Citation12 The other study, reported by Zhao et al, described the detection of E. coli O157:H7 in ground beef samples.Citation7 They reported a high intensity of fluorescence signals, a decrease in the amount of primary antibody, and detection of a single bacterium within 20 minutes as major advantages of this technique in their research.
It is noteworthy that the above studies were performed on standard strains, whereas in our study we used the FSNP assay in clinical specimens. Moreover, based on our data, the sensitivity and specificity values were both more than 90%, confirming the accuracy of this method.
In another study, these nanoparticles were used by Zhou et al as probes for DNA/microarray detection.Citation23 They reported a higher sensitivity for target DNA compared with fluorophores. The sensitivity and specificity of the molecular detection system and nanoparticle probe method is dependent on the amount of DNA or the primer sequence. DNA or primer sequences were not used as the template in our method, and thus did not play any role in sensitivity and specificity.
Despite the use of a definite filter for microscopic imaging, an interface by autofluorescent signals was exhibited by a few samples during our experiments (). All four samples that were negative by FSNP assay and positive by culture showed autofluorescent signals during microscopic examination. This problem has not been described in previous reports.Citation9,Citation7,Citation12 It seems that there is a specific problem when clinical samples are examined. In general, interface by autofluorescence can result from natural factors induced by some reagents and unspecific binding of antibodies to Fc receptors.Citation24,Citation25
In addition to increased sensitivity and specificity, another advantage of the FSNP assay compared with the conventional and PCR methods is the short duration of the procedure. Indeed, only 2–3 hours are needed to perform the FSNP assay compared with 6–8 weeks for the culture and 5 hours for the nested PCR assay.
The limit of the FSNP assay for detection of M. tuberculosis was performed on pure bacteria and found to be 100 colony forming units/mL. Less than 100 colony forming units/mL was not detected by either FSNP assay or culture. However, in clinical samples, we found some cases that were positive in culture but negative on FSNP assay, and the overall sensitivity of the FSNP assay versus culture was not 100% but 97%. The FSNP assay in comparison with PCR is cost-effective ($1.0–$1.5 per test) and the results come out in a shorter time. We believe the sensitivity and specificity of this method will enable it to become a routine technique for detection of M. tuberculosis in clinical specimens.
Conclusion
Compared with the gold standard method of culture, the total assay time and ease of performing the FSNP assay suggests that this technique can be of great use in the future. We performed the FSNP assay on a large number of clinical samples to confirm its diagnostic specificity and sensitivity, and consider it reliable enough for routine use in laboratories.
Acknowledgments
We are grateful to the members of our reference laboratory for their help. This study was supported by a research grant from the Infectious and Tropical Diseases Research Center, Ahvaz Jundishapour University of Medical Sciences, Ahvaz, Iran.
Disclosure
The authors report no conflicts of interest in this work.
References
- World Health OrganizationGlobal Tuberculosis Control: A Short Update to the 2009 ReportGenevaWorld Health Organization2009 Available from: http://whqlibdoc.who.int/publications/2009/9789241598866_eng.pdfAccessed September 9, 2011
- World Health OrganizationGlobal Tuberculosis Control 2010: WHO Report 2010GenevaWorld Health Organization2010 Available from: http://www.who.int/entity/tb/publications/global_report/2010/gtbr10_main.pdfAccessed September 9, 2011
- World Health OrganizationMultidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and responseGeneva, Switzerland2010 Available from: http://whqlibdoc.who.int/publications/2010/9789241599191_eng.pdfAccessed August 16, 2011
- TiruviluamalaPReichmanLBTuberculosisAnnu Rev Public Health20022340342611910069
- PalominoJCNon conventional and new methods in the diagnosis of tuberculosis: Feasibility and applicability in the fieldEur Respir J20052633935016055883
- PaiMFloresLLHubbardAReilyLWColfordJNucleic acid amplification tests in the diagnosis of tuberculosis pleuritis: A systemic review and meta analysisBMC Infect Dis2004411414720304
- ZhaoXJHilliardLRMecherySJWangYBagweRPA rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticlesProc Natl Acad Sci U S A2004101150271503215477593
- SalataOVApplications of nanoparticles in biology and medicineJ Nanobiotechnol2004238
- StöberWFinkABohnEControlled growth of monodisperse silica spheres in the micron size rangeJ Colloid Interface Sci1968266269
- QhobosheaneMSantraSZhangPTanWBiochemically functionalized silica nanoparticlesAnalyst20011261274127811534592
- WangLWangKSantraSZhaoXSmithJTanWWatching silica nanoparticles glow in the biological worldAnal Chem200678646654
- QinDHeXWangKZhaoXTanWChenJFluorescent nanoparticlesbased indirect immunofluorescence microscopy for detection of Mycobacterium tuberculosisJ Biomed Biotechnol200720078936418273415
- BuijtelsPPetitPComparison of NaOH/N-acetyl-L-cysteine (NALC) and sulfuric acid decontamination methods for recovery of Mycobacteria from clinical specimensJ Microbiol Methods200562838815823396
- World Health OrganizationLaboratory services in tuberculosis control. Part II: Microscopy Available from: http://whqlibdoc.who.int/hq/1998/WHO_TB_98.258_(part2).pdfAccessed August 16, 2011
- AmitaJVandanaTGuleriaRSVermaRKQuality evaluation of mycobacterial DNA extraction protocols for polymerase chain reactionMolecular Biology Today200234350
- EisenachKDCaveMDBatesJHCrawfordJTPolymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosisJ Infect Dis19901619779812109022
- WilsonSMMcNerneyRNyePMGodfrey-FaussettPDStokerNGVollerAProgress toward a simplified polymerase chain reaction and its application to diagnosis of tuberculosisJ Clin Microbiol199337767828463386
- SantraSZhangPWangKTapecRTanWConjugation of biomolecules with luminophore-doped silica nanoparticles for photostable biomarkersAnal Chem2001734988499311681477
- YavuzHDenizliAImmunoadsorption of cholesterol on protein A oriented beadsMacromol Biosci20055394815635714
- LangoneJJApplications of immobilized protein A in immunochemical techniquesJ Immunol Methods1982552772966762396
- BradfordMMA rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein dye bindingAnal Biochem197672248254942051
- SchlesingerLSBellinger-KawaharaCGPayneNRHorwitzMAPhagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3J Immunol1990144277127802108212
- ZhouXZhouJImproving the signal sensitivity and photostability of DNA hybridizations on microarrays by using dye-doped core-shell silica nanoparticlesAnal Chem2004765302531215362886
- ViegasMSMartinsTCSecoFCarmoAAn improved and cost- effective methodology for the reduction of autofluorescence in direct immunofluorescence studies on formalin-fixed paraffin-embedded tissuesEur J Histochem200751596617548270
- BillintonNKnightAWSeeing the wood through the tree: A review of techniques for distinguishing green fluorescence protein from endogenous autofluorescenceAnal Biochem20012917519711401292