Abstract
Background
Erythromycin, a hydrophobic antibiotic used to treat infectious diseases, is now gaining attention because of its anti-inflammatory effects and ability to inhibit osteoclasts formation. The aim of this study was to explore a cyclodextrin-erythromycin (CD-EM) complex for sustained treatment of orthopedic inflammation.
Methods and results
Erythromycin was reacted with β-cyclodextrin to form a nonhost-guest CD-EM complex using both kneading and stirring approaches. Physiochemical measurement data indicated that erythromycin and cyclodextrin formed a packing complex driven by intermolecular forces instead of a host-guest structure due to the limited space in the inner cavity of β-cyclodextrin. The CD-EM complex improved the stability of erythromycin in aqueous solution and had a longer duration of bactericidal activity than free erythromycin. Cytotoxicity and cell differentiation were evaluated in both murine MC3T3 preosteoblast cells and RAW 264.7 murine macrophage cells. The CD-EM complex was noncytotoxic and showed significant inhibition of osteoclast formation but had little effect on osteoblast viability and differentiation.
Conclusion
These attributes are especially important for the delivery of an adequate amount of erythromycin to the site of periprosthetic inflammation and reducing local inflammation in a sustained manner.
Introduction
Up to 20% of patients with total joint replacement will develop radiographic evidence of osteolysis and/or aseptic loosening.Citation1–Citation3 Aseptic loosening most likely results from an inflammatory response to billions of wear debris particles shed from the prosthesis during normal use. The interaction of macrophages with wear debris is the likely determinant of whether wear debris-induced inflammation will be resolved or progress to irreversible osteolysis/aseptic loosening.Citation4 A recent approach to limiting osteolysis/ aseptic loosening has focused on inhibiting periprosthetic inflammation by pharmaceutical intervention.Citation5–Citation7
Erythromycin, a 14-member lactone ring macrolide antibiotic, has been used for the treatment of infectious diseases for the past 50 years.Citation8 In the last decade, erythromycin has attracted a great deal of attention because of its additional anti-inflammatory effects at subantimicrobial doses.Citation9,Citation10 An example of erythromycin used as an anti-inflammatory drug has been demonstrated recently in Japan, where erythromycin was successful in treating diffuse panbronchiolitis.Citation11,Citation12 Our previous studies have indicated that erythromycin is a promising treatment for osteolysis/aseptic loosening. First, erythromycin inhibited wear debris-induced osteoclastogenesis by inhibition of NF-κB activity in both a RAW264.7 macrophage cell line and in mouse bone marrow progenitor cells.Citation13 Second, erythromycin inhibited wear debris-induced inflammatory osteolysis in a murine osteolysis model.Citation14 Third, oral erythromycin 600 mg/day reduced periprosthetic tissue inflammation in 32 patients with aseptic loosening who were candidates for surgical revision.Citation15 Although we have demonstrated the efficacy of orally administered erythromycin in improving periprosthetic tissue inflammation, long-term systemic treatment with erythromycin is not recommended in view of the potential side effects, such as bacterial resistance, nausea, and vomiting. Therefore, it would be advantageous to restrict delivery of erythromycin to the inflammatory periprosthetic site. However, delivery to and maintenance of prolonged erythromycin release at the site of periprosthetic inflammation presents a considerable challenge.Citation16 We propose that the efficiency of erythromycin delivered to a periprosthetic site can be increased by improving the chemical stability and water solubility of the drug.Citation17,Citation18
Cyclodextrins are cyclic oligomers of glucose that are molecular “carriers” with a hydrophilic exterior and a hydrophobic cavity. Cyclodextrins are biocompatible and do not elicit an immune response in humans.Citation19 Cyclodextrins can be divided into three types, ie, α, β, and γ cyclodextrins, based on their number of glucose monomers. Cyclodextrins are used to improve drug bioavailability by increasing solubility and/or dissolution through inclusion of water-insoluble drugs (like erythromycin) into their hydrophobic cavities,Citation20,Citation21 improving drug stability,Citation22–Citation24 increasing the permeability of water-insoluble drugs, and reducing drug toxicity by making the drug effective at lower doses.
Cyclodextrin-drug complexes can be prepared in several ways.Citation25 The most common procedure is to stir a mixture of cyclodextrins and drugs dissolved by two immiscible solvents vigorously. Another procedure is the so-called kneading method, which kneads cyclodextrins with drug components using a small amount of solvent. The physical force required to achieve the process of complex formation is favored energetically. Both methods could be used to prepare various kinds of water-insoluble drugs.Citation26
Complex formation between cyclodextrin monomers and drug molecules via dynamic noncovalent interactions using the host-guest model has been well described.Citation19 This model commonly involves using cyclodextrins as host molecules and attracting hydrophobic guest molecules, such as aromatic molecules, which are incorporated into the hydrophobic cavity of cyclodextrin through the formation of a hydrogen bond under thermodynamically favorable conditions.Citation25 However, the roles of cyclodextrin and the drug molecules could be reversed during complex formation when the drug molecules are too large to be incorporated.Citation27 Cyclodextrins act as nonhost molecules when forming complexes with cyclotetrachromotropyleneCitation28 and calcichrome.Citation29 The formation of a host-guest complex is dependent on the dimensional match between the cyclodextrin cavity and the size of the guest molecule.
In this paper, we describe the synthesis and characterization of a cyclodextrin-erythromycin (CD-EM) complex prepared using both the stirring and kneading methods. The physiochemical properties (water solubility, stability, and CD-EM interaction) and biological activity (cellular behavior and bacterial inhibition) of the complex are described. We suggest that a CD-EM complex might potentially be applied to a prosthetic surface, with sustained release of erythromycin to prevent orthopedic infection and aseptic loosening.
Materials and methods
Materials
Erythromycin, β-cyclodextrin, β-glycerophosphate, and a tartrate-resistant acid phosphatase (TRAP) assay kit were purchased from Sigma-Aldrich (St Louis, MO). An alkaline phosphatase (ALP) assay kit was obtained from BioVision (San Francisco, CA). Absolute alcohol (100%) was purchased from Deconlab Inc (King of Prussia, PA). Sodium phosphate (Na2CO3) and sodium hydroxide (NaOH) were purchased from Fisher Scientific (Fair Lawn, NJ). Deuterium oxide (D2O) and methanol D4 were obtained from Cambridge Isotope Laboratories Inc (Andover, MA), and an α-modified minimum essential medium, a live-dead cytotoxicity kit, and Dulbecco’s modified Eagle’s medium were purchased from Invitrogen (Camarillo, CA). Ascorbic acid was obtained from Sigma-Aldrich. Receptor activator of nuclear factor kappa-B ligand (RANKL) was purchased from Research Diagnostics Inc (Flanders, NJ). A lactate dehydrogenase assay kit was obtained from Roche Diagnostics GmbH (Mannheim, Germany). All chemicals used were of reagent grade.
Preparation of CD-EM complex
We used both a solvent evaporation method (MV) and a kneading method (MVI) to incorporate erythromycin into the hydrophobic cavity of β-cyclodextrin.Citation30 For the MV method, erythromycin was first dissolved in 100% alcohol (0.25 g/mL). Dropwise addition of erythromycin solution to the saturated CD-H2O (0.125 g/mL) solution was done under constant stirring at 800 rpm until the mixed solution was homogenous. Using the MVI method, the CD-EM complex was obtained by manually kneading a mixture of saturated cyclodextrin-H2O 0.125 g/mL solution and erythromycin-alcohol 0.25 g/mL at a molar ratio of 1:1 in a ceramic mortar for 3 hours. The complexes obtained by the MV and MVI methods were initially frozen at −20°C overnight and lyophilized at −30°C and 1.3 × 103 Pa overnight. Noninteracted erythromycin mixed with the complex was removed by rinsing twice with 100% alcohol. The CD-EM was collected by alcohol wash through a 0.8 μm membrane filter (Millipore, Bedford, MA). Finally, the CD-EM complex was sterilized by exposure to ultraviolet light overnight. The sterilized CD-EM mixture was resuspended in sterile distilled water (5 mg/mL) just before testing.
Measurement of erythromycin concentration
Briefly, 100 μL of the sample solution was mixed with 100 μL of alkaline solution (0.12 M/L, sodium phosphate; 0.25 M/L, sodium hydroxide) and incubated in a 60°C water bath for 15 minutes in order to cleave the macrocyclic lactone ring of erythromycin.Citation31 After cooling to room temperature, the samples were read using an ultraviolet-visible spectrophotometer (BioTek Synergy HT, Winooski, VT) at OD 235 nm. To calculate the erythromycin concentration, a standard erythromycin concentration curve was included in each sample measurement. The ratio of erythromycin and cyclodextrin incorporated into the CD-EM complex was reproducibly obtained and found to be about 1:3.3 (w/w) according to this quantification method.
1H-NMR and 13C-NMR spectra analysis
1H-nuclear magnetic resonance (NMR) experiments were performed on a Varian-500S spectrometer operating at a frequency of 500 mHz and 25°C using a 5 mm AutoX-probe. One-dimensional 1H-NMR spectra were recorded, with a flip angle of 90°, a spectral width of 4000 Hz, and 256 scans of 16 K points. Cyclodextrin 14 mg/mL and CD-EM 20 mg/mL samples were dissolved in D2O, while erythromycin 6 mg/mL was first dissolved in methanol D4 and then mixed with D2O. 13C-NMR experiments were also performed on the same instrument at a frequency of 500 mHz and 37°C for 20 hours. Cyclodextrin 30 mg/mL and CD-EM 30 mg/mL samples were dissolved in D2O at 37°C.
X-ray diffractometry
The crystalline structures and phases of cyclodextrin, erythromycin, and CD-EM (MV and MVI) were characterized using an x-ray diffractometer (Rigaku Smar tLab, Woodlands, TX). CuKα radiation (λ = 1.5418Å) was used to scan the diffraction angles (2θ) between 5° and 80° at a speed of 5°/minute. The spectra were compared with standard spectra from the JCPDS card via Jade 5 software in order to identify the sample phases and crystalline structures.
Ultraviolet spectrometry
Absorbance spectra of cyclodextrin, erythromycin, and CD-EM were obtained using an ultraviolet-visible spectrophotometer (BioTek Synergy HT). Briefly, 100 μL of cyclodextrin, erythromycin, and CD-EM water solution (10 mg/mL) were placed in a 96-well ultraviolet plate (Fisher Scientific, Pittsburgh, PA) and mounted onto the spectrophotometer. A scanning wavelength range of 200–300 nm and a step size of 1 nm were set.
Scanning electron microscopy
The morphologies of the cyclodextrin, erythromycin, and CD-EM samples were characterized by scanning electron microscopy (SEM, Hitachi S-2400, Japan). All samples were coated with gold (Gold Sputter, EFFA coater, Redding, CA) before SEM analysis. Morphologies were viewed at a 25 kV accelerating voltage.
Stability of CD-EM
Erythromycin was firstly dissolved in 100% of alcohol and then suspended in 5 mL of distilled water to make a concentration of 0.1 g/mL. CD-EM mixtures were also dissolved in 5 mL of distilled water to make a concentration of 0.43 g/mL. Both samples were incubated in a 37°C water bath for various time periods. A fraction of samples (100 μL) was collected at different time points (0, 3, 5, 7, 9, 12, 14, 16, 18, and 20 hours). These samples were filtered through syringe filters (Minisart, Sartorius Stedim North America Inc, Bohemia, NY) with a pore size of 0.45 μm. The filtrates (excluding the undissolved CD-EM complex) were then used to measure the erythromycin concentration. The percentage of measurable erythromycin was plotted against the predefined time points.
Antibacterial activity assay
A modified minimum inhibitory concentration assay was developed to measure the bactericidal activity of erythromycin released from the CD-EM conjugate. First, a Mueller-Hinton broth inoculated with Staphylococcus aureus at a concentration of 1–2 × 108 colony forming units/mL (OD reading at 625 nm around 0.08–0.1) was prepared. The bacterial suspension was then mixed with a known concentration of free erythromycin (64 μg/mL), free cyclodextrin (148 μg/mL), and CD-EM (cyclodextrin 212 μg/mL, erythromycin 64 μg/mL) separately and incubated at 37°C for approximately 160 hours. Bactericidal activity was evaluated by dynamic measurement of OD reading at different time points. Bacterial culture suspension in the absence of erythromycin was included as a positive control. In a separate experiment, the bactericidal activity of the CD-EM complex at different concentrations (0, 53, 106, 159, and 212 μg/mL) was evaluated for up to 24 hours in culture.
Effect of CD-EM complex on osteoblastic ALP activity
Mouse MC3T3-E1 preosteoblasts (American Type Culture Collection, CRL-2593) were plated at a density of 1 × 104 cells/well in a 12-well plate and cultured in α-modified minimum essential medium supplemented with 10% fetal bovine serum and a 1% (v/v) antibiotic mixture of penicillin and streptomycin at 37°C in a humidified incubator with 5% CO2 for 24 hours. The MC3T3-E1 cells were then cultured in cell differentiation medium (α-modified minimum essential medium supplemented with ascorbic acid 50 μg/mL and β-glycerophosphate 10 mM) for 16 days in the presence of erythromycin (1.6 mg/mL and 16 μg/mL), cyclodextrin (3.7 mg/mL and 37 μg/mL) and CD-EM (5.3 mg/mL and 53 μg/mL), respectively. Culture media and drugs were replaced every 3 days until the predetermined dates. The collected culture media were used for cytotoxicity assay.
Sixteen days after culture, the culture media was removed and the cell monolayer was gently washed twice with ice-cold phosphate-buffered saline. The cell lysates were homogenized with ALP assay buffer (200 μL/well), followed by centrifugation (14,000× g for 5 minutes) to remove insoluble material. ALP activity in the cell lysate samples was measured at OD 405 nm (UVmax colorimeter, Molecular Devices, Sunnyvale, CA) with an ALP assay kit. ALP activity was expressed in nanomoles of p-nitrophenol produced per minute per microgram of protein. All experiments were conducted in duplicate and repeated in three independent experiments.
Effects of CD-EM complex on osteoclast formation
Osteoclasts were generated from a murine RAW 264.7 macrophage cell line in the presence of RANKL.Citation32 RAW 264.7 cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and antibiotics (100 U/mL of penicillin G and 100 μg/mL of streptomycin) at 37°C in a humidified incubator with 5% CO2. To investigate the effects of erythromycin released from the CD-EM conjugate on osteoclast formation, RAW cells were cultured in the presence of RANKL 50 ng/mL for up to 7 days in the presence of erythromycin 16 μg/mL, cyclodextrin 37 μg/mL, and CD-EM 53 μg/mL. Culture media and drugs were replaced every 3 days. The collected cultured media at each time point were used for cytotoxicity assay.
TRAP staining was performed using a commercial kit as described elsewhere.Citation14 In brief, RAW cells were fixed in 4% paraformaldehyde, followed by incubation at 37°C for 1 hour in a solution of naphthol AS-BI phosphoric acid, fast garnet GBC, and sodium tartrate. Following a rinse in deionized water, images of the stained cells were captured at 10× by a microscope (Carl Zeiss MicroImaging, Thornwood, NY). The mean percentage of TRAP+ cells was calculated by reading the stained plate at 525 nm in the ultraviolet-visible spectrophotometer through the built-in protocol.
Cytotoxicity assay
Cell toxicity was determined by measuring the release of lactate dehydrogenase from dead or dying cells into the culture medium using a colorimetric method following the manufacturer’s instructions. Blank culture medium was used as a control. Lactate dehydrogenase (LDH) activity was expressed as absorbance (OD) per mg protein. In addition, the potential cellular toxicity in cells treated with higher doses of erythromycin (1.6 mg/mL), cyclodextrin (3.7 mg/mL), and CD-EM (5.3 mg/mL) were evaluated using the Live/Dead® viability/cytotoxicity staining kit (Invitrogen, Camarillo, CA).
Statistical analysis
The statistical analysis was performed using the analysis of variance test, with the Schafer formula for post hoc multiple comparisons using the SPSS software package version 7.5 (SPSS Inc, Chicago, IL). All treatments were performed in triplicate. Data were expressed as the mean ± standard error of the mean. A P value <0.05 was considered to be statistically significant.
Results and discussion
1H-NMR
The 1H-NMR spectrum results demonstrated the presence of erythromycin in the CD-EM composite (). Seven glucose units with identical conformations and hexagonal symmetry can be identified in by locating the integral peaks of 1–6 protons.Citation33 The 1H-NMR spectrum of erythromycin in can be identified, consistent with earlier observations.Citation34 The CD-EM complex prepared by both the MV and MVI methods has the same spectral pattern, as shown in . The 13C-NMR spectrum results also demonstrate the presence of erythromycin in the CD-EM complex (). 13C signals for cyclodextrin at δ = 101.884, 81.202, 73.150, 72.143, 71.906, and 60.394 ppm were the localization of C1, C2, C3, C4, C5, and C6, respectively, in D2O. The CD-EM spectra (MV and MVI) showed a similar pattern, with a minor chemical shift (0.008 ppm) to a lower field only from C1 and C6 and carbon peaks at δ = 57 and 16 ppm from erythromycin molecules.
Figure 1 500 mHz 1H-NMR spectra at 25°C of (A) cyclodextrin 14 mg (B) erythromycin 6 mg (C) cyclodextrin-erythromycin 20 mg (MV) (7/3, w/w) complex, (D) 20 mg of cyclodextrin-erythromycin (MVI) (7/3, w/w) complex in 750 μL of D2O ranging from 0.5 to 5.5 ppm. Important proton chemical shifts and peak integrations of solutes are labeled.
Abbreviations: MV, method V; MVI, method VI.
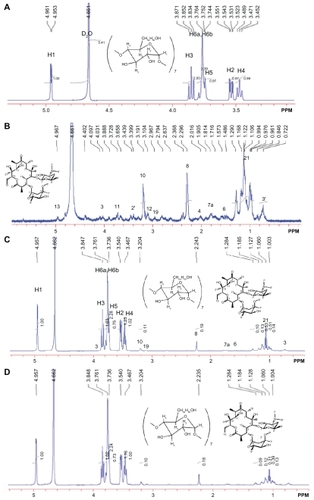
Figure 2 500 mHz 13C-NMR spectra at 37°C of 30 mg of cyclodextrin, 30 mg of cyclodextrin-erythromycin (MV) (7/3, w/w) complex, 30 mg of cyclodextrinerythromycin (MVI) (7/3, w/w) complex in 1 mL of D2O. Important carbons (C1–C6 and C from erythromycin) chemical shifts and peak integrations of solutes are labeled.
Abbreviations: CD, cyclodextrin; CD-EM, cyclodextrin-erythromycin; MV, method V; MVI, method VI.
The interaction between cyclodextrin and guest molecule occurs when guest molecule approaches the nonpolar cavity of cyclodextrin, which causes an upfield chemical shift (−δ) of guest molecule protons and a downfield chemical shift (δ) of host molecule carbons.Citation35 Steric hindrance is known to be the decisive factor for preventing formation of a cyclodextrin-drug complex as a host-guest model.Citation36 The large macrocyclic lactone ring structure in erythromycin is geometrically incompatible with the cavity dimensions of β-cyclodextrin. However, a nonhost-guest formation between cyclodextrin and erythromycin could be driven thermodynamically.
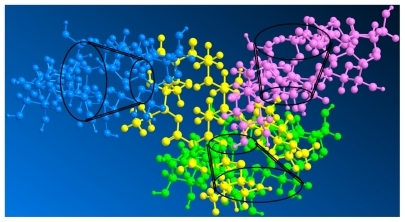
A selection of chemical shift variations obtained for erythromycin from its free form to a complex form with cyclodextrin is presented in . Chemical shift variations from protons of erythromycin in the presence of cyclodextrin indicate the formation of a complex, especially protons (7′, 8, and 21) at the edge of the erythromycin molecules with larger shifts, which are attributed to the anisotropic shielding effect of the cyclodextrin cavity. However, other unaffected protons (3′, 4, 6, 12, 10, and 19) of erythromycin indicate that the erythromycin molecule did not enter the cyclodextrin cavity. Instead, the shifts of protons (7′ and 8) directed outwards on two sugar units (L-cladinose and D-desosamine) indicate interaction of these two sugar units with the cyclodextrin molecules, while the shift of proton (21) from the diagonal position on the macrocyclic ring indicates the approach of another cyclodextrin molecule. Moreover, the shifts of C1 and C6, although minor, indicate a cyclodextrin molecule approaching erythromycin on the smaller side of the cavity. There is no evidence of inclusion of the macrolide in the cyclodextrin cavity because there is no chemical shift of C2, C3, C4, and C5 from the 13C-NMR spectra. The formation of an intermolecular hydrogen bond between cyclodextrin and the guest molecules would ordinarily cause larger chemical shift variations (δ > 0.05) of cyclodextrin protons, particularly the H3 and H5 directed inside the cyclodextrin cavity. However, the chemical shift for cyclodextrin after complex formation shows no significant variations (δ < 0.01), demonstrating the absence of hydrogen bond formation between cyclodextrin and erythromycin. Several assumptions have been made to interpret the interaction force during CD-drug complex formation, including Van der Waals interactions,Citation37 hydrogen bonding,Citation38 and release of strain energy in the ring of the cyclodextrin molecule.Citation39 Some aromatic molecules have been found to be able to be included in the cyclodextrin cavity by forming a hydrogen bond with H3 and H5.Citation35 However, the interactive force between nonaromatic molecules of supermolecular size and cyclodextrin when forming a nonhost-guest complex is still controversial. Intermolecular Van der Waals forces and hydrophobic interactions are hypothesized to be the dominant driving forces for the formation of a cyclodextrin- drug complex. Citation40 On the basis of the chemical shift of both cyclodextrin and erythromycin, we predict that the two sugar units and the diagonal position of erythromycin interacted with three cyclodextrin molecules via Van der Waals forces to form a packed complex. The three-dimensional topologic structure of the CD-EM complex is shown in .
Table 1A Proton chemical shift of cyclodextrin
Table 1B Proton chemical shift of erythromycin
Table 1C Carbon chemical shift of cyclodextrin
X-ray diffraction
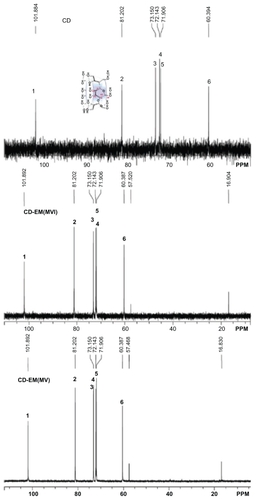
Formation of the complex was found to alter the original crystal lattice of cyclodextrin and subsequently the x-ray diffraction pattern.Citation41 The x-ray diffraction spectra of cyclodextrin, CD-EM (MV and MVI), and erythromycin are shown in . The spectrum of cyclodextrin indicates high crystallinity in cyclodextrin due to the characteristic sharp peaks at (041), (141), (180), (042), (162), (222), (223), and (044). The spectrum of erythromycin shows low crystallinity with numerous weaker intensity peaks and some amorphous curves, even though the (100), (021), (060), (070), and (121) planes were identified to match with the standard spectrum of erythromycin. Both spectra of CD-EM (MV and MVI) showed a pattern similar to cyclodextrin, but with distinct peaks at (220), (250), (231), and (062), and a red color different from that in the cyclodextrin spectrum. Moreover, the original peaks at (042), (251), (223), and (044) with a blue color had disappeared and the double peaks at (130) merged into a single peak in the CD-EM complex spectra. Some characteristic peaks of erythromycin [(060) and (121) with yellow color] were also seen in the CD-EM spectra. Generally, the spectra of CD-EM (MV) and CD-EM (MVI) were identical, with only some minor differences.
Figure 4 X-ray diffraction spectra for pure cyclodextrin, cyclodextrin-erythromycin (MV and MVI) before freeze-drying, and pure erythromycin. The Y axis (counts) of the diffraction pattern is vertically offset for clarity. Blue-peaks disappeared in spectra of CD-EM complex. Red-new peaks in spectra of CD-EM complex. Yellow-characteristic peaks of EM in in spectra of CD-EM complex.
Abbreviations: CD, cyclodextrin; EM, erythromycin; MV, method V; MVI, method VI.
As new crystalline peaks were shown in spectra of CD-EM, phase transition for CD-EM complex during preparation process could be predicted. The conversion of a crystalline type for cyclodextrin and erythromycin revealed the formation of new complex. The intermolecular interaction of erythromycin and cyclodextrin could interrupt the original organization of cyclodextrin molecules and reformed crystalline lattices.
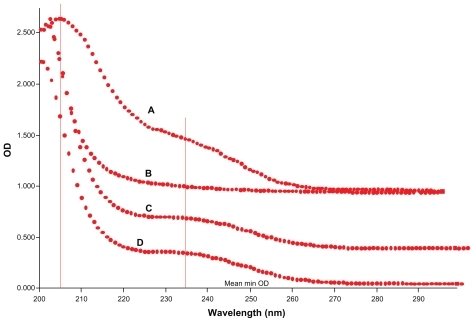
Ultraviolet-visible spectrometry
Formation of the cyclodextrin complex was also found to alter the original ultraviolet absorption spectrum.Citation25 The ultraviolet absorption spectral change on forming the CD-EM complex can be observed in . The absorption peak was detected at 205 nm for free erythromycin. Absorption at 230 nm is consistent with a literature report for macrocyclic lactone. For the CD-EM complex, the presence of these two absorption peaks indicates the erythromycin substrate. A slight shift of the absorption peak for CD-EM from 205 nm to 200 nm can be interpreted as the effect of partial shielding of the excitable electrons in the cavity of cyclodextrin, while the shift of absorption from 230 nm to 235 nm is a bathochromic shift that usually happens after formation of a cyclodextrin complex.Citation25
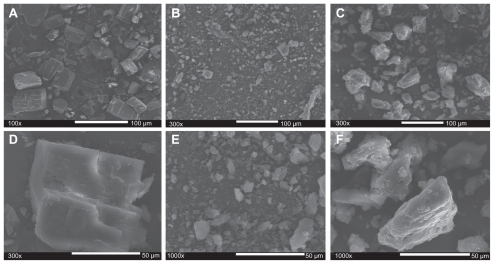
SEM analysis
To elucidate the formation of a CD-EM complex, morphologic observation was performed using high-resolution SEM. shows this for cyclodextrin (6A, D), erythromycin (6B, E), and CD-EM (6C, F). A monoclinic morphology of a cyclodextrin precipitate was observed under SEM () and this was consistent with analytic results from x-ray diffraction and literature reports,Citation42 while erythromycin powder was shown to be a semicrystalline precipitate (). Both the kneading and stirring processes facilitated interaction of cyclodextrin and erythromycin, altering their original crystalline structures and amorphization during freeze-drying, as shown in . For CD-EM prepared by both the MV and MVI methods, an appropriate water content initiated the CD-EM interaction as well as cyclodextrin matrix formation. The continuous phase of the cyclodextrin matrix stabilized the interactive erythromycin molecule, leading to aggregation of CD-EM precipitates.Citation31
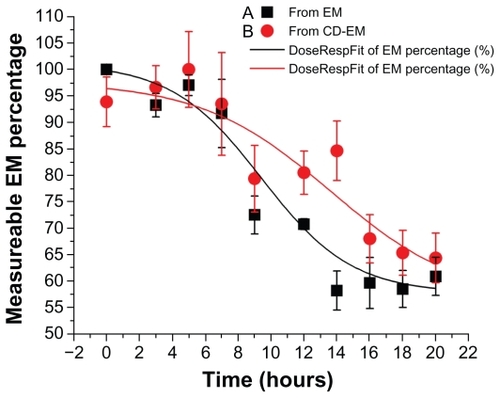
Drug stability assay
Decomposition of delivered drug, occurring within the initial period of implantation, is one of the main limitations of local drug delivery approaches, particularly for periprosthetic drug delivery.Citation43–Citation45 The formation of a cyclodextrin-drug complex aims to extend the duration of drug activity in an aqueous environment. In this assay, the concentration of erythromycin was dynamically decreased at 37°C over 20 hours for both pure erythromycin and CD-EM, as shown in . The dramatic decrease of measurable erythromycin from about 90% to 50% took place after incubation for 6 hours. However, the fitting curves indicated that the rate of decrease of CD-EM was slower than for pure erythromycin, especially after 6 hours of incubation. At each time point after 6 hours, the percentages of measurable erythromycin from CD-EM were all higher than from pure erythromycin, indicating better stability. The invalidation of erythromycin in aqueous solution was susceptible to decomposition of the large macrocyclic lactone ring structure during the hydrolytic process.Citation46 Protection of the integrity of the molecular structure of erythromycin in aqueous solution would help to increase its lifespan. By either the kneading or stirring method, formation of a CD-EM complex could provide a shielding effect for the erythromycin molecule from the surrounding cyclodextrin molecules, as described earlier. The packed cyclodextrin molecules improved the solubility of erythromycin in aqueous solution and stabilized its molecular structure by preventing attack from nucleophiles during hydrolysis. Nonetheless, the noncovalent interaction between cyclodextrin and erythromycin enables release of entrapped free erythromycin when the surrounding cyclodextrin becomes unstable.
Bactericidal activity
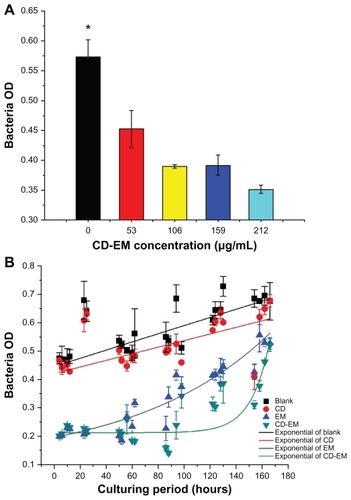
Erythromycin, one of the macrolide antibiotics, has been used to treat infectious diseases for more than 50 years. The molecular mechanism of its bactericidal activity is believed to involve binding to the 50s subunit of the bacterial 70s rRNA complex to interfere with aminoacyl translocation. Erythromycin is susceptible to loss of bactericidal activity in an acidic environment.Citation46 We aimed to extend the bactericidal activity of erythromycin by embedding the drug in cyclodextrin nanoparticles. As shown in , the results of a semiquantitative bactericidal activity assay show that bacterial growth was inhibited by treatment with the CD-EM complex in a dose-dependent manner (0–212 μg/ mL). This suggests that incorporation of erythromycin into cyclodextrin did not affect the bactericidal activity of erythromycin. To determine whether the CD-EM complex has longer bactericidal activity than free erythromycin, we repeated our bactericidal activity measurements in the presence of either the CD-EM complex (212 μg/mL, including erythromycin 64 μg/mL) or the same amount of free erythromycin for up to 170 hours (). We found that treatment with both erythromycin and CD-EM resulted in persistent bacterial growth inhibition for up to 170 hours. Based on an exponentially fitting curve analysis, the bactericidal activity of CD-EM was sustained for longer than for free erythromycin at the same dose. These data indicate that erythromycin has longer durability and bactericidal activity in aqueous solution when interacted with cyclodextrin, as compared with free erythromycin molecules.
Figure 8 Bactericidal effect assay. (A) Semiquantitative bacteria growth inhibition assay in 2 mL broth in the presence of cyclodextrin-erythromycin with gradient concentrations (53, 106, 159, and 212 μg/mL) for 24 hours, respectively. (B) Semiquantitative bacteria growth inhibition assay in 2 mL broth in the presence of erythromycin 64 μg/mL, cyclodextrin 148 μg/mL, and cyclodextrin-EM 212 μg/mL at predetermined time points, respectively.
Notes: All dots were exponentially fitted; P < 0.05 related to 0 versus 159 and 212 μg/mL; n = 4.
Abbreviations: CD, cyclodextrin; EM, erythromycin.
Effect of CD-EM complex on osteoblastic viability and differentiation
To assess the efficacy of the CD-EM complex, it was important to show that the complex was noncytotoxic and had no inhibitory effect on osteoblast differentiation. In addition, it was critical to define the safe and optimal dosage of erythromycin. The free cyclodextrin concentration was based on the amount of cyclodextrin present in the CD-EM complex at the indicated erythromycin concentration. For example, the highest concentration of erythromycin (1.6 mg/mL) is equivalent to approximately 5.3 mg/mL of CD-EM complex. Therefore, 3.7 mg/mL of free cyclodextrin was used as the control. As shown in , free cyclodextrin, erythromycin, and the CD-EM complex did not show appreciable cytotoxicity in MC3T3 cells at erythromycin concentrations up to 1.6 mg/mL for a treatment period of 16 days, based on the live/dead cell staining assay. Intracellular ALP activity was measured to determine the effect of the CD-EM complex on osteoblast differentiation. As shown in , treatment with erythromycin 1.6 mg/mL, cyclodextrin 3.7 mg/mL, and CD-EM 5.3 mg/mL all inhibited ALP activity significantly after 16 days in culture. We noticed that cyclodextrin 3.7 mg/mL achieved stronger inhibition of ALP activity than did erythromycin, while cells treated with the CD-EM complex showed the lowest ALP activity, although the difference was not statistically significant. Inhibition of ALP activity was closely related to the dose chosen. Our data demonstrated that MC3T3 cells treated with a lower concentration of cyclodextrin (37 μg/mL), erythromycin (16 μg/mL), or CD-EM complex (53 μg/mL) showed a slight increase in ALP activity compared with untreated cells (). This finding that a lower dose of CD-EM complex (with erythromycin 16 μg/mL) slightly increased ALP activity is important, because it will help us to design a safe and optimal range of drug loading for a periprosthetic drug delivery device. Our previous studies have indicated that erythromycin released from a periprosthetic device has bactericidal activity at a concentration of 10 μg/mL or less.Citation16 We have also noticed that treatment with erythromycin at a low concentration (5 μg/mL or less) leads to significant anti-inflammatory activity and inhibitory effect on osteoclast formation.Citation13
Figure 9 MC-3T3 cell activity assay. The live (green)-dead (red) staining (original magnification, ×50) of MC3T3 cells cultured for 16 days in the presence of (2) erythromycin 1.6 mg/mL, (3) cyclodextrin 3.7 mg/mL, and (4) cyclodextrin-erythromycin 5.3 mg/mL. ALP activity of MC-3T3 cells cultured under the same conditions in the presence of drugs with (A) higher concentrations of erythromycin (1.6 mg/mL), cyclodextrin (3.7 mg/mL), and cyclodextrin-erythromycin (5.3 mg/mL) and (B) lower concentrations of erythromycin (16 μg/mL), cyclodextrin (37 μg/mL), and cyclodextrin-erythromycin (53 μg/mL).
Note: *P < 0.05 related to control versus cyclodextrin and cyclodextrin-erythromycin; n = 6.
Abbreviations: ALP, alkaline phosphatase; CD, cyclodextrin; EM, erythromycin.
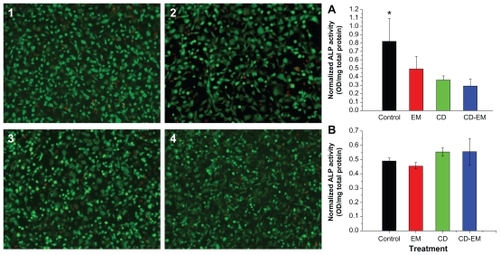
Considering these facts and the data presented here, a high concentration of CD-EM complex drug loading is unnecessary and should be avoided in future experiments. More efforts are required to optimize the drug loading dose and the dynamics of drug release.
Effect of the CD-EM complex on osteoclastogenesis
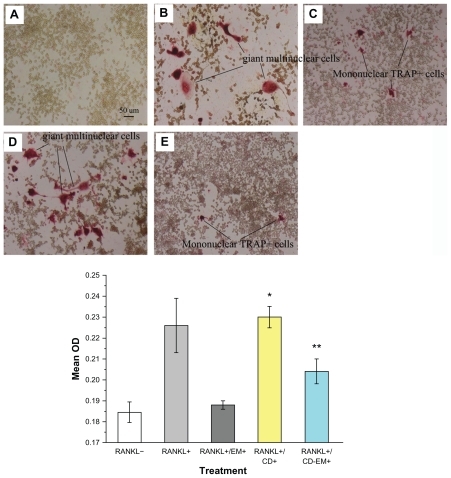
Recent studies have shown that RANKL and macrophage colony-stimulating factor are sufficient to induce differentiation of bone marrow osteoclast progenitors into mature osteoclasts in vitro in the absence of stromal cells.Citation47,Citation48 There is accumulating evidence to indicate that enhanced osteoclast formation and activity is a hallmark of aseptic loosening/ osteolysis.Citation49–Citation52 Our previous studies have indicated that erythromycin is a promising osteoclast inhibitor targeting the NF-kB signaling pathway.Citation13 To determine whether the CD-EM complex retains inhibitory activity against osteoclast formation, we cultured RAW 264.7 osteoclast precursor cells in the continuous presence of RANKL with or without CD-EM complex for 7 days. As seen in , TRAP+ cells were developed in the presence of RANKL. Multinucleated (more than three nuclei) TRAP+ cells were counted as osteoclasts (). Mononuclear TRAP+ cells were counted as preosteoclasts ().Citation13 Treatment with both erythromycin 16 μg/mL and an equivalent CD-EM complex significantly decreased the number of TRAP+ cells (). However, treatment with free cyclodextrin had no inhibitory effect on the formation of TRAP+ cells (). Quantitative analysis of the percentage of TRAP+ cells in each well is summarized in the histogram shown in . Through quantification results, we noticed that free erythromycin showed a better (albeit not statistically significant) inhibitory effect on the formation of TRAP+ cells than did the CD-EM complex, which had a significant inhibitory effect on osteoclastogenesis. However, inhibition of formation of giant multinuclear cells by erythromycin and CE-EM showed a negligible difference. The mechanism involved needs to be investigated further. Nevertheless, our data indicate that the inhibitory effects of erythromycin are well preserved in the CD-EM complex.
Figure 10 Erythromycin inhibits the amount of RANKL-induced TRAP+ cell formation in murine RAW 264.7 cells. (A–E) Representative TRAP staining (original magnification, ×100): (A) RANKL (−); (B) RANKL (+); (C) RANKL (+) + erythromycin (16 μg/mL); (D) RANKL (+) + cyclodextrin (37 μg/mL); and (E) RANKL (+) + cyclodextrin-erythromycin (53 μg/mL). TRAP+ cells were quantified by Image Analysis software (bottom panel). The value represents TRAP+ cells location quantified as area percentage of TRAP+ cells (%). Values represent averages of triplicate cultures from one representative experiment.
Notes: Using analysis of variance, *P < 0.05 refers to RANKL (+) + cyclodextrin 37 μg/mL versus RANKL (−), RANKL (+) + erythromycin 16 μg/mL, and RANKL (+) + cyclodextrin-erythromycin 53 μg/mL; **P < 0.05 related to RANKL (+) + cyclodextrin-erythromycin 53 μg/mL versus RANKL (−); n = 3.
Abbreviations: CD, cyclodextrin; EM, erythromycin; RANKL, receptor activator of nuclear factor kappa-B ligand.
Cytotoxicity of CD-EM complex in MC3T3 and RAW 264.7 cells
Although cyclodextrins are pharmaceutical excipients approved by the US Food and Drug Administration and expected to be safe for use,Citation53 it is important to show that the CD-EM complex is not cytotoxic. Measurement of lactate dehydrogenase activity in culture medium is an indicator of increased permeability of the plasma membrane as a result of cytotoxicity. As shown in , treatment with free CD 3.7 mg/mL, erythromycin 1.6 mg/mL, or CD-EM 5.3 mg/mL did not show noticeable cytotoxicity in MC3T3 cells for up to 16 days. In addition, no significant increase in lactate dehydrogenase activity was found in RAW 264.7 cells with the same treatment as described for the MC3T3 cells up to 7 days (). The mechanism of the slight increase in lactate dehydrogenase activity observed in all RANKL-treated cells at day 4 is not clear. After 7 days in culture, we could not find any difference in lactate dehydrogenase activity between cells treated and not treated with CD-EM, indicating that the CD-EM complex is not cytotoxic to this cell line, even at a very high concentration.
Figure 11 Lactate dehydrogenase activity in (A) culture medium of MC-3T3 cells after drug treatment [higher concentrations of erythromycin (1.6 mg/mL), cyclodextrin (3.7 mg/mL), and cyclodextrin-erythromycin (5.3 mg/mL)] for 6, 11, and 16 days, n = 6; (B) in culture medium of RAW 264.7 cells after drug treatment [lower concentrations of erythromycin (1.6 ug/mL), cyclodextrin (3.7 ug/mL), and cyclodextrin-erythromycin (5.3 ug/mL)] for 4 and 7 days, n = 3.
Note: The values shown are the mean ± standard error of the mean from six observations.
Abbreviations: CD, cyclodextrin; EM, erythromycin; LDH, lactate dehydrogenase.
![Figure 11 Lactate dehydrogenase activity in (A) culture medium of MC-3T3 cells after drug treatment [higher concentrations of erythromycin (1.6 mg/mL), cyclodextrin (3.7 mg/mL), and cyclodextrin-erythromycin (5.3 mg/mL)] for 6, 11, and 16 days, n = 6; (B) in culture medium of RAW 264.7 cells after drug treatment [lower concentrations of erythromycin (1.6 ug/mL), cyclodextrin (3.7 ug/mL), and cyclodextrin-erythromycin (5.3 ug/mL)] for 4 and 7 days, n = 3.Note: The values shown are the mean ± standard error of the mean from six observations.Abbreviations: CD, cyclodextrin; EM, erythromycin; LDH, lactate dehydrogenase.](/cms/asset/71aa9266-28ec-4940-a735-1e925c86545a/dijn_a_23530_f0011_c.jpg)
Conclusion
Our study results indicate that the hydrophobic erythromycin molecule can interact with β-cyclodextrin via either the kneading or cogrinding method to form a nonhost-guest CD-EM complex. We found that the intermolecular packing structure of the CD-EM complex enabled extended stability in aqueous solution, durable bactericidal activity, an inhibitory effect on osteoclastogenesis with little effect on osteoblastic viability and differentiation, and safety with no cytotoxicity. These attributes are especially important in delivering a sufficient amount of erythromycin to the site of periprosthetic inflammation and reducing local inflammation in a sustained manner.
Acknowledgments
This work was supported by an institutional research grant from Sinai-Grace Hospital, Detroit Medical Center, and the National Natural Science Foundation of China (81071487). We would like to acknowledge Sunxi Wang and Dr Mao for sample characterization.
Disclosure
The authors report no conflicts of interest in this work.
References
- BerryDJHarmsenWSCabanelaMEMorreyBFTwenty-five-year survivorship of two thousand consecutive primary Charnley total hip replacements: Factors affecting survivorship of acetabular and femoral componentsJ Bone Joint Surg Am200284217117711861721
- KeenerJDCallaghanJJGoetzDDPedersonDRSullivanPMJohnstonRCTwenty-five-year results after Charnley total hip arthroplasty in patients less than fifty years old: a concise follow-up of a previous reportJ Bone Joint Surg Am200385A61066107212784004
- FenderDHarperWMGreggPJOutcome of Charnley total hip replacement across a single health region in England: the results at five years from a regional hip registerJ Bone Joint Surg Br199981457758110463724
- SethiRKNeavynMJRubashHEShanbhagASMacrophage response to cross-linked and conventional UHMWPEBiomaterials200324152561257312726710
- SchwarzEMWhat potential biologic treatments are available for osteolysis?J Am Acad Orthop Surg200816 Suppl 1S727518612019
- PollicePFRosierRNLooneyRJPuzasJESchwarzEMO’KeefeRJOral pentoxifylline inhibits release of tumor necrosis factor-alpha from human peripheral blood monocytes: a potential treatment for aseptic loosening of total joint componentsJ Bone Joint Surg Am200183A71057106111451976
- ChildsLMGoaterJJO’KeefeRJSchwarzEMEfficacy of etanercept for wear debris-induced osteolysisJ Bone Miner Res200116233834711204434
- YatsunamiJHayashiSFourteen-membered ring macrolides as antiangiogenic compoundsAnticancer Res2001216B4253425811908678
- CervinAThe anti-inflammatory effect of erythromycin and its derivatives, with special reference to nasal polyposis and chronic sinusitisActa Otolaryngol20011211839211270500
- Giamarellos-BourboulisEJMacrolides beyond the conventional antimicrobials: a class of potent immunomodulatorsInt J Antimicrob Agents2008311122017935949
- KudohSErythromycin treatment in diffuse panbronchiolitisCurr Opin Pulm Med1998421161219612675
- KudohSAzumaAYamamotoMIzumiTAndoMImprovement of survival in patients with diffuse panbronchiolitis treated with low-dose erythromycinAm J Respir Crit Care Med19981576 Pt 1182918329620913
- RenWPLiXYChenBDWooleyPHErythromycin inhibits wear debris-induced osteoclastogenesis by modulation of murine macrophage NFkB activityJ Orthop Res2004221212914656655
- RenWPBinWMaytonLWooleyPHErythromycin (EM) inhibits wear debris-induced inflammatory osteolysis in a murine modelJ Orthop Res200624228029016435361
- RenWBlasierRPengXShiTWooleyPHMarkelDEffect of oral erythromycin therapy in patients with aseptic loosening of joint prosthesesBone200944467167719154802
- RenWZhangRHawkinsMShiTMarkelDCEfficacy of periprosthetic erythromycin delivery for wear debris-induced inflammation and osteolysisInflamm Res201059121091109720607583
- KhabrievRUPopkovVAReshetnyakVYKrasnyukIISolubility of erythromicin from solid dispersionsPharm Chem J200943113643
- JayaramanSCRamachandranCWeinerNTopical delivery of erythromycin from various formulations: an in vivo hairless mouse studyJ Pharm Sci19968510108210848897275
- DavisMEBrewsterMECyclodextrin-based pharmaceutics: past, present and futureNat Rev Drug Discov20043121023103515573101
- ShimpiSChauhanBShimpiPCyclodextrins: application in different routes of drug administrationActa Pharm200555213915616179128
- LiuXMWiswallATRutledgeJEOsteotropic beta-cyclodextrin for local bone regenerationBiomaterials200829111686169218199479
- FerrariFSorrentiMRossiSVancomycin-triacetyl cyclodextrin interaction products for prolonged drug deliveryPharm Dev Technol2008131657318300101
- SonglXDuFYGuoXQPanSZFormation, characterization, and thermal degradation behavior of a novel tricomponent aggregate of beta-cyclodextrin, ferrocene, and polypropylene glycolJ Phys Chem B201011441738174420055361
- MatilainenLMaunuSLPajanderJThe stability and dissolution properties of solid glucagon/gamma-cyclodextrin powderEur J Pharm Sci2009364–541242019059479
- SzejtliJCyclodextrins and Their Inclusion Complex2nd edBudapest, HungaryAkademiai Kiado1982
- LafontFTran VanNGHanadaKSansonettiPvan der GootFGInitial steps of Shigella infection depend on the cholesterol/sphingolipid raft-mediated CD44-IpaB interactionEMBO J200221174449445712198147
- PohBLTheeKKNo distinction between host and guest – the case of cyclodextrins complexing with disodium 1,8-disulfonato- 3,4,5,6-acridinetetracarboxylic acidTetrahedron Lett1996374174337436
- PohBLTanCMComplexation of amino-acids by cyclotetrachromotropylene in aqueous-solution – importance of ch-pi and pi-pi interactionsTetrahedron1994501134533462
- PohBLCyclodextrins (alpha-, beta-, and gamma-) as guests to calcichrome (calcion) in waterTetrahedron199652331097910984
- WongmekiatATozukaYMoribeKOguchiTYamamotoKPreparation of drug nanoparticles by co-grinding with cyclodextrin: formation mechanism and factors affecting nanoparticle formationChem Pharm Bull (Tokyo)200755335936317329871
- FloreyCAnalytical Profiles of Drug SubstancesNew York, NYAcademic Press Inc1979
- YanTRiggsBLBoyleWJKhoslaSRegulation of osteoclastogenesis and RANK expression by TGF-beta1J Cell Biochem200183232032511573248
- WoodDJHruskaFESaengerWH-1 NMR-study of inclusion of aromatic-molecules in alpha-cyclodextrinJ Am Chem Soc197799617351740
- EverettJRHattonIKTylerJWConformational-analysis of erythromycin a derivatives – a predictive method using NMR chemical-shift and coupling-constant dataMagn Reson Chem1990282114118
- BenderMLKomiyamaMCyclodextrin ChemistryNew York, NYSpringer-Verlag1978
- LachJLCohenJInteraction of pharmaceuticals with schardinger dextrins II. Interaction with selected compoundsJ Pharm Sci196352213714213927899
- VanEttenRLSebatianJFClowesGABenderMLAcceleration of phenyl ester cleavage by cycloamyloses: a model for enzymatic specificityJ Am Chem Soc1967891332423253
- CramerFHettlerHInclusion compounds of cyclodextrinsNaturwissenschaften196754246256324298643
- ManorPCSaengerWWater molecule in hydrophobic surroundings: structure of a-cyclodextrin-hexahydrateNature1972237392393
- GuoQXLiuLCaiWSJiangYLiuYCDriving force prediction for inclusion complexation of alpha-cyclodextrin with benzene derivatives by a wavelet neural networkChem Phys Lett19982904–6514518
- TakeoKKugeTComplexes of starchy materials with organic compounds part III. X-ray studies on amylose and cyclodextrin complexesAgric Biol Chem19693311741180
- AreeTJacobJSaengerWHoierHCrystal structure of alpha- cyclodextrin- acetonitrile-hexahydrateCarbohydr Res19983073–41911979675363
- StigterMBezemerJdeGKLayrollePIncorporation of different antibiotics into carbonated hydroxyapatite coatings on titanium implants, release and antibiotic efficacyJ Control Release200499112713715342186
- ZhangRXuDLanderyouTEctopic bone formation using osteogenic protein-1 carried by a solution precipitated hydroxyapatiteJ Biomed Mater Res A200471341241815472924
- ZhangRAnYTothCADraughnRADimaanoNMHawkinsMVOsteogenic protein-1 enhances osseointegration of titanium implants coated with peri-apatite in rabbit femoral defectJ Biomed Mater Res B Appl Biomater200471240841315389505
- ButlerMNWeberWJAccelerated transformation and deactivation of erythromycin in superheated water. 1. Temperature effects, transformation rates, and the impacts of dissolved organic matterEnviron Sci Technol20053972294230015871267
- KongYYYoshidaHSarosiIOPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesisNature199939767173153239950424
- LaceyDLTimmsETanHLOsteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activationCell19989321651769568710
- RenWWuBPengXHuaJHaoHNWooleyPHImplant wear induces inflammation, but not osteoclastic bone resorption, in RANK(−/−) miceJ Orthop Res20062481575158616779834
- GehrkeTSersCMorawietzLReceptor activator of nuclear factor kappaB ligand is expressed in resident and inflammatory cells in aseptic and septic prosthesis looseningScand J Rheumatol200332528729414690142
- GreenfieldEMBiYRagabAAGoldbergVMvan de MotterRRThe role of osteoclast differentiation in aseptic looseningJ Orthop Res20022011811855378
- SabokbarAFujikawaYNealeSMurrayDWAthanasouNAHuman arthroplasty derived macrophages differentiate into osteoclastic bone resorbing cellsAnn Rheum Dis19975674144209486003
- IrieTUekamaKPharmaceutical applications of cyclodextrins. 3. Toxicological issues and safety evaluationJ Pharm Sci19978621471629040088